Abstract
In mammalian cells, newly synthesized mRNAs undergo a pioneer round of translation that is important for mRNA quality control. Following maturation of messenger ribonucleoprotein particles during and after the pioneer round, steady-state cycles of mRNA translation generate most of the cell’s proteins. Translation factors, RNA binding proteins and targets of signaling pathways that are particular to newly synthesized mRNAs regulate critical functions of the pioneer round.
Introduction
In mammalian cells, newly synthesized transcripts are subject to a series of nuclear processing steps to become mature templates for protein synthesis (Moore and Proudfoot, 2009). The 5’-ends of these transcripts acquire a 5’-m7GpppN cap structure (where N is the first transcribed nucleotide) while being elongated by RNA polymerase II. The cap first binds to the cap binding protein (CBP) heterodimer CBP80-CBP20 (CBC), which supports the pioneer round of mRNA translation (Isken and Maquat, 2008). This round involves the loading of one or more ribosomes, depending on the efficiency of translation initiation and the length of the open translational reading frame (Isken and Maquat, 2008; Isken et al., 2008). The cap subsequently binds to the eukaryotic translation initiation factor 4E (eIF4E), which directs steady-state rounds of mRNA translation (Isken and Maquat, 2008).
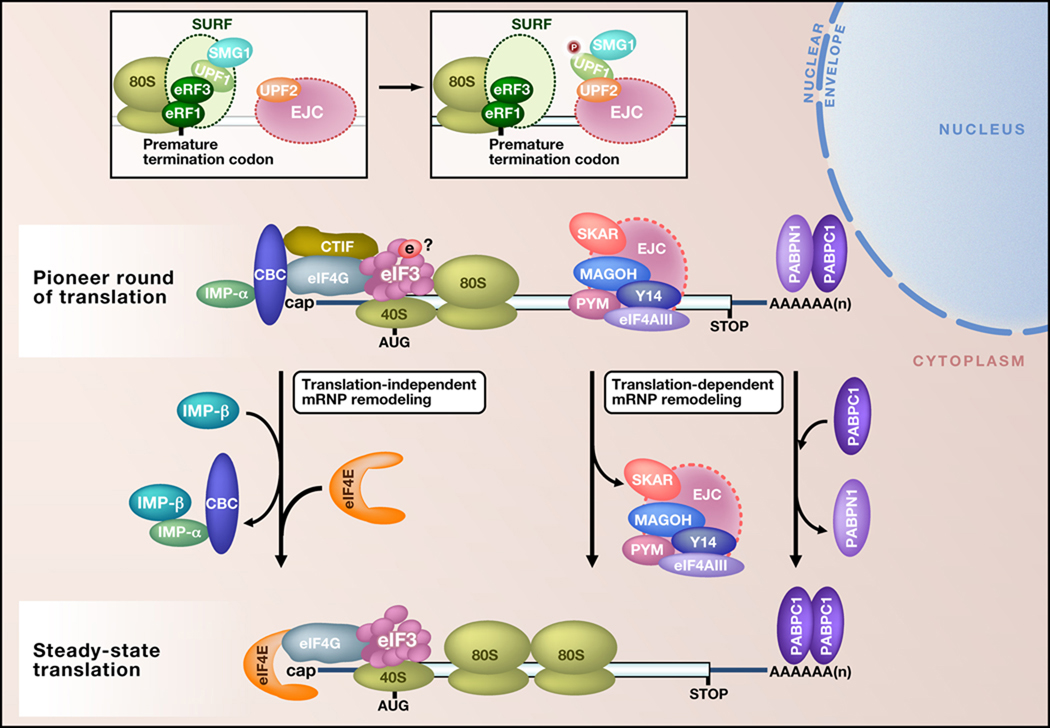
Although CBC-bound mRNAs are precursors to eIF4E-bound mRNAs, the two messenger ribonucleoprotein particles (mRNPs) differ in significant ways (Figure 1). For example, spliced CBC-bound mRNAs differ from the eIF4E-bound mRNAs that derive from them because they are associated with one or more exon-junction complexes (EJCs) of proteins. By the time eIF4E replaces CBC at the mRNA cap, EJCs are no longer detectable, largely because most reside within the coding region of mRNAs and therefore are displaced by translating ribosomes during the pioneer round (Gehring et al., 2009; Sato and Maquat, 2009). As another example, the poly(A) tails of CBC-bound mRNAs are associated with the mostly nuclear but shuttling poly(A)-binding protein N1 (PABPN1) and the primarily cytoplasmic but likewise shuttling PABPC1; in contrast, eIF4E-bound mRNAs do not detectably bind to PABPN1, the replacement of which by PABPC1 is promoted by the pioneer round of translation (Sato and Maquat, 2009).
Figure 1. Pioneer and steady-state translation initiation complexes.
Shown is a CBP80-CBP20 (CBC)-bound mRNP from the pioneer round of translation and an eIF4E-bound mRNP from steady-state translation. CBC-bound mRNAs direct pioneer rounds of translation, whereas eIF4E-bound mRNAs, which derive from the remodeling of CBC-bound mRNAs, support the bulk of cellular protein synthesis. CBC-bound mRNAs are associated with at least one exon-junction complex (EJC) (provided they are the product of pre-mRNA splicing) and the poly(A) binding proteins PABPN1 and PABPC1. PYM, which interacts with EJC components and the small 40S ribosomal subunit (not shown), and SKAR (S6 kinase 1 ALY-REF-like target), a component of EJCs, may help to activate the pioneer round of translation. eIF3e, a non-core subunit of the eukaryotic translation initiation factor eIF3, may regulate the translation of a specific set of CBC-bound mRNAs. CTIF (CBP80-CBP20-dependent translation initiation factor) interacts directly with CBP80, as does eIF4G. It is currently unclear whether CTIF is the sole eIF4G-like molecule or if eIF4G also functions during pioneer rounds. eIF4G has been proposed to form a complex with poly(A)-bound PABPC1 to circularize and promote the translation of CBC-bound mRNAs, similar to how poly(A)-bound PABPC1 circularizes and promotes the translation of eIF4E-bound mRNAs. Importin (IMP)-β binds to IMP-α (which is a stable constituent of cap-bound CBC) and augments the translation-independent replacement of CBC by eIF4E. In contrast, the pioneer round of translation promotes the removal of EJCs and of associated RNA-binding proteins such as SF2/ASF (which also activates the pioneer round; not shown), and the replacement of PABPN1 by PABPC1. The insets depict how the SURF complex (comprising the PIK-related protein kinase SMG1, UPF1, eRF1 and eRF3) assembles together with an 80S stalled ribosome at a premature termination codon of CBC-bound mRNA. SMG1 subsequently phosphorylates UPF1 upon UPF1 and SMG1 binding to a downstream EJC during the process of nonsense-mediated mRNA decay. AUG, translation initiation codon; STOP, normal termination codon.
Despite these and other differences (see below), both CBC-bound and eIF4E-bound mRNAs most likely engage in similar mechanisms of translation initiation, elongation and termination. Therefore, it is not surprising that both CBC-bound and eIF4E-bound mRNAs use many of the same translation initiation factors. These factors include not only PABPC1, which data indicate is important for activating the translation of both mRNPs, but also eIF4G, eIF3, eIF4B, eIF4A and eIF2 (Figure 1, 2; Isken and Maquat, 2008), and undoubtedly many other factors that work in conjunction with ribosomes to synthesize proteins. Although both mRNPs support protein synthesis and can be targeted for translational activation or repression, the purpose for so doing is distinct: the translation of CBC-bound mRNAs provides a means to control the quality of gene expression; in contrast, the translation of eIF4E-bound mRNAs generates the bulk of cellular proteins (Isken and Maquat, 2008). Here, we discuss our growing understanding of how cells maintain the specialized functions of each mRNP via associations with particular translation factors, RNA binding proteins and signaling targets.
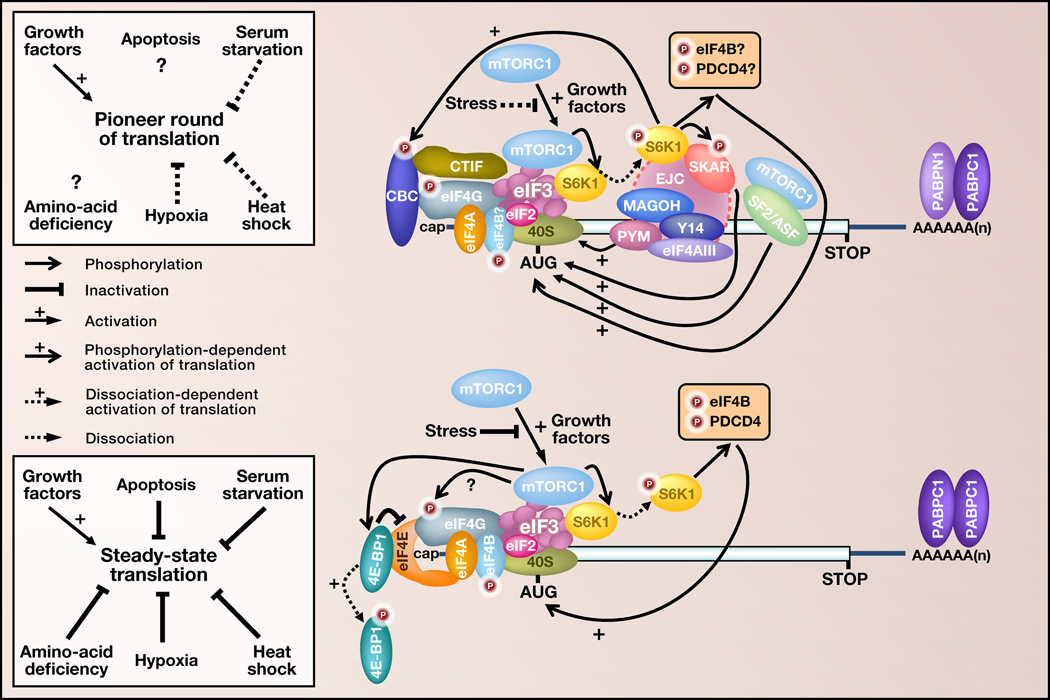
Figure 2. Signaling pathways and translation.
Shown are the signaling pathways that regulate the pioneer round of translation (top) and subsequent steady-state translation (bottom). Under conditions that activate the mTOR signaling pathway, mTORC1 becomes activated and may bind to the translation initiation factor eIF3 associated with CBP80-CBP20 (CBC)-bound mRNA resulting in the phosphorylation and dissociation of activated S6 kinase 1 (S6K1). The EJC component SKAR (S6 kinase 1 ALY-REF-like target) then recruits phosphorylated (i.e., activated) S6K1. Activated S6K1 next mediates the phosphorylation of SKAR and other downstream effectors, including possibly eIF4B and PDCD4 (programmed cell death factor 4), that also associate with CBC-bound mRNA and thereby promote the pioneer round of translation for spliced mRNAs. The Cdc42-dependent phosphorylation of S6K1 also may promote the binding of CBC to cap structures, which would further stimulate the pioneer round. Another CBC-bound mRNA constituent, SF2/ASF, binds to the RNA export receptor TAP and also triggers S6K1 signaling via mTORC1 in ways that promote CBC-bound mRNA translation. Moreover, PYM interacts with the EJC core proteins Y14-MAGOH and with the 40S ribosomal subunit to somehow promote translation of spliced mRNAs presumably during and certainly beyond the pioneer round. (Left) The boxes summarize the translational regulation of CBC-bound or eIF4E-bound mRNAs under various cell-growth conditions. Under conditions of heat shock, hypoxia or serum starvation, the pioneer round of translation is favored over steady-state translation; steady-state translation is largely inhibited by phosphorylation of eIF2α (not shown) or by binding of 4E-BP1 to eIF4E.
The Pioneer Round of Translation Supports Nonsense-mediated Decay
In higher eukaryotes, the vast majority of genes contain multiple introns that are removed from the primary transcript by the process of splicing. Splicing may be accompanied by routinely made mistakes so as to result in mRNAs that contain a premature termination codon (McGlincy and Smith, 2008). Premature termination codons can also arise during splicing as a consequence of a conditionally regulated process as exemplified by those pre-mRNAs whose splice site usage is influenced by the encoded RNA binding protein (McGlincy and Smith, 2008). Nonsense-mediated decay is a translation-dependent mRNA surveillance pathway that detects and eliminates transcripts containing premature termination codons, which have the potential to be deleterious by virtue of the truncated proteins they encode (see, e.g., Rebbapragada and Lykke-Andersen, 2009).
During the pioneer round of translation, nonsense-mediated decay is thought to be triggered by the first ribosome that translates newly processed CBC-bound mRNAs and arrives at a premature termination codon (or a normal termination codon) that is situated more than ~ 50–55-nucleotides upstream of an EJC-bearing exon-exon junction, although there are exceptions to this rule. The CBC plays a critical role in nonsense-mediated decay not only because it comprises the mRNP that harbors EJCs but also because CBP80 interacts directly with the nonsense-mediated decay factor, up-frameshift 1 (UPF1), enhancing the efficiency of this process (Isken and Maquat, 2008). In short, nonsense-mediated decay is thought to involve recognition of premature termination codons by the SURF complex, which consists of the UPF1 phosphoinositide 3-kinase (PIK)-related protein kinase called SMG1, UPF1, and the two translation termination factors referred to as eukaryotic release factor (eRF)1 and eRF3 (Figure 1; Kashima et al., 2006; Isken and Maquat, 2008). After recognition of the premature termination codon, SMG1 and UPF1 join the EJC that resides downstream of the premature termination codon. Notably, the interaction between CBP80 and UPF1 promotes nonsense-mediated decay at two sequential steps (Hwang et al., 2010). The first is the association of SMG1-UPF1 with eRF1–eRF3 at a premature termination codon to form the SURF complex. The second is the association of SMG1-UPF1 with a downstream EJC, which results in SMG1-mediated UPF1 phosphorylation. Subsequently, phosphorylated UPF1 elicits translational repression by binding to the eIF3 constituent of the 43S preinitiation complex that is poised at the translation initiation codon (Isken et al., 2008). Phosphorylated UPF1 also promotes the recruitment of mRNA decay activities (Isken et al., 2008). The importance of the CBC to nonsense-mediated decay is corroborated by the finding that nonsense-mediated decay is still restricted to CBC-bound mRNAs in the case of mRNAs containing premature termination codons that initiate translation in a cap-independent mode from an internal ribosome entry site (IRES), provided that IRES function depends on eIF3 (Isken and Maquat, 2008).
CBC-bound mRNAs form polysomes that are smaller than the polysomes associated with their eIF4E-bound counterparts, consistent with the replacement of CBC by eIF4E taking place on polysomes during the pioneer round of translation. This finding, and data demonstrating that nonsense-mediated decay involves a step of translational repression that targets eIF3, indicate that CBC-bound mRNAs, like eIF4E-bound mRNAs, are generally loaded with more than one ribosome, eIF3 and other well-characterized translation initiation factors.
Specialized factors for the pioneer round of translation
The process of translation can be divided into four phases – initiation, elongation, termination and ribosome recycling. The majority of regulation occurs during the initiation phase (Sonenberg and Hinnebusch, 2009). There is considerably more known about the translation of eIF4E-bound mRNAs than the translation of CBC-bound mRNAs, which is a relatively recent discovery. Nevertheless, there are likely to be many more similarities than differences between the translation of the two mRNPs, as available data have borne out. Factors that specifically regulate the pioneer round of translation will likely target, directly or via another protein, the CBC, the EJC, PABPN1 or other constituents that do not typify eIF4E-bound mRNA, just as factors that specifically regulate steady-state translation often target eIF4E.
PYM
Compared to non-spliced mRNA, it is well established that the process of pre-mRNA splicing endows spliced mRNA with a higher translational capacity during both pioneer and steady-state rounds of translation (Moore and Proodfoot, 2009). The ability of splicing to promote translation appears to be mediated at least in part by EJCs, which data indicate are dynamic rather than either static or homogenous complexes. Different EJC constituents may augment different steps of translation initiation (Lee et al., 2009). For example, PYM may move between mRNA-bound EJCs and translationally active ribosomes: PYM can interact with the EJC core proteins Y14-MAGOH and, via a separate domain, with the 40S ribosomal subunit and the 48S preinitiation complex to somehow promote the translation of spliced mRNAs during and beyond the pioneer round (Diem et al., 2007). The finding that PYM detectably co-immunoprecipitates with CBP80 but not eIF4E is not only consistent with the idea that PYM is present during, and gone after, the pioneer round of translation but also raises the possibility that PYM constitutes a fraction of ribosomes that may specifically function during pioneer rounds. For example, by coupling ribosome recruitment to EJC disassembly, PYM may ensure the preferential recruitment of newly synthesized CBC-bound mRNAs over eIF4E-bound mRNAs to the translational machinery. Alternatively, translation factors that are unique to CBC-bound mRNAs and/or EJCs could recruit ribosomes bound by PYM for the pioneer round. Along these lines, PYM is recruited to those newly exported mRNAs of Kaposi’s sarcoma-associated herpesvirus that are intronless and thus devoid of EJCs, by the viral open reading frame 57 protein (ORF57). ORF57 interacts with PYM and enhances viral mRNA translation via the PYM-mediated recruitment of pre-initiation complexes (Boyne et al., 2010). Notably, the recent report that PYM is an EJC disassembly factor that inhibits nonsense-mediated decay when overexpressed, i.e., when a fraction of PYM exists free of an association with 40S ribosomal subunits so as to promiscuously dissociate EJCs (Gehring et al., 2009), is compatible with the idea that EJC constituents can activate translation.
SKAR
The EJC constituent called SKAR, for S6 kinase 1 (S6K1) ALY-REF-like target, may also mark spliced CBC-bound mRNAs with a translational status that is distinct from the translational status of eIF4E-bound mRNAs and provides another example of how EJCs positively regulate the pioneer round of translation. The mammalian target of the rapamycin (mTOR) signaling pathway promotes cellular translation depending on the presence of growth factors, the absence of stress, and the availability of nutrients or other energy sources (Ma and Blenis, 2009). mTOR is a PIK-related protein kinase that, together with raptor and LST8, form the mTOR complex 1 (mTORC1). Interestingly, signaling through mTORC1 modulates the translation of both CBC-bound mRNAs and eIF4E-bound mRNAs by distinct but undoubtedly overlapping mechanisms (Figure 2).
In the case of eIF4E-bound mRNAs, the recruitment of activated mTORC1 by eIF3 into the proximity of the eIF4E binding protein 1 (4E-BP1) activates translation by mediating the phosphorylation of 4E-BP1 and S6K1. 4E-BP1 phosphorylation results in 4E-BP1 dissociation from mRNA-bound eIF4E, which activates translation by allowing eIF4G to interact with and thereby stabilize the association of eIF4E, eIF4A and PABPC1 with mRNA. S6K1 phosphorylation results in the dissociation of activated S6K1 from mRNA-bound eIF3. Released S6K1 promotes scanning of the 43S preinitiation complex to the AUG translation initiation codon and, therefore, translation by phosphorylating the eIF4A activator eIF4B, and the eIF4A inhibitor and tumor suppressor programmed cell death 4 (PDCD4).
In the case of CBC-bound mRNAs, activated mTORC1 may be recruited by eIF3 and, additionally, SF2/ASF (see below), promoting S6K1 phosphorylation-induced activation and then dissociation of activated phosphorylated S6K1. EJC-bound SKAR then recruits activated S6K1 to CBC-bound spliced mRNAs, and SKAR-bound S6K1 promotes the pioneer round of translation by allowing for the phosphorylation of SKAR and other downstream effectors that could include eIF4B and PDCD4, which also associate with CBC-bound mRNAs (Ma et al., 2008). The finding that SKAR regulates cell growth (Richardson et al., 2004) raises the question of how the mTORC1-induced translation of all or a subset of CBC-bound spliced mRNAs is transduced to a functionally significant increase in total-cell protein synthesis.
SF2/ASF
The serine-arginine-rich protein SF2/ASF plays a central role in recruiting the splicing machinery to pre-mRNA splice sites and can either enhance or inhibit splicing. Like PYM, SF2/ASF co-immunoprecipitates with CBP80 but not detectably with eIF4E (Sato et al., 2008). Data indicate that SF2/ASF, when bound to exonic sequences, can travel as a constituent of CBC-bound mRNPs from nuclei into the cytoplasm, where it has the ability to promote the pioneer round of translation (Sato et al., 2008). Evidence indicates that SF2/ASF recruits a number of translational activators to mRNAs, including the RNA export receptor TAP (Sato et al., 2008) and mTORC1 (Michlewski et al., 2008). Also, like PYM, SF2/ASF promotes mRNA translation beyond the pioneer round (Sato et al., 2008; Michlewski et al., 2008).
CBP80
Another effector of the pioneer round could be CBP80 as the binding of CBC to cap structures is stimulated by growth factors during the G1/S phase of the cell cycle, and by activated forms of the Ras proto-oncogene protein and the Rho-GTPase Cdc42. Cdc42 has been shown to transduce extracellular signals from G-coupled protein receptors, integrins and growth factor receptors at least in part by binding to S6K1 in a GTP-dependent manner. Binding facilitates recognition of the Cdc42-S6K1 complex by upstream S6K-activating kinases (Chou and Blenis, 1996; Wilson and Cerione, 2000). Moreover, signaling from Cdc42 to CBP80 is transmitted in part by the S6K-catalyzed phosphorylation of CBP80, although the biological consequence remains unknown (Ma and Blenis, 2009).
As exemplified above, it appears that an efficient pioneer round of translation is followed by efficient steady-state cycles of translation. Growth factors may augment cellular protein synthesis not only by promoting CBC binding to caps but also by expediting the replacement of CBC by eIF4E (see below). Despite not yet knowing mechanistic details, it is clear that activating or inhibiting pioneer rounds of translation depending on cellular growth conditions is likely to provide an important checkpoint mechanism for the bulk of cellular protein synthesis.
Importin-α
Regulated binding of CBC to mRNA caps is additionally influenced by importin-α, which stably associates with cap-bound CBC (Sato and Maquat, 2009). Cytoplasmic importin-β interacts directly with importin-α and CBP20, thereby promoting the replacement of CBC by eIF4E at mRNA caps (Figure 1; Dias et al., 2009; Sato and Maquat, 2009). In view of the recent structure of an importin-α-CBC-cap complex and a structural model of the importin-β-importin-α-CBC complex (Dias et al., 2009), it will be possible to study how signaling pathways, and the resulting post-translational modifications of importin-α, importin-β and/or CBC, might regulate importin-α or importin-β binding to cap-associated CBC to either stabilize or disrupt the interaction of CBC with mRNA caps. When considering the critical role of CBC in assuring the quality of gene expression through nonsense-mediated decay, and that CBC can be replaced by eIF4E even when the pioneer round of translation is inhibited (Sato and Maquat, 2009), it is important that timing the pioneer round relative to the replacement of CBC by eIF4E be coordinated so that the pioneer round of translation largely takes place first.
eIF3e
Mammalian eIF3, which is a large protein complex consisting of ~13 subunits, has multiple functions in stimulating translation initiation (Sonenberg and Hinnebusch, 2009). The eIF3 subunit eIF3e (also called p48/INT6) co-immunoprecipitates with CBP80 as well as the EJC constituent and nonsense-mediated decay factor UPF2, but it does not detectably co-immunoprecipitate with eIF4E and, like all eIF3 subunits that have been tested, is essential for nonsense-mediated decay (Morris et al., 2007; Isken et al., 2008). eIF3e, which is a core subunit of mammalian eIF3 that is not present in all species, appears to contribute to the recruitment of ribosome-bound eIF3 to mRNA by directly associating with eIF4G (Sonenberg and Hinnebusch, 2009). However, at least in fission yeast, eIF3e is not critical for global protein synthesis, and there exist distinct eIF3 complexes that differ in their non-core subunits. Interestingly, those complexes that contain eIF3e associate with a restricted set of mRNAs, some of which are implicated in the response to cell stress (Zhou et al., 2005). Therefore, it is possible that a specialized eIF3e-containing eIF3 complex, if present in mammals, could specifically function during pioneer rounds of translation by recruiting a defined set of newly synthesized mRNAs to the translational machinery. By so doing, this specialized complex could route those mRNAs that are targets of nonsense-mediated decay to this pathway, and activate the steady-state translation of those that are not nonsense-mediated decay targets.
CTIF
Another initiation factor that is central to steady-state translation is eIF4G, which binds directly to cap-associated eIF4E during steady-state translation and serves critical roles as a scaffold for eIF3, PABPC1 and eIF4A. eIF4G, which also binds directly to CBC, is thought to function in a similar fashion during pioneer rounds of translation. Recently, an eIF4G-like protein called CBP80-CBP20-dependent translation initiation factor (CTIF) has been implicated specifically during pioneer rounds. CTIF interacts directly with CBP80 as well as pre-mRNAs, co-immunoprecipitates with eIF3 in an RNase-resistant manner, and functions in the translation of CBC-bound mRNAs; however, it plays no detectable role in the translation of eIF4E-bound mRNAs (Kim et al., 2009). CTIF could promote the recruitment of ribosomes to newly synthesized CBC-bound mRNAs. As one possibility, CTIF, which contains a middle domain of eIF4G (MIF4G) but lacks the eIF4E binding domain of eIF4G, may functionally replace eIF4G during pioneer rounds if, for example, it (possibly together with CBP80, which contains three MIF4Gs) serves as an eIF4G-like scaffold for CBC-bound mRNPs. Alternatively, CTIF may function in addition to eIF4G considering that CTIF lacks the PABPC1 and eIF4A binding domains that typify eIF4G and that another MIF4G-containing protein, called SLIP, interacts directly with eIF4G to promote the translation of histone mRNA (Cakamakic et al., 2008). Moreover, data indicate that eIF4G functions during pioneer rounds of translation. Regardless, as is possible for eIF3, the use of translation initiation factors that are unique to and activate pioneer rounds could be advantageous by shortening the time span between transcriptional activation and the start of protein production.
The pioneer round during cell stress
The bulk of cellular cap-dependent translation is rapidly downregulated by signaling pathways in response to most physiological and environmental stressors as part of a repertoire of events that mitigate cellular damage and promote cell survival (Figure 2). This global downregulation during, for example, viral infection, amino-acid starvation, hypoxia or heat shock, is often accompanied by eIF2α phosphorylation, which decreases the translation of the majority of CBC-bound as well as eIF4E-bound mRNAs by limiting ternary complex abundance while mediating the selective translation of a subset of cellular mRNAs that initiate translation in a mechanism that depends on an upstream open reading frame. These mRNAs encode specific transcription factors that help the cell adapt to stress and ultimately restore translation (Holcik and Sonenberg, 2005). In addition to eIF2α phosphorylation, stress-induced proteolysis of translation initiation factors can also compromise cellular translation.
There are variations to this theme that result in the differential regulation of pioneer rounds of translation relative to subsequent rounds of translation. For example, the preferential translation of CBC-bound mRNAs is maintained during serum starvation and heat shock, when the translation of eIF4E-bound mRNAs is compromised (Oh et al., 2007b; Marín-Vinader et al., 2006). Serum starvation does not result in eIF2α phosphorylation, but instead activates 4E-BP1.
As another example, during early phases of hypoxia, activation of the endoplasmic reticulum kinase PERK leads to eIF2α phosphorylation and the translational inhibition of both CBC-bound and eIF4E-bound mRNAs (Oh et al., 2007a). However, during late stages of hypoxia, eIF2α dephosphorylation allows for the resumption of CBC-bound mRNA translation (Oh et al., 2007a). Concomitantly, in an eIF2α-independent pathway, disruption of eIF4E-eIF4G-eIF4A and sequestration of eIF4E by dephosphorylated 4E-BP1 and the eIF4E-transporter (which moves eIF4E into the nucleus and processing bodies) maintain the repression of eIF4E-bound mRNA translation (Koritzinski et al., 2006; Lee et al., 2008). Conceivably, the restoration of CBC-bound mRNA translation as a first step toward translational normalcy promotes cell survival after stress by allowing for the surveillance of newly synthesized mRNAs. Furthermore, the renewed engagement of newly made mRNAs with the translational machinery could speed up the cellular response after stress.
Spatial regulation of the pioneer round of translation
Proper embryonic patterning and development as well as neuronal function often involve proteins that bind to the 3’-untranslated regions (3’-UTRs) of particular mRNAs in a sequence-specific manner to couple mRNA translational activation and mRNA localization (Sonenberg and Hinnebusch, 2009). By so doing, an expression gradient of the encoded morphogens, in the case of oocytes, or of proteins that maintain synaptic function or plasticity, in the case of neurons, is spatially established.
There have been many reports of translational repression occurring prior to mRNA localization so as to inhibit ectopic protein production from unlocalized mRNAs. If the sole mechanism of translational repression targets eIF4E, then an mRNA should have undergone the pioneer round of translation prior to repression although, as noted above, it is possible for eIF4E to replace CBC without a pioneer round. In theory, other mechanisms of repression might target both CBC-bound mRNA and eIF4E-bound mRNA.
Examples of translational repression include: sequestrating mRNAs into translationally silenced particles, as Bruno does for oskar mRNA in Drosophila; recruiting the CCR4 deadenylase to shorten poly(A) tail length, as Bicaudal-C does for its own mRNA and certain other germline mRNAs in Drosophila; and blocked joining of the 60S ribosomal subunit to the 48S preinitiation complex, as Zipcode-binding protein 1 (ZBP1, also called IMP1) does for β-actin mRNA at the leading lamellipodia of fibroblasts or neurite growth cones (Besse and Ephrussi, 2008; Sonenberg and Hinnebusch, 2009). Notably, the findings that ZBP1 associates co-transcriptionally with β-actin mRNA (Besse and Ephrussi, 2008), and RNP granules undergoing ZBP1-mediated transport contain CBP80 and EJC constituents but lack detectable eIF4E (Jønson et al., 2007), suggest that ZBP1-mediated translational repression targets CBC-bound transcripts.
To date, studies of arc mRNA and other specific mRNAs in mammalian neurons provide the best-studied examples of translational repression that targets CBC-bound mRNAs (Giorgi et al., 2008). arc mRNA, which harbors two 3’-UTR introns, is a natural target of nonsense-mediated decay: When the pioneer round of translation terminates normally, arc mRNA undergoes nonsense-mediated decay. The arc gene is transcriptionally induced upon intense synaptic activation, and the resulting mRNA is translated and thus targeted for nonsense-mediated decay, once it localizes to activated synapses in a mechanism that depends on glutamatergic receptors. By essentially limiting ARC protein synthesis to the pioneer round of translation at activated synapses, improper protein synthesis at inopportune times and cellular locations is prevented. These findings are consistent with data indicating that CBP80-bound mRNAs migrate to spines when glutamatergic receptors of rat dendrites are stimulated (di Penta et al., 2009), and thus have yet to undergo the pioneer round of translation. At present, which signaling factors control these pioneer rounds of translation in neurons remain unknown.
Features of the pioneer translation initiation complex also appear to be important for proper localization of oskar mRNA to the posterior pole in Drosophila oocytes. For example, splicing of the first intron of oskar pre-mRNA (or another intron at the site of the first intron) is required for oskar localization, presumably by virtue of the resulting EJC in conjunction with nearby mRNA sequences (Hatchet and Ephrussi, 2004). In fact, demonstrated roles for the EJC constituents eIF4AIII, Tsunami-Magonashi and Barentz (the latter two of which are orthologous to mammalian Y14-MAGOH and MLN51, respectively) in oskar mRNA localization, and their co-localization with oskar mRNA at the posterior pole (Besse and Ephrussi, 2008) indicate that the pioneer round of translation and removal of the first EJC occur after the regulatory steps of translational repression and localization. As noted above, these regulatory steps may involve the RNA binding protein Bruno. As oskar mRNA is also translationally repressed by Bruno-mediated recruitment of the Cup protein, which targets eIF4E (Besse and Ephrussi, 2008), there appear to be multiple pathways to translationally silence oskar mRNA. In fact, a number of mRNAs are repressed at different steps of translation when bound by CBC or eIF4E.
MicroRNAs
It was recently reported that the microRNA (miRNA) mi-R2 mediates translational repression, which interferes with the interaction between eIF4E and eIF4G in a way that is predicted to have no consequence to the pioneer round of translation (Zdanowicz et al., 2009). Therefore, mRNA caps bound by eIF4E could represent another signature target used by cells to differentially regulate the translation of CBC-bound mRNA relative to eIF4E-bound mRNA. Nevertheless, other mechanisms of miRNA-mediated translational repression can function simultaneously to repress the translation of CBC-bound mRNA. This became evident through the recent demonstration that (i) Ago2, which is known to inhibit the translation of its target mRNAs, is loaded onto both CBC-bound mRNAs and eIF4E-bound mRNAs, (ii) the loading of endogenous microRNA-induced silencing complexes (miRISCs) onto the 3’-UTRs of CBC-bound mRNAs that contain a premature termination codon abrogates nonsense-mediated decay, and (iii) several natural substrates of nonsense-mediated decay are stabilized by miRISC-mediated translational repression (Choe et al., 2010). It will be interesting to determine if factors that are specific for CBC-bound mRNA and the pioneer round of translation are recognized during miRNA-mediated translational repression.
Future Directions
Mammalian cells have evolved two mRNP templates for protein synthesis that manifest a precursor-product relationship and serve distinct functions. Although these two templates share many constituent proteins that homogenize their regulation, at least some unique features impart a means for their differential control. Examples also exist of distinct regulatory mechanisms that coordinate CBC-bound mRNA translation and eIF4E-bound mRNA translation by targeting factors that are particular to each mRNP. As eIF4E-bound mRNA supports the bulk of cellular protein synthesis, it is the logical target for a rapid response to changes in physiological conditions. However, CBC-bound mRNA holds the unique capability of enhancing subsequent rounds of translation, downregulating further translation, or completely stopping not only CBC-supported mRNA surveillance but also eIF4E-supported protein production. Future studies are expected to illuminate molecular aspects of known and unforeseen regulatory mechanisms.
Acknowledgements
We thank Chenguang Gong and Hanae Sato for helpful comments and acknowledge NIH R01 grants GM074593 and GM59614 (L.E.M), the National Science Council of Taiwan (W.-Y. T.) and start-up funds from the University of Lübeck (O.I.) for support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Besse F, Ephrussi A. Translational control of localized mRNAs: restricting protein sythesis in space and time. Nat. Rev. Mol. Cell. Biol. 2008;9:971–980. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- Boyne JR, Jackson BR, Taylor A, Macnab SA, Whitehouse A. Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with PYM to enhance translation of viral intronless mRNAs. EMBO J. 2010;29:1851–1864. doi: 10.1038/emboj.2010.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakmakci NG, Lerner RS, Wagner EJ, Zheng L, Marzluff WF. SLIP1, a factor required for activation of histone mRNA translation by the stem-loop binding protein. Mol. Cell. Biol. 2008;28:1182–1194. doi: 10.1128/MCB.01500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou MM, Blenis J. The 70S kDa S6 kinase complexes with and is activated by the Rho family of G proteins Cdc42 and Rac1. Cell. 1996;85:573–583. doi: 10.1016/s0092-8674(00)81257-x. [DOI] [PubMed] [Google Scholar]
- Choe J, Cho H, Lee HC, Kim YK. microRNA/Argonaute 2 regulates nonsense-mediated messenger RNA decay. EMBO Rep. 2010;11:380–386. doi: 10.1038/embor.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias SM, Wilson KF, Rojas KS, Ambrosio AL, Cerione RA. The molecular basis for the regulation of the cap-binding complex by the importins. Nat. Struct. Mol. Biol. 2009;16:930–937. doi: 10.1038/nsmb.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Penta A, Mercaldo V, Florenzano F, Munck S, Ciotti MT, Zalfa F, Mercanti D, Molinari M, Bagni C, Achsel T. Dendritic LSm1/CBP80-mRNPs mark the early steps of transport commitment and translational control. J. Cell Biol. 2009;184:423–435. doi: 10.1083/jcb.200807033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diem MD, Chan CC, Younis I, Dreyfuss G. PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat. Struct. Mol. Biol. 2007;14:1173–1179. doi: 10.1038/nsmb1321. [DOI] [PubMed] [Google Scholar]
- Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW. Disassembly of exon junction complexes by PYM. Cell. 2009;137:536–548. doi: 10.1016/j.cell.2009.02.042. [DOI] [PubMed] [Google Scholar]
- Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC Factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Hatchet O, Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428:959–963. doi: 10.1038/nature02521. [DOI] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- Hwang J, Sato H, Tang Y, Matsuda D, Maquat LE. UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol. Cell. 2010 doi: 10.1016/j.molcel.2010.07.004. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat. Rev. Genet. 2008;9:699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jønson L, Vikesaa J, Krogh A, Nielsen LK, Hansen TO, Borup R, Johnsen AH, Christiansen J, Nielsen FC. Molecular composition of IMP1 ribonucleoprotein granules. Mol. Cell. Proteomics. 2007;6:798–811. doi: 10.1074/mcp.M600346-MCP200. [DOI] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes & Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KM, Cho H, Choi K, Kim J, Kim BW, Ko YG, Jang SK, Kim YK. A new MIF4G domain-containing protein, CTIF, directs nuclear cap-binding protein CBP80/20-dependent translation. Genes & Dev. 2009;23:2033–2045. doi: 10.1101/gad.1823409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koritzinsky M, Magagnin MG, van den Beucken T, Seigneuric R, Savelkouls K, Dostie J, Pyronnet S, Kaufman RJ, Weppler SA, Voncken JW, Lambin P, Koumenis C, Sonenberg N, Wouters BG. Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. EMBO J. 2006;25:1114–1125. doi: 10.1038/sj.emboj.7600998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Cho H, Kim YK. Ectopic expression of eIF4E-transporter triggers the movement of eIF4E into P-bodies, inhibiting steady-state translation but not the pioneer round of translation. Biochem. Biophys. Res. Commun. 2008;369:1160–1165. doi: 10.1016/j.bbrc.2008.03.017. [DOI] [PubMed] [Google Scholar]
- Lee HC, Choe J, Chi SG, Kim YK. Exon junction complex enhances translation of spliced mRNAs at multiple steps. Biochem. Biophys. Res. Commun. 2009;384:334–340. doi: 10.1016/j.bbrc.2009.04.123. [DOI] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell. Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Ma XM, Yoon SO, Richardson CJ, Jülich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133:303–313. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- Marín-Vinader L, van Genesen ST, Lubsen NH. mRNA made during heat shock enters the first round of translation. Biochim. Biophys. Acta. 2006;1759:535–542. doi: 10.1016/j.bbaexp.2006.10.003. [DOI] [PubMed] [Google Scholar]
- McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem. Sci. 2008;3:385–393. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Michlewski G, Sanford JR, Cáceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol. Cell. 2008;30:179–189. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–670. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Morris C, Wittmann J, Jäck M, Jalinot P. Human INT6/eIF3e is required for nonsense-mediated mRNA decay. EMBO Rep. 2007;8:596–602. doi: 10.1038/sj.embor.7400955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh N, Kim KM, Choe J, Kim YK. Pioneer round of translation mediated by nuclear cap-binding proteins CBP80/20 occurs during prolonged hypoxia. FEBS Lett. 2007a;581:5158–5164. doi: 10.1016/j.febslet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Oh N, Kim KM, Choe J, Kim YK. Pioneer round of translation occurs during serum starvation. Biochem. Biophys. Res. Commun. 2007b;362:145–151. doi: 10.1016/j.bbrc.2007.07.169. [DOI] [PubMed] [Google Scholar]
- Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr. Opin. Cell. Biol. 2009;21:394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Richardson CJ, Bröenstrup M, Fingar DC, Jülich K, Ballif BA, Gygi S, Blenis J. SKAR is a specific target of S6 kinase 1 in cell growth control. Curr Biol. 2004;14:1540–1549. doi: 10.1016/j.cub.2004.08.061. [DOI] [PubMed] [Google Scholar]
- Sato H, Hosoda N, Maquat LE. Efficiency of the pioneer round of translation affects the cellular site of nonsense-mediated mRNA decay. Mol. Cell. 2008;29:255–262. doi: 10.1016/j.molcel.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Sato H, Maquat LE. Remodeling of the pioneer translation initiation complex involves translation and the karyopherin importin β. Genes & Dev. 2009;23:2537–2550. doi: 10.1101/gad.1817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnesbusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KF, Cerione RA. Signal transduction and post-transcriptional gene expression. Biol Chem. 2000;381:357–365. doi: 10.1515/BC.2000.048. [DOI] [PubMed] [Google Scholar]
- Zdanowicz A, Thermann R, Kowalska J, Jemielity J, Duncan K, Preiss T, Darzynkiewicz E, Hentze MW. Drosophila miR2 primarily targets the m7GpppN cap structure for translational repression. Mol Cell. 2009;35:881–888. doi: 10.1016/j.molcel.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Zhou C, Arslan F, Wee S, Krishnan S, Ivanov AR, Oliva A, Leatherwood J, Wolf DA. PCI proteins eIF3e and eIF3m define distinct translation initiation factor 3 complexes. BMC Biol. 2005;3:14. doi: 10.1186/1741-7007-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]