Abstract
Ethanolamine is a compound readily derived from cell membranes that certain bacteria can utilize as a source of carbon and/or nitrogen. The complex biology and chemistry of this process has been investigated since the 1970’s, primarily in one or two species. However, recent investigations into ethanolamine utilization have revealed significant and intriguing differences in gene content and regulatory mechanisms among the bacteria that harbor this catabolic capability. Additionally, many reports have connected this process to bacterial pathogenesis. The recent information from divergent sources regarding the phylogeny, regulation, and possible roles of ethanolamine utilization in pathogenesis is compiled in this Progress article.
Introduction
Phosphotidylethanolamine is an abundant phospholipid in both mammalian and bacterial cell membranes1,2, which can be readily broken down into glycerol and ethanolamine by phosphodiesterases3,4. The host diet and the bacterial and epithelial cells in the intestine are thought to provide a rich source of ethanolamine in nature1,2,5. A variety of bacteria, including species of Salmonella, Enterococcus, Arthrobacter, Erwinia, Flavobacterium, Klebsiella,, Mycobacterium, Pseudomonas, Achromobacter, Corynebacterium, Clostridium and Escherichia, can use ethanolamine as a sole source of carbon and/or nitrogen6-8. The process involves splitting ethanolamine into acetaldehyde and ammonia by an ethanolamine ammonia lyase9,10. The ammonia can serve as a cellular supply of reduced nitrogen, and the acetaldehyde can be converted into the metabolically useful compound acetyl-CoA11.
The central genes in this process are eutB and eutC, the protein products of which form the ethanolamine ammonia lyase. Additional accessory genes, as many as sixteen, can be associated with eutBC. An initial understanding of the chemistry and genetics behind ethanolamine utilization was elucidated in Enterobacteriaceae such as Salmonella typhimurium and Escherichia coli. Recently, the gene content, organization and regulation of the eut operon were discovered to vary significantly between species7,12,13. Additionally, many reports have identified an association between ethanolamine utilization and virulence in various pathogens. Despite the long history of research on this topic and an emerging, renewed interest, no review articles have been written. Herein, a synthesis of ethanolamine utilization and the important unanswered questions in terms of the catabolic and structural components, regulatory mechanisms and potential roles in pathogenesis is provided to spur further investigations in this area.
Overview of ethanolamine catabolism
S. typhimurium: a well-studied example
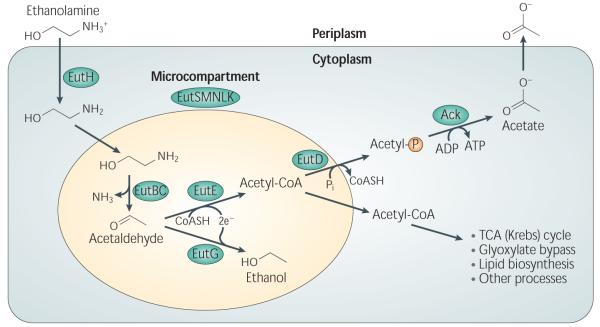
Ethanolamine utilization has been studied extensively in S. typhimurium. In this organism, 17 genes encoding the proteins involved in ethanolamine catabolism are located together on the chromosome in the ethanolamine utilization operon (eut) (Table 1)5,14-17. One characteristic of ethanolamine utilization is that all the essential enzymes reside in a multi-protein complex termed the carboxysome17, due to its similarity to organelles found in cyanobacteria that concentrate CO2 for fixation18; such organelles are alternately referred to as metabolosomes, enterosomes, polyhedral bodies, or bacterial microcompartments (Box 1). In ethanolamine utilization, the formation of a microcompartment is thought to help retain the volatile intermediate acetaldehyde to prevent the loss of this source of carbon and/or protect the cell from its potential toxic effects19,20. The ethanolamine-specific microcompartment comprises five different Eut proteins encoded by the eutS, L, K, M, and N genes17. Inside, as indicated schematically in Figure 1, the ethanolamine ammonia lyase EutBC breaks down ethanolamine into acetaldehyde and ammonia5,14. This requires AdoCbl (adenosylcobalamin) as a cofactor21, which is produced from cobalamin by a corrinoid cobalamin adenosyltransferase encoded by eutT15,22. The acetaldehyde produced is converted to acety-CoA by the acetaldehyde dehydrogenase, EutE5,14. Acetyl-CoA is subsequently used in a variety of different metabolic processes such as the TCA cycle, the glyoxylate cycle, or lipid biosynthesis. Acetyl-CoA can also be converted into acetylphosphate by the phosphotransacetylase, EutD. The housekeeping acetate kinase Ack can then carry out substrate level phosphorylation to create acetate from acetylphosphate, concomitantly generating ATP23,24. Alternatively, acetaldehyde can be converted to ethanol by the alcohol dehydrogenase, EutG, a function found necessary for the use of ethanolamine as a carbon source in S. typhimurium, perhaps because it prevents the buildup of toxic levels of acetaldehyde16.
Table 1.
Function of ethanolamine genes
| Gene | Function |
|---|---|
| eutS | Putative carboxysome structural protein |
| eutP | Ethanolamine utilization protein |
| eutQ | Ethanolamine utilization protein |
| eutT | Cobalamin adenosyltransferase |
| eutD | Phosphate acetyltransferase |
| eutM | Carboxysome structural protein |
| eutN | Carboxysome structural protein |
| eutE | Aldehyde oxidoreductase |
| eutJ | Putative chaperonin |
| eutG | Alcohol dehydrogenase |
| eutH | Transport protein |
| eutA | Reactivating factor |
| eutB | Ethanolamine ammonia-lyase large subunit |
| eutC | Ethanolamine ammonia-lyase small subunit |
| eutL | Carboxysome structural protein |
| eutK | Carboxysome structural protein |
| eutR | Regulator |
| eat | Ethanolamine permease |
| eutV | Sensor histidine kinase |
| eutW | Response regulator |
Data from Tsoy et al.13
Box1: Carboxysomes and related microcompartments.
Carboxysomes and other types of bacterial microcompartments are organelle-like structures distinguished by being enclosed with a protein shell rather than a lipid bilayer as found in eukaryotic organelles. Specific metabolic pathways are sequestered in these organelles including ethanolamine utilization, propanediol utilization, and carbon fixation in cyanobacteria. Carbon-fixing microcompartments, from which the name “carboxysome” originates, are the best understood. In these organelles, ribulose-1,5-bipshosphate carboxylase/oxygenase (RuBisCO) is exposed to elevated levels of CO2 enabling effective carbon fixation (reviewed in 18). It is postulated that the ethanolamine microcompartment may play a similar role by concentrating the intermediate acetaldehyde with the catabolic enzymes. In support of this idea, the need for the microcompartment can be abolished in S. typhimurium growing on ethanolamine by overproducing the catabolic enzymes19. However, profound structure/function questions remain such as how microcompartments allow relatively large compounds and proteins to enter, while at the same time retaining small gases.
Structural studies of the carboxysome indicate its shape to be icosahedral (20 sides) in shape with flat, triangular facets. The main shell proteins form hexamers consisting of six subunits surrounding a central pore. These hexagonal units pack together tightly to form the flat facets of the icosahedral structure whereas different shell proteins that form pentamers make up the 12 vertices (reviewed in 18). The structures of the shell proteins EutS, EutK, EutM, EutN and EutL that constitute part of the ethanolamine-degrading microcompartment have recently been elucidated53-55. Other than EutK, they all form hexagonal units like the previously elucidated shell proteins of the carboxysome. EutL has been crystallized with the pore both open and closed suggesting that these pores are gated55. Gated pores could explain how these protein organelles allow in certain large molecules (the co-factor Ado-B12 for example) while retaining small substrates and intermediates such as CO2 and acetaldehyde.
Figure 1.
Model for ethanolamine catabolism. Adapted from Brinsmade et al. 19. Ethanolamine enters the cell through diffusion or with the help of EutH. Within the ethanolamine-specific microcompartment, ethanolamine is degraded to acetaldehyde and ammonia by EutBC, which requires the co-factor AdoCbl. Acetaldehyde can be catabolized to alcohol by EutG or the metabolically useful compound, acetyl-CoA, by EutE. Acetyl-CoA can be used in a variety of metabolic processes or made into acetylphosphate by EutD. By substrate level phosphorylation, Ack can generate ATP and acetate from acetylphosphate.
Several other proteins encoded in the eut locus play a more indirect role in the utilization of ethanolamine in S. typhimurium. EutA is a reactivating factor for the ethanolamine ammonia lyase25, , EutJ may be a chaperone for EutG and EutE16,26. Ethanolamine generally diffuses freely across membranes, but at low pH EutH facilitates diffusion across the cell envelope26. Two genes, eutP and eutQ, have not yet been assigned a function19, though EutP is postulated to have GTPase activity and may be involved in reactivating the AdoCbl cofactor13. Finally, a single, DNA-binding protein belonging to the AraC family, EutR, positively regulates the transcription of the eut operon of S. typhimurium5,27.
eut gene organization in other species
A recent study focused on the comparative genomics of ethanolamine utilization discovered that almost 100 fully sequenced bacterial genomes contain eut operons13. The authors determined the content and organizational differences among the different phylogenetic groups of bacteria that contain these genes. Among the interesting findings, Actinobacteria and most Proteobacteria have short eut operons containing eutBC, frequently with a transporter encoded by eat, which is a functional, non-homologous equivalent to eutH. Some of the Proteobacteria also contain eutR at a different genomic location from eutBC. In contrast, members of the proteobacterial family Enterobacteriaceae, which includes S. typhimurium and E. coli, have long eut operons, as do members of the phylum Firmicutes. Among these long operon-containing species, there are considerable differences in the exact gene content and organization. Interestingly, some species, such as Klebsiella pneumoniae and Pseudomonas fluorescens, contain both a long and a short operon. A closer look at some of the data presented in this study reveals the following. The microcompartment structural components are not present in those species containing the short operons suggesting that this organelle’s possible roles in concentrating acetaldehyde or protecting the rest of the cell from its toxic effects are expendable. Additionally, almost all the species that contain the long operons are facultative anaerobes that live either as commensals and/or pathogens in the gut and/or mouth. In contrast, it appears that many or most of the organisms that have short operons are obligate aerobes. Therefore it is possible that the microcompartment is necessary for ethanolamine utilization in the GI tract and/or under anaerobic conditions. Perhaps the organisms in this environment are more dependent on ethanolamine as an energy source and have been under selective pressure to evolve and maintain the accessory proteins because they promote efficient utilization of this compound. Indeed it has been shown S. typhimurium can still utilize ethanolamine in the absence of the microcompartment proteins as long as the catalytic enymes are overproduced, suggesting that the microcompartment increases efficiency19. A deeper understanding of the biological role(s) of the microcompartment will require further study.
In addition to this analysis, the gene organization and content of the eut operon in Enterococcus faecalis was recently described in detail. It contains homologues to most of the genes found in S. typhimurium with a few exceptions, the most notable of which is the lack of the gene encoding the EutR regulator and the inclusion of a two-component system designated EutW (sensor kinase) and EutV (response regulator)7,12. Tsoy et al. found that, like E. faecalis, all other Firmicutes that contain eut operons have the EutVW regulatory system, whereas all Enterobacteriaceae have the EutR regulator. Together, these observations have been taken to suggest that the earliest version of the eut operon contained eutBC and the eat genes. eutR and the other components then appeared in the branch leading to the Enterobacteriaceae, while eutA and the genes encoding the two-component system were independently acquired in Firmicutes. The other genes contained in the long operons are postulated to have arisen in Firmicutes and been acquired by Enterobacteriaceae by horizontal transfer events13. Therefore study of the regulatory systems, EutR and EutVW, offers the chance to analyze two different regulatory solutions for a common task.
Regulatory mechanisms controlling eut gene expression
The EutR system
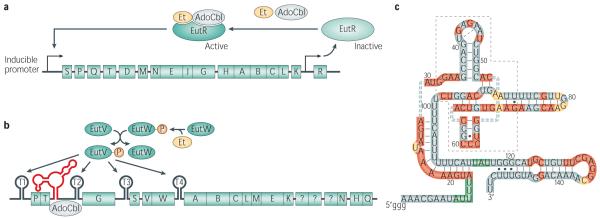
Early work established that both ethanolamine and AdoCbl are required for synthesis of EutBC in E. col28. The mechanism by which the “concerted induction” of the ethanolamine ammonia lyase is achieved was not studied until a decade later. Working in S. typhimurium, the Roth laboratory first surmised the existence of EutR, a global positive regulator of eut gene expression, from a genetic screen for mutants unable to utilize ethanolamine12,24. Further studies, that continued to rely almost exclusively on the use of bacterial genetics, established the following facts about the system as illustrated in Figure 2A. The primary eut promoter at the beginning of the operon is inducible in the presence of ethanolamine and AdoCbl. A second promoter, in front of the eutR gene, provides a low-level of constitutive expression. Induction of the operon via EutR requires the presence of both ethanolamine and AdoCbl, but high-levels of EutR can cause partial induction with just one inducer and increased maximal expression in the presence of both inducers27. An interesting feature of this regulatory system is that EutR induces the expression of its gene in a positive regulatory loop. Increasing the levels of EutR may be necessary to maintain the induction since one of the inducing agents, AdoCbl, is also required as a co-factor for the catalytic activity of EutBC. Using strains in which EutR and EutBC are produced at different levels, the Roth group presented evidence that autoregulation of eutR helps maintain induced expression despite competition between EutR and EutBC for AdoCbl. The model is as follows – uninduced cells producing only a small amount of EutR can sense low levels of AdoCbl. Once EutBC is produced, it competes with EutR for AdoCbl but because more EutR is produced in addition to EutBC upon system induction, EutR is still able to sense low levels of AdoCbl and maintain induced expression29.
Figure 2.
Regulatory features of the eut operons in S. typhimurium (A) and E. faecalis (B). (A) The message encoding EutR is produced at a low-level from constitutive promotor P2. The inactive regulator is thought to sense the presence of ethanolamine and AdoCbl by direct binding which changes the conformation to the active form, promoting transcription of the entire eut operon from promoter P1. (B) Upon ethanolamine binding, the sensor kinase EutW autophosphorylates followed by phosphotransfer to the response regulator EutV. Activation of the two-component system by phosphorylation leads to disruption of terminators present in the message, likely by direct binding, allowing transcriptional read-through. The untranslated region proximal to the T2 terminator also contains a riboswitch, which upon binding to AdoCbl additionally promotes antitermination. P = promoter, T = terminator, blue boxes with letters represent the various eut genes mentioned in this review. The question marks represent genes whose identity is still controversial.
The exact mechanism by which AdoCbl and ethanolamine activate EutR remains unknown. The genetic and in vivo work suggest EutR directly binds these compounds, presumably causing a conformational change that allows for DNA binding and transcriptional activation. However, direct biochemical evidence for binding of EutR to the inducing compounds or to DNA is still lacking. In fact, no work has been done on this regulator in over 15 years. The only new information has been provided by a bioinformatics analysis that identified potential EutR binding sites in the promoter regions of eut operons in species that contain this regulatory system13. Although questions remain regarding EutR regulation in proteobacterial species, recent data have revealed a unique regulatory mechanism in Firmicutes.
The EutVW system
As mentioned, in common with the Enterobacteriacae, many of the Firmicutes that encode eut genes do so in a “long” eut operon. However, the Firmicutes do not have the EutR regulator and instead encode the EutVW two-component system (Figure 2B). Histidine kinases of two-component systems generally autophosphorylate upon sensing a specific signal and subsequently transfer the phosphoryl group to a dedicated response regulator. Phosphorylation of the response regulator typically occurs at a conserved aspartate residue in the receiver domain and causes a conformational change that influences the activity of a second, output domain, which carries out the regulatory response30. Most output domains modulate the activity of various processes via protein-DNA or protein-protein interactions, however, it has been suggested that a small percentage (< 1.0%) may regulate gene expression through RNA-binding activity31. Specifically, the output domains of these response regulators are predicted to have antitermination activity that is believed to be mediated by ANTAR (AmiR and NasR Transcriptional Antiterminator Regulators) domains32. Studies on two ANTAR-containing response regulators, AmiR and NasR, demonstrated that these proteins interact with their target mRNA transcripts and prevent formation of a transcriptional terminator to induce gene expression31-35.
The eut-associated response regulator, EutV, like AmiR and NasR, has an ANTAR domain7,12, indicating that it may also bind RNA and disrupt terminators to affect gene expression. Recent data show that the long operons of Firmicutes species that contain this regulator have up to four putative stem-loop structures that resemble intrinsic transcriptional terminators, located at varying positions within the operon12. Depending on the operon organization, stem-loops usually precede the eutP, eutV, eutG, eutS and eutA genes. In E. faecalis, intrinsic transcription terminators have been identified in front of eutP, eutG, eutS and eutA (Figure 2B). Although most of the primary sequence of the stem-loops was not highly conserved, a 13-nucleotide patch of sequence conservation overlapping the 5′ proximal portion of the terminators was identified. Not only was this sequence conserved among the terminators found in eut operons, it was also visible in the primary sequence of the terminator recognized by AmiR. These data suggest the straightforward hypothesis that this sequence element comprises the antiterminator recognition element, and that binding of the protein to this site in the RNA prevents terminator formation. Preliminary gel shift experiments indicated that EutV is able to interact with the eutP 5′ UTR (untranslated region) that contains this terminator12.
The predicted effect of antitermination by EutV is positive regulation of eut gene expression. Indeed, deletion of the two-component system prevented expression from a lacZ gene when fused to the upstream regulatory region of eutP12. Moreover, it has also been shown that, like S. typhimurium, E. faecalis can grow on ethanolamine as a sole carbon source as long as AdoCbl is provided. (Interestingly and unlike S. typhimurium, E. faecalis also requires anaerobic conditions to grow on ethanolamine. Since no alternative electron acceptor is provided in the minimal medium, the growth conditions suggest that E. faecalis is fermenting ethanolamine). However, a strain containing a deletion in eutV cannot, presumably because the gene product is required for induction of the catabolic machinery7. Together, these data suggest that in the Firmicutes, in addition to the two-component system, ethanolamine and AdoB12 are required to activate eut gene expression.
How does this system sense and respond appropriately to the presence of ethanolamine and AdoCbl? The sensor kinase, EutW, has a unique sensing domain that is not homologous to the sensing domains of other histidine kinases. Using in vitro kinase assays it was shown that ethanolamine activates autophosphorylation of EutW. When purified EutV was additionally added, phospho-transfer to EutV was also observed 7,12. A simple model can be proposed in which direct binding of ethanolamine to EutW causes auto-phosphorylation followed by phospho-transfer and activation of EutV. EutV binds to the untranslated regions in the eut operon and prevents terminator formation, allowing transcriptional readthrough. How does this system also sense the presence of AdoCbl? (AdoCbl does not affect the efficiency of autophosphorylation and phosphorylation of EutW and EutV, at least in vitro (A. Ramesh, personal communication)).
In addition to the stem-loop structures indicating putative terminators that were found in the untranslated regions of the E. faecalis eut operon, a possible AdoCbl riboswitch was also identified7,12,36. Riboswitches are cis-acting regulatory components located in untranslated regions of mRNAs. In bacteria, they frequently function by affecting the transcription or translation of downstream genes. They comprise an aptamer domain that directly interacts with various metabolites and an expression platform, which undergoes structural changes upon metabolite binding to influence gene expression. In Gram-positive bacteria, the expression platform mostly consists of terminator/antiterminator structures that affect transcription elongation, whereas in Gram-negative bacteria it is more common to find stem-loop structures affecting the availability of a ribosome binding site, thereby modulating translation37.
In E. faecalis, the potential AdoCbl riboswitch was discovered in an intergenic region after the first two genes (eutP and eutT) and before a terminator in front of eutG (Figure 2B). AdoCbl increases eut expression through an antitermination mechanism exerted via an AdoCbl-sensing riboswitch12. As summarized in Figure 2B, these data indicate that Firmicutes have evolved posttranscriptional regulatory mechanisms for sensing ethanolamine and AdoCbl in lieu of transcriptional regulation by EutR, as occurs in proteobacterial species. This suggests that, in certain instances, posttranscriptional regulatory networks are likely to be functionally equivalent to transcriptional counterparts.
Many important questions remain. Interestingly, not all the Firmicutes have an AdoCbl riboswitch; Clostridium species, for example, appear to be missing this feature (K. Fox, personal communication). Do these bacteria still actively sense AdoCbl, and if so, how? How and why are these operons regulated by up to four different terminators? One can imagine a mechanism by which the rate at which transcripts from a given gene are produced is influenced by the sensitivity of the proximal terminator to antitermination by the two-component system. Such a mechanism could adjust the final ratios of the ethanolamine components produced. How do ANTAR-domain response regulators such as EutV bind RNA to affect antitermination? Some structure/function studies have been carried out in the NasR and AmiR systems by introducing mutations into the predicted stem loops of the terminators, but this has not led to a clear hypothesis for the mechanism33-35. AmiR and another putative antiterminator from Mycobacterium tuberculosis have been crystallized38,39, but a co-crystal of an antiterminator bound to RNA could potentially reveal much about the mechanism of antitermination in these systems.
Possible roles of ethanolamine utilization in bacterial pathogenesis
Many of the bacterial species that harbor eut genes are found in the intestines of mammals including species of Listeria, Salmonella, Escherichia, Enterococcus and Clostridium. Indeed, the intestinal tract provides a rich source of phosphatidylethanolamine due to its presence in bacterial and eukaryotic cell membranes, and in the host diet 1,2,5. Even in fasting animals, a significant amount of phosphatidylethanolamine is found in the intestine simply due to turnover and exfoliation of intestinal cells40. There is now evidence that the bacterial genes encoding proteins for ethanolamine utilization are highly expressed in the gut. For example, a recent study examining gene expression in L. monocytogenes under a variety of conditions demonstrated that all of the eut genes are upregulated in the intestine of a mouse infection model41.
The ability to use ethanolamine could contribute to the pathogenesis of these species in an indirect way by providing a useful source of carbon and/or nitrogen that promotes successful colonization of the intestine. An alternative to this “nutrition hypothesis” would be if the breakdown of ethanolamine contributed to disrupting gut functions, particularly innate immune functions. Although there is no direct evidence for such a mechanism occurring during intestinal infections, a related example is found in lung infections. The pathogen Pseudomonas aeruginosa uses a phospholipase C enzyme (PlcH) to digest phosphatidylcholine, which is among the phospholipids that constitute lung surfactant42. The resulting phosphorylcholine is further catabolized and is a potentially useful nutritional source of phosphate, carbon and nitrogen. However, the breakdown of phosphatidylcholine also changes the properties of the pulmonary surfactant such that it is unable to maintain alveolar stability and respiratory function, contributing to disease in the P. aeruginosa-infected lung43. The ability to breakdown phosphatidylethanolamine, a prevalent phospholipid in the gut, could contribute to disease by a similar mechanism if this lipid was necessary for proper gut functioning and/or mucosal innate immunity. Another possibility is that the breakdown of ethanolamine could modulate innate immunity via the production of acetate. Recall that during the breakdown of ethanolamine, acetate can be formed from acetyl-CoA by a pathway involving the phosphotransacetylase EutD24. Recent data reveal that in the gut, acetate interacts with a G-protein coupled receptor (GPR43) to modulate innate immunity and inflammation44.
In support of a role for ethanolamine utilization in bacterial pathogenesis, various expression studies in Salmonella typhimurium have linked eut expression to the activity of global regulators of virulence. For example, CsrA, a global regulator important for the expression of the virulence genes located in SPI-I (Salmonella pathogenicity island 1) and flagellar genes, also upregulates the eut genes. In a microarray analysis of a csrA mutant, all of the eut genes were expressed at levels two- to 10-fold less than wild type45. Fis, another global regulator of Salmonella typhimurium virulence determinants, including SPI genes (specifically those found in SPI-1, SPI-2, SPI-3 and SPI-5) and flagellar genes, also positively regulates the eut genes, as discovered by microarray analysis46. One study examined the effects of mutations in eut genes on a mouse model of S. typhimurium infection and found a 5-10-fold increase in the 50% lethal dose16.
In contrast to Salmonella, a global regulator of virulence in E. faecalis called the Fsr system was found by microarray analysis to strongly downregulate genes in the eut operon during stationary phase between 8- and almost 300-fold47. Perhaps this difference can be attributed to the role of E. faecalis as a commensal organism rather than a pathogen in the mammalian intestine where ethanolamine utilization may help E. faecalis colonize and/or survive rather than cause infection. Interestingly, in the nematode Caenorhabditis elegans, E. faecalis causes a persistent and deadly infection in the intestine48. In this context, a transposon insertion mutant in the eut operon was identified in a screen for mutants attenuated in killing C. elegans, suggesting that ethanolamine utilization promotes pathogenesis in the worm49.
Upregulation of eut genes during a variety of host-pathogen interactions provides additional evidence supporting a role for ethanolamine utilization in bacterial pathogenesis. For example, in an extensive expression study using tiling microarrays to look at gene expression in L. monocytogenes under a variety of conditions, all of the eut genes were strongly upregulated in the intestines of infected mice and many were also expressed at slightly higher levels in human blood41. It was also found that the eut operon was upregulated in Listeria monocytogenes during intracellular growth in the cytosol of human colon epithelial cells (Caco-2) cells by microarray analysis. When a strain containing a deletion in eutB was studied, it was defective in intracellular growth, reaching only 10% of the level of the control group after being allowed to replicate within Caco-2 cells for seven hours. These data suggest that EutB positively contributes to the ability of L. monocytogenes to grow effectively in the intracellular environment50. Photorhabdus luminescens is a Gram-negative bacterium that lives as a symbiote in the gut of the nematode Heterorhabditis bacteriophora. When this nematode invades insects such as the wax moth caterpillar (Galleria mellonella), the bacteria are regurgitated into the insect where they cause a lethal infection, thereby providing a meal for their nematode host. Interestingly, in a study using a promoter-trap library, eutABC were among the genes upregulated in the hemolymph of the infected insect, suggesting a possible role for ethanolamine utilization during this host-pathogen interaction51. The upregulation of eut genes during host-pathogen interactions in plants has also been observed. A GFP-based in vivo expression technology (IVET) leaf assay identified eutR among the genes upregulated in a screen of the Gram-negative plant pathogen Erwinia chrysanthemi during infection of spinach. A strain with a mutation in eutR was subsequently found to still be capable of creating a local maceration, but was unable to cause systematic infection in the African violet. These data suggest that the ability to use ethanolamine contributes in some way to the ability of E. chrysanthemi to metastasize within the plant52.
In conclusion, a variety of evidence from a diverse range of pathogens and different types of infection links ethanolamine utilization to bacterial pathogenesis. However, to date, many of these connections are indirect and remain speculative. It is unknown whether ethanolamine is simply a valuable nutritional source of carbon and/or nitrogen in a range of host environments, or if the ability to digest this compound contributes to host invasion and/or immune evasion by a more specific mechanism.
Summary
Aspects of ethanolamine utilization have been studied since the 1970s in E. coli and S. typhimurium with a variety of interesting and important findings, including the characterization of the enzymatic and structural components as well as the genetic locus that encodes these functions. More recent studies that encompass bacterial species other than E. coli and S. typhimurium have demonstrated that there is wide variation in the gene content, organization and regulation of eut operons7,12,13. Investigations into the regulation have revealed novel mechanisms of post-transcriptional regulation, but much remains to be learned. Another future challenge will be to elucidate the relationship between ethanolamine utilization and bacterial pathogenesis. Despite a preponderance of recent data suggesting a connection, no clear understanding of the role of ethanolamine utilization in virulence has emerged and this remains a ripe topic for future study.
Acknowledgements
I thank W.C. Winkler and K.A. Fox for critical reading of the manuscript. This work was supported by award R21AI078104 to D.A.G. from the National Institute of Allergy and Infectious Diseases.
References
- 1.Randle CL, Albro PW, Dittmer JC. The phosphoglyceride composition of Gram-negative bacteria and the changes in composition during growth. Biochim Biophys Acta. 1969;187:214–20. doi: 10.1016/0005-2760(69)90030-7. [DOI] [PubMed] [Google Scholar]
- 2.White DA. Phospholipid composition of mammalian tissues. In: Ansell GB, Hawthorne JN, Dawson RMC, editors. Form and function of phospholipids. Elsevier Publishing Company; New York: 1973. pp. 441–482. [Google Scholar]
- 3.Larson TJ, Ehrmann M, Boos W. Periplasmic glycerophosphodiester phosphodiesterase of Escherichia coli, a new enzyme of the glp regulon. J Biol Chem. 1983;258:5428–32. [PubMed] [Google Scholar]
- 4.Proulx P, Fung CK. Metabolism of phosphoglycerides in E. coli. IV. The positional specificity and properties of phospholipase A. Can J Biochem. 1969;47:1125–8. doi: 10.1139/o69-181. [DOI] [PubMed] [Google Scholar]
- 5.Roof DM, Roth JR. Ethanolamine utilization in Salmonella typhimurium. J Bacteriol. 1988;170:3855–63. doi: 10.1128/jb.170.9.3855-3863.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang GW, Chang JT. Evidence for the B12-dependent enzyme ethanolamine deaminase in Salmonella. Nature. 1975;254:150–1. doi: 10.1038/254150a0. [DOI] [PubMed] [Google Scholar]
- 7.Del Papa MF, Perego M. Ethanolamine activates a sensor histidine kinase regulating its utilization in Enterococcus faecalis. J Bacteriol. 2008 doi: 10.1128/JB.00952-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackwell CM, Scarlett FA, Turner JM. Ethanolamine catabolism by bacteria, including Escherichia coli. Biochem Soc Trans. 1976;4:495–7. doi: 10.1042/bst0040495. [DOI] [PubMed] [Google Scholar]
- 9.Bradbeer C. The clostridial fermentations of choline and ethanolamine. II. Requirement for a cobamide coenzyme by an ethanolamine deaminase. J Biol Chem. 1965;240:4675–81. [PubMed] [Google Scholar]
- 10.Bradbeer C. The clostridial fermentations of choline and ethanolamine. 1. Preparation and properties of cell-free extracts. J Biol Chem. 1965;240:4669–74. [PubMed] [Google Scholar]
- 11.Jones PW, Turner JM. Interrelationships between the enzymes of ethanolamine metabolism in Escherichia coli. J Gen Microbiol. 1984;130:299–308. doi: 10.1099/00221287-130-2-299. [DOI] [PubMed] [Google Scholar]
- 12.Fox KA, et al. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proc Natl Acad Sci U S A. 2009;106:4435–40. doi: 10.1073/pnas.0812194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsoy O, Ravcheev D, Mushegian A. Comparative genomics of ethanolamine utilization. J Bacteriol. 2009 doi: 10.1128/JB.00838-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roof DM, Roth JR. Functions required for vitamin B12-dependent ethanolamine utilization in Salmonella typhimurium. J Bacteriol. 1989;171:3316–23. doi: 10.1128/jb.171.6.3316-3323.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheppard DE, Penrod JT, Bobik T, Kofoid E, Roth JR. Evidence that a B12-adenosyl transferase is encoded within the ethanolamine operon of Salmonella enterica. J Bacteriol. 2004;186:7635–44. doi: 10.1128/JB.186.22.7635-7644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stojiljkovic I, Baumler AJ, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357–66. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kofoid E, Rappleye C, Stojiljkovic I, Roth J. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J Bacteriol. 1999;181:5317–29. doi: 10.1128/jb.181.17.5317-5329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat Rev Microbiol. 2008;6:681–91. doi: 10.1038/nrmicro1913. [DOI] [PubMed] [Google Scholar]
- 19.Brinsmade SR, Paldon T, Escalante-Semerena JC. Minimal functions and physiological conditions required for growth of salmonella enterica on ethanolamine in the absence of the metabolosome. J Bacteriol. 2005;187:8039–46. doi: 10.1128/JB.187.23.8039-8046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Penrod JT, Roth JR. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J Bacteriol. 2006;188:2865–74. doi: 10.1128/JB.188.8.2865-2874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scarlett FA, Turner JM. Microbial metabolism of amino alcohols. Ethanolamine catabolism mediated by coenzyme B12-dependent ethanolamine ammonia-lyase in Escherichia coli and Klebsiella aerogenes. J Gen Microbiol. 1976;95:173–6. doi: 10.1099/00221287-95-1-173. [DOI] [PubMed] [Google Scholar]
- 22.Buan NR, Suh SJ, Escalante-Semerena JC. The eutT gene of Salmonella enterica Encodes an oxygen-labile, metal-containing ATP:corrinoid adenosyltransferase enzyme. J Bacteriol. 2004;186:5708–14. doi: 10.1128/JB.186.17.5708-5714.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brinsmade SR, Escalante-Semerena JC. The eutD gene of Salmonella enterica encodes a protein with phosphotransacetylase enzyme activity. J Bacteriol. 2004;186:1890–2. doi: 10.1128/JB.186.6.1890-1892.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Starai VJ, Garrity J, Escalante-Semerena JC. Acetate excretion during growth of Salmonella enterica on ethanolamine requires phosphotransacetylase (EutD) activity, and acetate recapture requires acetyl-CoA synthetase (Acs) and phosphotransacetylase (Pta) activities. Microbiology. 2005;151:3793–801. doi: 10.1099/mic.0.28156-0. [DOI] [PubMed] [Google Scholar]
- 25.Mori K, Bando R, Hieda N, Toraya T. Identification of a reactivating factor for adenosylcobalamin-dependent ethanolamine ammonia lyase. J Bacteriol. 2004;186:6845–54. doi: 10.1128/JB.186.20.6845-6854.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Penrod JT, Mace CC, Roth JR. A pH-sensitive function and phenotype: evidence that EutH facilitates diffusion of uncharged ethanolamine in Salmonella enterica. J Bacteriol. 2004;186:6885–90. doi: 10.1128/JB.186.20.6885-6890.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roof DM, Roth JR. Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella typhimurium. J Bacteriol. 1992;174:6634–43. doi: 10.1128/jb.174.20.6634-6643.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blackwell CM, Turner JM. Microbial metabolism of amino alcohols. Formation of coenzyme B12-dependent ethanolamine ammonia-lyase and its concerted induction in Escherichia coli. Biochem J. 1978;176:751–7. doi: 10.1042/bj1760751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheppard DE, Roth JR. A rationale for autoinduction of a transcriptional activator: ethanolamine ammonia-lyase (EutBC) and the operon activator (EutR) compete for adenosyl-cobalamin in Salmonella typhimurium. J Bacteriol. 1994;176:1287–96. doi: 10.1128/jb.176.5.1287-1296.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–45. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 31.Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol. 2006;188:4169–82. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shu CJ, Zhulin IB. ANTAR: an RNA-binding domain in transcription antitermination regulatory proteins. Trends Biochem Sci. 2002;27:3–5. doi: 10.1016/s0968-0004(01)02036-9. [DOI] [PubMed] [Google Scholar]
- 33.Chai W, Stewart V. NasR, a novel RNA-binding protein, mediates nitrate-responsive transcription antitermination of the Klebsiella oxytoca M5al nasF operon leader in vitro. J Mol Biol. 1998;283:339–51. doi: 10.1006/jmbi.1998.2105. [DOI] [PubMed] [Google Scholar]
- 34.Chai W, Stewart V. RNA sequence requirements for NasR-mediated, nitrate-responsive transcription antitermination of the Klebsiella oxytoca M5al nasF operon leader. J Mol Biol. 1999;292:203–16. doi: 10.1006/jmbi.1999.3084. [DOI] [PubMed] [Google Scholar]
- 35.Wilson SA, Wachira SJ, Norman RA, Pearl LH, Drew RE. Transcription antitermination regulation of the Pseudomonas aeruginosa amidase operon. Embo J. 1996;15:5907–16. [PMC free article] [PubMed] [Google Scholar]
- 36.Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 38.Morth JP, Feng V, Perry LJ, Svergun DI, Tucker PA. The crystal and solution structure of a putative transcriptional antiterminator from Mycobacterium tuberculosis. Structure. 2004;12:1595–605. doi: 10.1016/j.str.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 39.O’Hara BP, et al. Crystal structure and induction mechanism of AmiC-AmiR: a ligand-regulated transcription antitermination complex. Embo J. 1999;18:5175–86. doi: 10.1093/emboj/18.19.5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cotton PB. Non-dietary lipid in the intestinal lumen. Gut. 1972;13:675–81. doi: 10.1136/gut.13.9.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toledo-Arana A, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009 doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 42.Berka RM, Vasil ML. Phospholipase C (heat-labile hemolysin) of Pseudomonas aeruginosa: purification and preliminary characterization. J Bacteriol. 1982;152:239–45. doi: 10.1128/jb.152.1.239-245.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lema G, Dryja D, Vargas I, Enhorning G. Pseudomonas aeruginosa from patients with cystic fibrosis affects function of pulmonary surfactant. Pediatr Res. 2000;47:121–6. doi: 10.1203/00006450-200001000-00021. [DOI] [PubMed] [Google Scholar]
- 44.Maslowski KM, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–6. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lawhon SD, et al. Global regulation by CsrA in Salmonella typhimurium. Mol Microbiol. 2003;48:1633–45. doi: 10.1046/j.1365-2958.2003.03535.x. [DOI] [PubMed] [Google Scholar]
- 46.Kelly A, et al. A global role for Fis in the transcriptional control of metabolism and type III secretion in Salmonella enterica serovar Typhimurium. Microbiology. 2004;150:2037–53. doi: 10.1099/mic.0.27209-0. [DOI] [PubMed] [Google Scholar]
- 47.Bourgogne A, Hilsenbeck SG, Dunny GM, Murray BE. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J Bacteriol. 2006;188:2875–84. doi: 10.1128/JB.188.8.2875-2884.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garsin DA, et al. A simple model host for identifying Gram-positive virulence factors. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10892–7. doi: 10.1073/pnas.191378698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maadani A, Fox KA, Mylonakis E, Garsin DA. Enterococcus faecalis Mutations Affecting Virulence in the C. elegans Model Host. Infect Immun. 2007 doi: 10.1128/IAI.01372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joseph B, et al. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol. 2006;188:556–68. doi: 10.1128/JB.188.2.556-568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Munch A, Stingl L, Jung K, Heermann R. Photorhabdus luminescens genes induced upon insect infection. BMC Genomics. 2008;9:229. doi: 10.1186/1471-2164-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang S, et al. Genome-wide identification of plant-upregulated genes of Erwinia chrysanthemi 3937 using a GFP-based IVET leaf array. Mol Plant Microbe Interact. 2004;17:999–1008. doi: 10.1094/MPMI.2004.17.9.999. [DOI] [PubMed] [Google Scholar]
- 53.Forouhar F, et al. Functional insights from structural genomics. J Struct Funct Genomics. 2007;8:37–44. doi: 10.1007/s10969-007-9018-3. [DOI] [PubMed] [Google Scholar]
- 54.Sagermann M, Ohtaki A, Nikolakakis K. Crystal structure of the EutL shell protein of the ethanolamine ammonia lyase microcompartment. Proc Natl Acad Sci U S A. 2009;106:8883–7. doi: 10.1073/pnas.0902324106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tanaka S, Sawaya MR, Yeates TO. Structure and mechanisms of a protein-based organelle in Escherichia coli. Science. 2010;327:81–4. doi: 10.1126/science.1179513. [DOI] [PubMed] [Google Scholar]