Abstract
DDA3 is a microtubule-associated protein that controls chromosome congression and segregation by regulating the mitotic spindle. Depletion of DDA3 alters spindle structure, generates unaligned chromosomes at metaphase, and delays the mitotic progression. Through a mass spectrometry analysis, we found that DDA3 is phosphorylated on Ser225 during mitosis. Phosphorylation of this residue is important for the mitotic function of DDA3, as the phospho-mimicking DDA3-S225D variant, but not the nonphosphorable DDA3-S225A mutant, rescues the DDA3-knockdown phenotype. We conclude that the mitotic function of DDA3 is regulated by phosphorylation on the Ser225 residue.
Keywords: DDA3, phosphorylation, spindle, chromosome congression, mitosis
INTRODUCTION
Cell cycle progression is regulated by protein kinases, such as cyclin-dependent kinase 1 (Cdk1), Aurora A kinase, and Polo-like kinase 1 (Plk1), which regulate the biochemical activity and cellular localization of substrate proteins through phosphorylation [1; 2; 3]. For example, both Plk1 and Aurora A phosphorylate the microtubule (MT) depolymerase Kif2a [4]. While phosphorylation of Kif2a by Plk1 enhances its depolymerase activity and its association with the mitotic spindle, phosphorylation of Kif2a by Aurora A suppresses its depolymerase activity and its association with MTs. This antagonistic regulation confers differential stability to MTs in the spindle vs. at the pole vs. in cytosol, which is important for spindle assembly and function [4].
In mitosis, chromosome movement is mediated by the dynamic assembly of MTs into a mitotic spindle [5]. A driving force for the congression and segregation of chromosomes is the dynamic turnover of MTs controlled by MT-associated proteins, such as DDA3 [6], and by MT depolymerases, such as Kif2a [7; 8]. DDA3 was initially discovered as a cell cycle regulator activated by p53 [9; 10]. We recently showed that DDA3 is a regulator of the spindle essential for mitotic progression [6]. Depletion of DDA3 results in a high frequency of unaligned chromosomes, a substantial reduction in tension across sister kinetochores at metaphase, and a decrease in the velocity of chromosome segregation at anaphase. DDA3 directly associates with the mitotic spindle. Furthermore, DDA3 interacts with the MT depolymerase Kif2a and recruits Kif2a to the mitotic spindle and spindle poles to control MT dynamics. Thus, DDA3 is a novel mitotic regulator essential for the high fidelity of chromosome segregation [6].
DDA3 is phosphorylated in mitosis [6]. To understand the function and regulation of DDA3 in mitosis, we analyzed its phospho-regulation. We report here the mapping of a phosphorylation site in DDA3 and the characterization of its physiological importance in mitosis.
Materials and Methods
Antibodies
Rabbit antibodies against Mad2, DDA3 and GFP were described previously [6; 11]. Anti-β-tubulin E7 monoclonal antibody was obtained from the Developmental Studies Hybridoma Bank and anti-cyclin B and anti-p38MAPK antibodies from Santa Cruz Biotechnology.
Cell culture, siRNAs and Transfection
HeLa S3 and HeLa cells were cultured in Dulbecco’s modified Eagles Medium containing 10% fetal bovine serum (Invitrogen) and antibiotics. HeLa S3 cells were synchronized at prometaphase by a thymidine-nocodazole treatment (an 18-h thymidine arrest and then a 6-h release, followed by a 14-h nocodazole arrest) [12].
siRNAs were synthesized by Dharmacon, Inc, using sequences extensively characterized and validated previously [6]. Sequence targeting DDA3 was 5′-AAGCAAGACTTCAGTAGCATT-3′. This siRNA targets the 3′ untranslated region of DDA3 and therefore does not block the expression of transfected DDA3 genes. The control siRNA (siGL2) was 5′-CGTACGCGGAATACTTCGATT-3′. siRNAs were transfected into HeLa cells using DharmaFect 1 (Dharmacon, Inc.). DNA transfection was performed using Effectene (Qiagen) as instructed by the manufacturer.
In experiments examining the effects of ectopic expression of DDA3, cells with a broad spectrum of expression were analyzed in order to generate representative data.
Mapping the phosphorylation sites in DDA3
Purification of the DDA3 protein
The DDA3 gene was fused to the GFP and S-peptide tags in the pBabe-Puro vector and subsequently transfected into Phoenix-Ampho packaging cells. The viral supernatant was used to infect HeLa S3 cells to isolate stable clones after selection with 0.4 μg/ml puromycin for 2 weaks. A clone stably expressing DDA3-S-GFP gene at a level comparable to that of endogenous DDA3 was used to purify the DDA3 protein by two-step affinity columns as described previously [3].
Liquid chromatography-tandem mass spectrometry
An 100-μm i.d. capillary with a 5-μm pulled tip was packed with 10 cm of 5-μm Aqua C18 material (Phenomenex). The desalting column was then equilibrated for 30 min with Buffer A (5% acetonitrile/0.1% formic acid) and the protein digest was pressure loaded onto it. The column was placed inline with a quaternary HPLC (Agilent Technologies) and analyzed by an one-step separation that consisted of a 120-min gradient from 0 to 100% Buffer B (80% acetonitrile/0.1% formic acid). As peptides eluted from the microcapillary column, they were electrosprayed directly onto an LTQ mass spectrometer (Thermo Fisher Scientific) with the application of a distal 2.4-kV spray voltage. A cycle of one full-scan mass spectrum (400 – 1,400 m/z) was followed by three data-dependent tandem mass spectrometry spectra at a 35% normalized collision energy.
Analysis of tandem mass spectrometry
Tandem mass spectra for phospho-peptides was analyzed by using the following software analysis protocol. Poor-quality spectra were removed from the dataset by using an automated spectral quality assessment algorithm. Tandem mass spectra remaining after filtering were searched with the SEQUEST algorithm against a human database concatenated to a decoy database in which the sequence for each entry in the original database was reversed [13]. The search parameters include a static cysteine modification of 57 and differential modification on serine, threonine and tyrosine residues of 80. Trypsin specificity was required for all peptides. The database search results were assembled and filtered using the DTASelect program with a spectra level false positive rate of less than 0.5%. The validity of the phosphopeptides was confirmed using the software DeBunker [14] and the exact residue of phosphorylation was confirmed using in house software Pinpointer and by manual validation of tandem mass spectra.
Immunofluoresence
HeLa cells on coverglasses were fixed with −20°C methanol for 5 min. Subsequently, cells were permeabilized and blocked with PBS-BT (1× PBS, 3% BSA, and 0.1% Triton X-100) for 30 min at room temperature. Coverslips were then incubated in primary and secondary antibodies diluted in PBS-BT. Images were acquired with Openlab 5.2 (Improvision) under a Zeiss Axiovert 200M microscope using a 1.4NA Plan-Apo 100× oil immersion lens. Z-stacks of fluorescence images were deconvolved using AutoDeblur v9.1 and AutoVisualizer v9.1 (AutoQuant Imaging) and the 2-D sum-projections of deconvolved z-stacks were shown.
RESULTS AND DISCUSSION
DDA3 is phosphorylated at Ser225 during mitosis
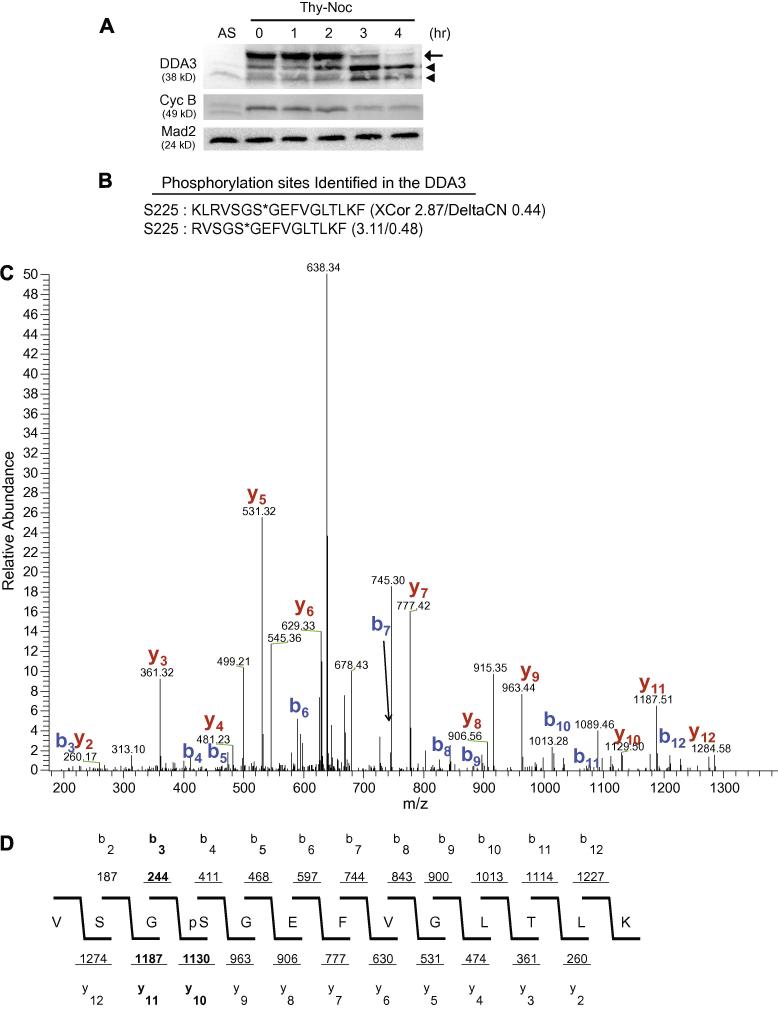
We have previously reported that DDA3 is a cell cycle-regulated phospho-protein, as treatment of mitotic DDA3 with λ-phosphatase increases its mobility in SDS-PAGE [6]. To analyze the phosphorylation of DDA3 in mitosis, HeLa S3 cells were synchronized at prometaphase by a thymidine-nocodazole (Thy-Noc) treatment and then released from mitosis into G1 [12]. During mitotic exit, the abundance of hyper-phosphorylated DDA3 decreased, while the abundance of hypo-phosphorylated DDA3 increased (Fig 1A). We next determined the site of mitotic phosphorylation on DDA3 by mass spectrometry analysis. We established a HeLa S3 cell line stably expressing tagged DDA3 at a level comparable to that of the endogenous DDA3. Cells were synchronized at prometaphase by a thymidine-nocodazole treatment and the tagged DDA3 was tandem-affinity-purified from nocodazole-arrested prometaphase cells [6]. Analysis of purified DDA3 by mass spectrometry indicated that Ser225 was phosphorylated in mitotic DDA3 (Fig 1B-D). Two phospho-Ser225-containing peptides were independently identified, both with high confidence, as reflected by its high scores for XCorr and DeltaCN (Fig 1 B-D). Although Ser225 is phosphorylated in mitotic cells, we cannot exclude the possibility that this residue may also be phosphorylated in interphase cells.
Figure 1. DDA3 is phosphorylated at Ser225 in mitosis.
(A) HeLa cells were synchronized by a thymidine-nocodazole (Thy-Noc) block, released into fresh media and harvested at indicated time. The arrow points to phosphorylated mitotic DDA3 and arrowheads point to two forms of interphase DDA3, as characterized previously [6]. AS, unsynchronized cells. (B-D) DDA3-S-GFP cells were synchronized at prometaphase by a thymidine-nocodazole arrest and the DDA3 protein was tandem-affinity-purified from the mitotic DDA3-S-GFP cells and analyzed by mass spectrometry. Peptides derived from phosphorylated DDA3 are listed in (B) with XCorr and DeltaCN scores indicated (*, phosphorylation site). Shown in (C) was the actual spectrum for the second peptide listed in (B) with signals for the b-ion and y-ion series indicated. The spectrum data was summarized in (D). The validity of the phospho-peptide VSGS*GEFVGLTLK (C & D) was confirmed by the software DeBunker [14]. Manual annotation of the tandem mass spectrum also confirmed the phosphorylation at S225. Of the four site-determining ions for Ser225, b3, y10 and y11 [highlighted in (D)] were observed in the tandem mass spectra, providing great confidence on the validity of this phosphorylation site. The fourth site-determining ion b2 was not within the scanning range of the instrument.
Phosphorylation of S225 does not affect the localization of DDA3
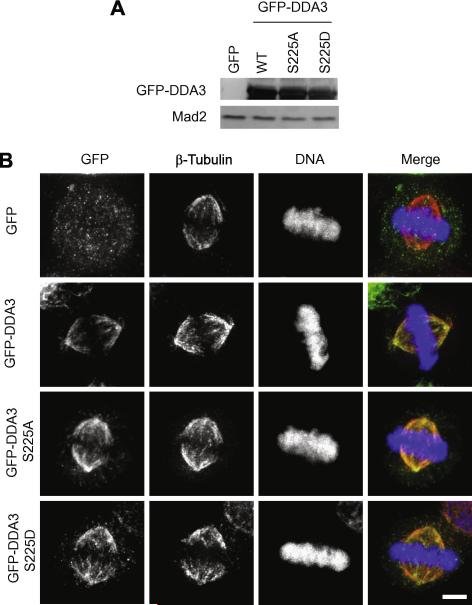
To investigate the function of phosphorylation on DDA3-Ser225 in mitosis, we mutated Ser225 to Ala (DDA3-S225A, nonphosphorable) or to Asp (DDA3-S225D, phospho-mimicking) and analyzed the effect of phosphorylation on localization of DDA3 in mitosis. Wild-type, S225A, and S225D GFP-DDA3 were expressed in HeLa cells at comparable levels by transient transfection (Fig 2A). Quantitative Western blot analysis indicated that the expression levels of the fusion proteins were 50 fold above the endogenous DDA3 (data not shown). When analyzed by immuno-fluorescence staining in metaphase cells, all three fusion proteins localized to the mitotic spindle in the same pattern as the endogenous DDA3 (Fig 2B) [6], suggesting that DDA3 localization is independent of phosphorylation on Ser225. Furthermore, in the presence of endogenous wild-type DDA3, over-expression of neither DDA3-S225A nor DDA3-S225D at this level affected chromosome congression, alignment, or segregation in mitosis (data not shown).
Figure 2. Phosphorylation of S225 does not affect the localization of DDA3.
HeLa cells were transfected with GFP, GFP-DDA3, GFP-DDA3-S225A, and GFP-DDA3-S225D. Lysates of transfected cells were analyzed by Western blotting with antibodies against GFP and against Mad2 (a loading control) (A). Cells were fixed at 36 hr post-transfection and stained. Shown in (B) are maximum projections from deconvolved z stacks of cells stained for GFP (green), β-tubulin (red), and DNA (blue). These images are from representative cells analyzed in three independent repeat experiments. Scale bar, 5 μm.
Phosphorylation of S225 is important for chromosome congression
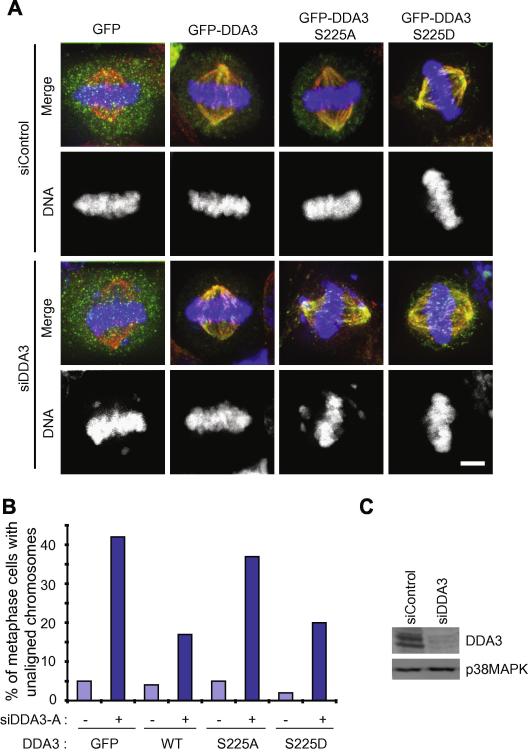
Next, we determined whether phosphorylation on Ser225 is required for the mitotic function of DDA3 in chromosome congression. HeLa cells were transfected with a control siRNA or a siRNA targeting the 3′ untranslated region of DDA3 (siDDA3) and the knockdown efficiency was analyzed by Western blotting (Fig 3C) [6]. Twenty-four hours post-transfection, cells from each siRNA transfection were split into four dishes and transfected with GFP, GFP-DDA3, GFP-DDA3-S225A, and GFP-DDA3-S225D, respectively. Knockdown of DDA3 resulted in a high frequency of unaligned chromosomes in metaphase cells [6] and this phenotype was rescued by the expression of wild-type GFP-DDA3 and GFP-DDA3-S225D, but not by the expression of GFP alone or GFP-DDA3-S225A (Fig 3A-B), indicating that phosphorylation of Ser225 is important for the mitotic function of DDA3. Both wild-type GFP-DDA3 and GFP-DDA3-S225D only partially rescued the DDA3-knockdown phenotype, suggesting that the precise level of DDA3 expression is important for its full physiological function. On the other hand, the fact that GFP-DDA3 and GFP-DDA3-S225D rescued the DDA3-knockdown phenotype to the same extent indicates that DDA3-S225D is fully active in mitosis.
Figure 3. Phosphorylation of S225 is essential for chromosome congression.
HeLa cells were transfected with a control or a previously characterized siRNA targeting the 3′ untranslated region of DDA3 [6]. Twenty-four hours later, cells were transfected again with GFP-DDA3 (wild type or mutants). At 54 hours post-transfection of siRNAs, cells were fixed and stained. Shown in (A) are maximum projections from deconvolved z stacks of cells stained for GFP (green), β-tubulin (red), and DNA (blue). These images are from representative cells analyzed in three independent repeat experiments. The percentage of metaphase cells with unaligned chromosomes over total metaphase cells were quantified and plotted (n=100 cells from three independent experiments) (B). As a control, the degree of DDA3-knockdown was analyzed in GFP-transfected cells by Western blotting against DDA3 and p38MAPK (a loading control) (C).
A key unresolved question on phospho-regulation of DDA3 is the kinase responsible for phosphorylation of Ser225. Multiple kinases, including cyclin B/Cdk1, Aurora A, and Plk1, all can phosphorylate DDA3 in vitro (data not shown). However, the sequence around Ser225, V-S-G-pS225-G-E-F, does not resemble a consensus recognition motif for any of these characterized mitotic kinases. Furthermore, all three kinases phosphorylate DDA3-S225A as efficiently as the wild-type DDA3 (data not shown). Thus, the physiological kinase for this serine and the regulation of this phosphorylation in the cell cycle remain to be characterized. On the other hand, mutating Ser225 to Ala did not abolish the retarded mobility in SDS-PAGE of the mutant DDA3 protein from mitotic cells, indicating that additional sites of phosphorylation exist in mitotic DDA3. We speculate that phosphorylation on one of these other sites may play a role in mitotic-specific association of DDA3 with the spindle, given that DDA3 only associates with MTs in mitosis, but phosphorylation of Ser225 is not required for spindle localization of DDA3.
How does phosphorylation of DDA3 confer its mitotic function? DDA3 interacts with Kif2a and controls the density and dynamics of MTs in the mitotic spindle [6]. We found that DDA3, DDA3-S225A, and DDA3-S225D all interact with Kif2a in co-immunoprecipitation experiments and that neither the ability of DDA3-S225D nor the inability of DDA3-S225A to rescue chromosome congression defects in siDDA3 cells correlates with the extent of recruitment of Kif2a to the spindle poles in these cells (data not shown). Thus, DDA3 acts in chromosome congression, likely independent of or in parallel to its recruitment of Kif2a. The exact cellular pathway regulated by the phosphorylation of DDA3-S225 and required for chromosome congression remains to be fully characterized.
In summary, we have identified a phospho-regulatory mechanism for DDA3 that is important for its function in mediating chromosome congression in mitosis. As we have not been able to generate an antibody specific to the phosphorylated state of DDA3, the important question of spatial and temporal phospho-regulation of DDA3 in the cell cycle remains to be analyzed in the future.
Acknowledgement
We are grateful to members of the Fang lab for discussions. This work was supported by a postdoctoral fellowship from the Korea Research Foundation of the Korean Government (MOEHRD) (KRF-2006-352-C00057) (C.J.), by a Burroughs-Wellcome Career Award in Biomedical Research (G.F.) and by grants from National Institutes of Health (GM062852 to G.F. and HL079442 and RR11823-10 to J.Y.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Barr AR, Gergely F. Aurora-A: the maker and breaker of spindle poles. J Cell Sci. 2007;120:2987–96. doi: 10.1242/jcs.013136. [DOI] [PubMed] [Google Scholar]
- [2].Murray AW. Recycling the cell cycle: cyclins revisited. Cell. 2004;116:221–34. doi: 10.1016/s0092-8674(03)01080-8. [DOI] [PubMed] [Google Scholar]
- [3].Seki A, Coppinger JA, Jang CY, Yates JR, Fang G. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008;320:1655–8. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jang CY, Coppinger JA, Seki A, Yates JR, 3rd, Fang G. Plk1 and Aurora A regulate the depolymerase activity and the cellular localization of Kif2a. J Cell Sci. 2009;122:1334–41. doi: 10.1242/jcs.044321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gadde S, Heald R. Mechanisms and molecules of the mitotic spindle. Curr Biol. 2004;14:R797–805. doi: 10.1016/j.cub.2004.09.021. [DOI] [PubMed] [Google Scholar]
- [6].Jang CY, Wong J, Coppinger JA, Seki A, Yates JR, 3rd, Fang G. DDA3 recruits microtubule depolymerase Kif2a to spindle poles and controls spindle dynamics and mitotic chromosome movement. J Cell Biol. 2008;181:255–67. doi: 10.1083/jcb.200711032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kline-Smith SL, Walczak CE. Mitotic spindle assembly and chromosome segregation: refocusing on microtubule dynamics. Mol Cell. 2004;15:317–27. doi: 10.1016/j.molcel.2004.07.012. [DOI] [PubMed] [Google Scholar]
- [8].Wordeman L. Microtubule-depolymerizing kinesins. Curr Opin Cell Biol. 2005;17:82–8. doi: 10.1016/j.ceb.2004.12.003. [DOI] [PubMed] [Google Scholar]
- [9].Lo PK, Chen JY, Lo WC, Chen BF, Hsin JP, Tang PP, Wang FF. Identification of a novel mouse p53 target gene DDA3. Oncogene. 1999;18:7765–74. doi: 10.1038/sj.onc.1203167. [DOI] [PubMed] [Google Scholar]
- [10].Hsieh WJ, Hsieh SC, Chen CC, Wang FF. Human DDA3 is an oncoprotein down-regulated by p53 and DNA damage. Biochem Biophys Res Commun. 2008;369:567–72. doi: 10.1016/j.bbrc.2008.02.047. [DOI] [PubMed] [Google Scholar]
- [11].Wong J, Fang G. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J Cell Biol. 2006;173:879–91. doi: 10.1083/jcb.200511132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998;12:1871–83. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–6. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- [14].Lu B, Ruse C, Xu T, Park SK, Yates J., 3rd Automatic validation of phosphopeptide identifications from tandem mass spectra. Anal Chem. 2007;79:1301–10. doi: 10.1021/ac061334v. [DOI] [PMC free article] [PubMed] [Google Scholar]