Abstract
Actin filament nucleators initiate polymerization in cells in a regulated manner. A common architecture among these molecules consists of tandem W domains that recruit three to four actin subunits to form a polymerization nucleus. We describe a low-resolution crystal structure of an actin dimer assembled by tandem W domains, where the first W domain is crosslinked to Cys-374 of the actin subunit bound to it, whereas the last W domain is followed by the C-terminal pointed end-capping helix of Tβ4. While the arrangement of actin subunits in the dimer resembles that of a long-pitch helix of the actin filament, important differences are observed. These differences result from steric hindrance of the W domain with inter-subunit contacts in the actin filament. We also determined the structure of the first W domain of Vibrio parahaemolyticus VopL crosslinked to actin Cys-374, and show it to be nearly identical to non-crosslinked W-actin structures. This result validates the use of crosslinking as a tool for the study of actin nucleation complexes, whose natural tendency to polymerize interferes with most structural methods. Combined with a biochemical analysis of nucleation, the structures may explain why nucleators based on tandem W domains with short inter-W linkers have relatively weak activity, cannot stay bound to filaments after nucleation, and are unlikely to influence filament elongation. The findings may also explain why Nucleation Promoting Factors of the Arp2/3 complex, which are related to tandem W domain nucleators, are ejected from branch junctions after nucleation. We finally show that the simple addition of the C-terminal pointed end-capping helix of Tβ4 to tandem W domains can change their activity from actin filament nucleation to monomer sequestration.
Keywords: Actin nucleation, W domain, Nucleation Promoting Factors, Tβ4, actin monomer sequestration
INTRODUCTION
The nucleation of actin filaments in cells is kinetically unfavorable because of the instability of polymerization intermediates (dimers, trimers and tetramers) and the actions of actin monomer binding proteins such as profilin and thymosin-β4 (Tβ4) 1; 2. This creates an opportunity for cells to use molecules known as actin filament nucleators to initiate the formation of actin polymerization nuclei in a spatially and temporally controlled manner.
The actin filament can be described as either a single left-handed short-pitch helix, where consecutive subunits are staggered with respect to one another by half a monomer length, or two right-handed long-pitch helices of head-to-tail bound actin subunits 3; 4; 5. Different nucleators work by different mechanisms, stabilizing small actin oligomers along either the long- or the short-pitch helices of the actin filament 6; 7.
Most actin filament nucleators use the WASP-Homology 2 (WH2 or W) domain for interaction with actin. The W domain has a short length (17–27aa) and is extremely abundant and functionally versatile 7; 8; 9. The N-terminal portion of the W domain forms a helix that binds in the hydrophobic (or target-binding) cleft 10 formed between subdomains 1 and 3 at the barbed end of the actin monomer 11; 12; 13. After this helix, the W domain presents an extended region that is directed towards the pointed end of the actin monomer (formed by subdomains 2 and 4 of actin). This region is variable in length and sequence, but comprises the conserved four residue motif LKKT(V), which is critical for the interaction with actin 11.
Filament nucleators are characterized by the presence of multiple actin-binding sites. The simplest and most common architecture consists of tandem repeats of the W domain, occurring in the proteins Spire 14, Cobl 15 and VopL/VopF 16; 17. The W domain also participates in filament nucleation through the Nucleation Promoting Factors (NPFs) of the Arp2/3 complex, which can have between one and three W domains 18; 19; 20. The muscle-specific nucleator Lmod also contains one W domain 21. The nucleation activities of tandem W domain-based nucleators vary widely. At least in part, the reason for these differences may lie in the highly variable linkers between W domains. When the linkers are short, as in the relatively weak nucleator Spire 14, only actin subunits along the long-pitch helix of the actin filament can be connected. In contrast, the brain-enriched protein Cobl is a strong nucleator, featuring three W domains with a long linker between its second and third W domains, and is thought to stabilize a short-pitch actin trimer for nucleation 15. The examples of Cobl, the Arp2/3 complex, and formins suggest that stabilization of a short-pitch actin nucleus is a more effective way to promote polymerization than stabilization of a long-pitch actin nucleus 6; 7. However, the structural bases for this observation are not well understood.
In an attempt to understand the nucleation mechanism of tandem W domain-based nucleators, we recently reported a solution study, using Small Angle X-ray Scattering (SAXS), of an actin dimer and a trimer stabilized by tandem W domain constructs 22. These complexes, referred to as 2W-Actin and 3W-Actin, and containing respectively two and three W domains, were capped at the barbed end by structure-based crosslinking of the first W domain to Cys-374 of the first actin subunit and at the pointed end by addition of the C-terminal helix of Tβ4. Constructs 2W and 3W are based on the W domain repeat present in the NPF protein N-WASP, which like Spire presents short inter-W linkers. The SAXS study suggested that the actin subunits in the complexes adopted an elongated conformation similar to that of the long-pitch helix of the actin filament. However, the resolution of this study was insufficient to establish a direct comparison between the longitudinal contacts of actin subunits in the complexes and in the actin filament.
Here we report the crystal structure of 3W-Actin at 7 Å resolution. Only two actin subunits are present in the structure, indicating that one of the actin subunits is released during crystallization. Despite its low resolution, this structure, obtained by fitting high-resolution structures of W-Actin complexes into the low-resolution data, offers a clearer picture of the relative disposition of actin subunits bound to tandem W domains that are separated by Spire-like short inter-W linkers. While the longitudinal arrangement of actin subunits in the structure is somewhat related to that of the long-pitch helix of the actin filament 3; 4, important differences are observed. These differences probably result from steric hindrance of the W domain with inter-subunit contacts in the filament. The determination of the structure of 3W-Actin was aided by determination of the 2.9 Å resolution crystal structure of the first W domains of VopL 16 crosslinked to actin Cys-374 (hereafter referred to as WxActin). The structure of WxActin is nearly undistinguishable from non-crosslinked W-Actin structures determined previously 11; 12; 13, thus validating the use of crosslinking as a tool to stabilize actin polymerization complexes for structural investigation. The structures, and a biochemical analysis of nucleation, reveal important clues about the existing disparities in the nucleation activities of tandem W domain-based nucleators.
RESULTS AND DISCUSSION
Crystal structure of crosslinked WxActin
In two previous studies, we reported low-resolution SAXS structures of actin nucleation complexes formed by the Arp2/3 complex and tandem W domains 22; 23. Barbed end polymerization in these studies was blocked by crosslinking of the W domain to Cys-374 of the actin subunit located at the barbed end of the complexes. This approach was based on analysis of the structures of various W-actin complexes 11; 12; 13, which placed the N-terminus of the W domain within disulfide bond distance to actin Cys-374. In each case, a Cys residue was introduced into the W domain at the most favorable position for crosslinking to actin Cys-374. Here, this approach was used again to obtain the low-resolution crystal structure of 3W-Actin. However, it remained unclear whether the crosslink altered the structure of actin and/or the W domain in a significant way, which prompted us to pursue the determination of a crosslinked WxActin structure. It later became apparent that this structure also provided the best molecular replacement model for determination of the structure of 3W-Actin.
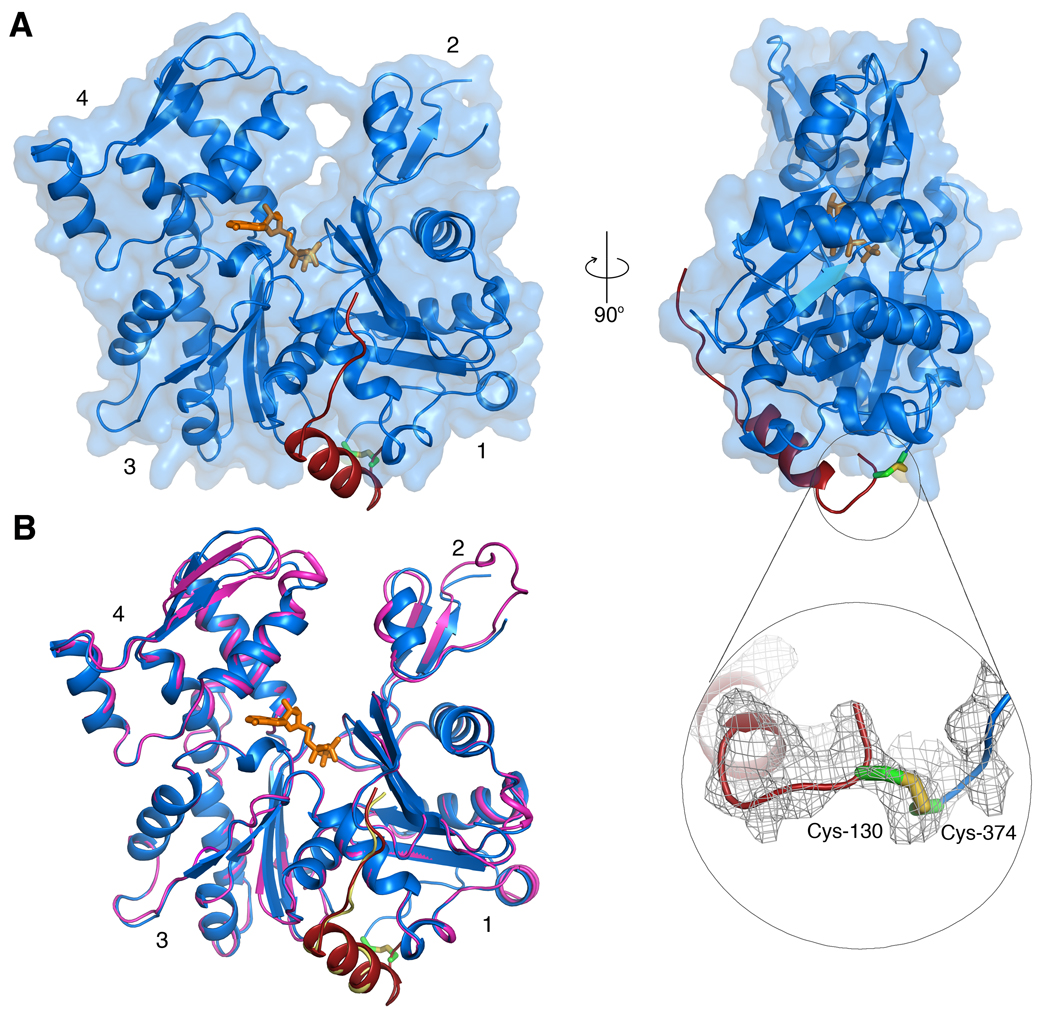
After testing crystallization with various W domains, good diffracting crystals were obtained of the crosslinked complex of actin with a synthetic peptide corresponding to the first W domain (amino acids 130–160) of Vibrio parahemolyticus VopL. During synthesis, residue Val-131 of this W domain was replaced by Cys and crosslinked to actin Cys-374 (Materials and Methods). The crystal structure of WxActin was determined by molecular replacement to 2.9 Å resolution (Fig. 1A and Table 1).
FIG. 1.
Structure of WxActin. (A) Two perpendicular views of the structure of WxActin. The inset shows the 2Fo-Fc electron density map (contoured at 1σ) in the region around the crosslink. Although the crosslink was visualized, this is one of less well-defined regions of the map. (B) Superimposition of the structures of WxActin (blue, actin; red, W domain) and the non-crosslinked complex of actin with the W domain of WASP (pink, actin; yellow, W domain), showing the similarity of the structures. (Two Column Figure).
Table 1.
Crystallographic Data and Refinement Statistics
| WxActin | 3W-Actin | |
|---|---|---|
| Diffraction data | ||
| Wavelength (Å) | 1.0 | 1.0 |
| Space group | P 212121 | P 65 2 2 |
| Unit cell a/b/c (Å) | 66.6 / 76.4 / 86.1 | 100.7 / 100.7 / 458.8 |
| Unit cell α/β/γ (°) | 90.0 / 90.0 / 90.0 | 90.0 / 90.0 / 120.0 |
| Resolution (Å) | 50.0-2.89 (2.99–2.89) | 50.0-7.0 (7.9–7.0) |
| Unique reflections | 10207 | 2182 |
| Completeness (%) | 99.2 (92.5) | 90.0 (70.1) |
| Redundancy | 12.9 (6.4) | 20.5 (6.0) |
| Rmergea (%) | 16.8 (46.1) | 8.6 (37.3) |
| I/σ | 16.3 (1.8) | 16.5 (2.1) |
| Refinement | ||
| Resolution (Å) | 37.51–2.89 | |
| Atoms used in refinement | 3058 | |
| Rfactorb (%) | 21.2 | |
| Rfreec (%) | 26.5 | No atomic refinement was performed |
| Rmsd bond lengths (Å) | 0.011 | |
| Rmsd bond angles (°) | 1.910 | |
| Average B factors (Å2) | ||
| All atoms | 62.90 | |
| Protein atoms | 62.92 | |
| Solvent | 58.57 | |
| Residues in Ramachandran plot | ||
| Most favored regions (%) | 90.2 | |
| Other allowed regions (%) | 9.8 | |
| PDB accession code | 3M1F | 3M3N |
Values in parentheses correspond to highest resolution shell
Rmerge=Σhkl(I-<I>)/ΣI, where I and <I> are the observed and mean intensities of all the observations of reflection hkl, including its symmetry-related equivalents
Rfactor=Σhkl∥Fobs| - |Fcalc∥/Σ|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors of reflection hkl
Rfree, Rfactor calculated for a randomly selected subset of reflections (5%) that were not used in refinement
The structure of WxActin is very similar to those of non-crosslinked W-Actin complexes determined with bound DNase I 11; 13 and that of Drosophila ciboulot bound to actin-latrunculin A 12. Figure 1B shows a comparison of the structure of WxActin with that of the non-crosslinked complex of actin with the W domain WASP (PDB code 2A3Z). The two structures superimpose with r.m.s deviation of 0.66 Å for 358 equivalent Cα atoms. The most important differences occur in regions that were visualized in one of the structures but not the other, including the DNase I-binding loop (D-loop), the C-terminus of actin, and the N-terminus of the W domain. The D-loop is disordered in most structures of actin, as well as in the structure of WxActin described here, but forms an extended β-sheet with β-strands of DNase I in the non-crosslinked structure. The C-terminus of actin is also disordered in most crystal structures, except complexes with profilin, which interacts with the C-terminus of actin 24; 25; 26. In the non-crosslinked W-Actin complex, the last 10 amino acids of actin (Gly-366 to Phe-375) are disordered and the W domain is only visualized starting from WASP residue Arg-431 (corresponding to VopL Asn-132). In contrast, in the crosslinked structure only the last amino acid of actin (Phe-375) is unresolved in the electron density map, whereas the W domain of VopL is visualized from residue 130 to 151, i.e. the last nine amino acids of the synthetic peptide were not resolved. The disulfide bond between actin Cys-374 and VopL Cys-131 is visualized in the electron density map (inset in Fig. 1A), although it is poorly defined compared to the rest of the structure.
The similarity of the structures suggests that the crosslinking approach used here and in previous studies 22; 23 as a tool to cap the barbed end of actin polymerization nuclei for structural investigation does not introduce significant structural distortions. Furthermore, as we show next, the availability of the structure of WxActin aided the determination of the structure of 3W-Actin.
Crystal structure of 3W-Actin
The solution SAXS study of 3W-Actin revealed an elongated molecule, consistent with the presence of three actin subunits, somewhat similar to the long-pitch helix of the actin filament 22. However, the nature of actin-actin contacts in the complex could not be determined. We had suggested that subdomain 2 of actin could move slightly, which combined with a helical conformation in the D-loop, would make the binding of tandem W domains fully compatible with intersubunit contacts in the actin filament 3; 4. Other investigators had suggested that the W domain would probably interfere with intersubunit contacts in the filament 27; 28. Knowing which proposal is correct is important, because it may shed light on the mechanism of nucleation, and possibly explain why tandem W domain-based nucleators do not influence elongation the way formins do. It may also answer important questions about differences in the activities of tandem W domain-based nucleators, and the mechanism of action of NPFs of the Arp2/3 complex, which also contain tandem W domains 7. Therefore, we set out to crystallize the complexes of 2W-Actin and 3W-Actin. While both complexes were crystallized readily, the crystals did not diffract the X-rays. Additional search for conditions led to the identification of additives, such as RbCl and polyvinylpyrrolidone K15, which improved diffraction somewhat. After several attempts, the best result consisted of a rather complete and highly redundant X-ray dataset collected from crystals of 3W-Actin to 7 Å resolution. While we initially considered not reporting this structure, we later recognized that significant information could be obtained by positioning high-resolution W-Actin structures into the unit cell of the 3W-Actin crystals by molecular replacement. Because the individual structures are known at high-resolution, this approach overcomes some of the typical limitations of low-resolution structures in which the content of the unit cell is totally unknown. The limitations, however, are that individual atomic positions cannot be refined and the inter-W linkers cannot be visualized.
Consistent with the design and mass measurements in solution of the complex of 3W-Actin 22, three copies of the W-Actin basic unit were expected in the asymmetric unit of the crystal. The volume of the asymmetric unit was also compatible with it containing three copies of the W-Actin unit (corresponding to a solvent content of 43%). However, weak diffraction is typically consistent with a higher solvent content. Not surprisingly, the molecular replacement solution, performed independently with the programs Phenix 29; 30 and AMoRe 31, located only two W-Actin complexes in the asymmetric unit, for a solvent content of 62% (see Material and Methods and detailed description in Supplementary Material). We do not understand why one of the actin molecules dissociates during crystallization, although it could simply be that this molecule is bound loosely and is therefore displaced by favorable crystal contacts. Analysis of the crystal packing demonstrates why a third actin molecule was never found. Consecutive actin dimers are stacked head-to-tail, forming a helix along the crystallographic c axis (Movie S1). Two such helices assemble tightly in anti-parallel fashion (see Movie S2). Each anti-parallel pair comprises 24 actin subunits along the length of the c axis, which constitutes the basic building block of the crystal lattice. Adjacent pairs of helices crossover twice in a repeat (or helical turn), corresponding to the length of the c axis (see Movies S3 and S4), thus assuring the connectivity of the crystal lattice and leaving no extra-space for the missing third actin subunit (or rather 12 actin subunits, when the P6522 symmetry of the crystal is taken into consideration).
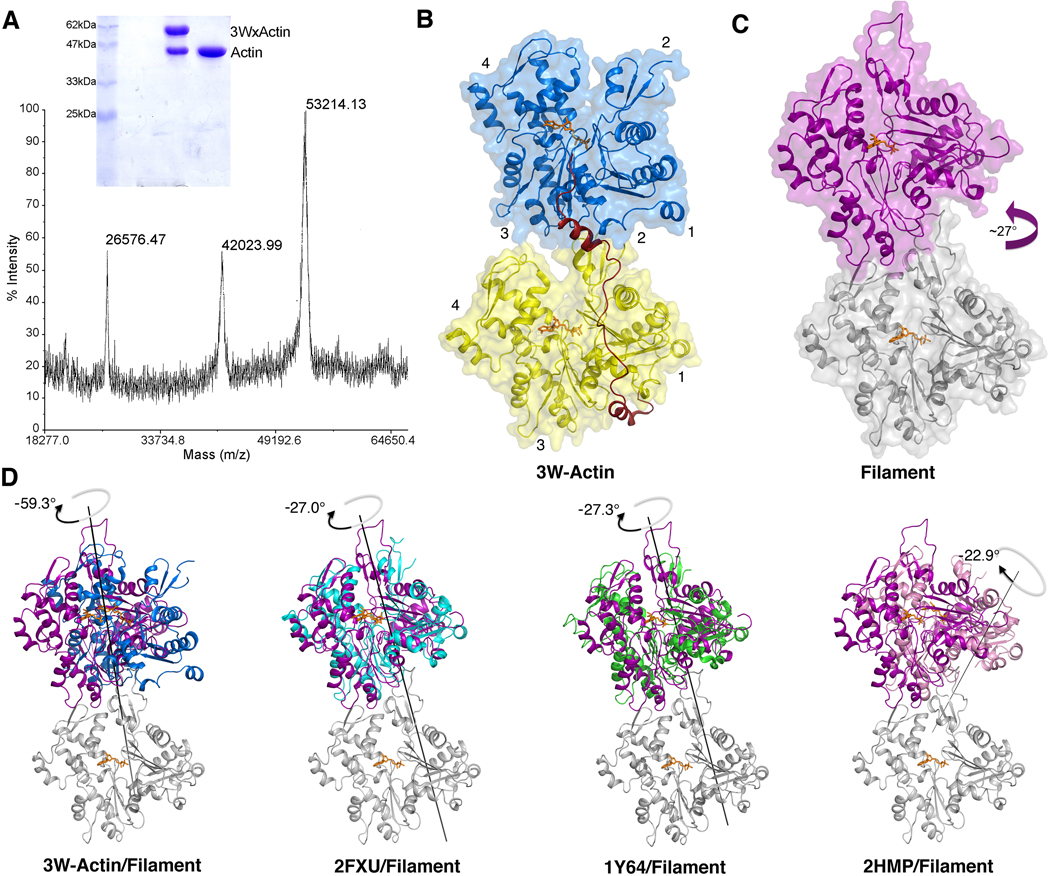
Because of the limited resolution, we could not identify which of the actin subunits is lost during crystallization, or whether the crystals consist solely of the actin subunit crosslinked to the long 3W polypeptide. Note that any non-crosslinked actin dissociated from the complex would be expected to polymerize during crystallization, and would therefore not be present in the crystals. To address this question, a large number of crystals were collected, washed multiple times in the crystallization solution by transferring them with a cryo-loop, and then dissolved in water for analysis by non-reducing gel electrophoresis and mass spectrometry (Materials and Methods). The results clearly illustrate that the crystals consist of a 50/50 mixture of actin crosslinked to construct 3W and non-crosslinked actin (Fig. 2A). Therefore, we conclude that one of the non-crosslinked actins was lost during crystallization which, based on the arrangement of actin subunits in the asymmetric unit, is most likely that bound to the last W domain.
FIG. 2.
Structure of 3W-Actin. (A) Non-reducing gel electrophoresis and mass spectrometry analysis indicate that the crystals of 3W-Actin consist of a 50/50 mixture of actin crosslinked to construct 3W (expected mass 53,021 Da) and non-crosslinked actin. Actin is also shown in the gel as a control. (B) Illustration of the actin dimer in the structure of 3W-Actin. The linker between W domains was modeled. (C) Illustration of a longitudinal actin dimer from the actin filament model 4. (D) Comparisons of the relative rotations between actin subunits in the actin filament model (gray and magenta) and the structures of 3W-Actin and three representative actin dimers observed in crystal structures (including non-crystallographic and symmetry-related dimers, see also Table 2). For this comparison, the structures were superimposed using as reference the lower actin subunit (gray), which is only shown for the filament model. Note that compared to a long-pitch dimer of the actin filament, in which subunits are rotated by ~27° (magenta arrow), there is a −60° rotation between the two crystallographically independent actin subunits in the structure of 3W-Actin. Other dimers observed in crystal structures tend to be flat due to symmetry constraints and are therefore rotated −27° relative to a longitudinal dimer of the filament. The relative rotations between actin subunits were calculated with the program DynDom 44. (Two Column Figure)
The disposition of the actin subunits in the structure of 3W-Actin (Fig. 2B) is somewhat similar to the longitudinal arrangement of actin subunits in the long-pitch helix of the actin filament model 3; 4 (Fig. 2C). However, important differences are observed. To better understand these differences, it is important to discuss what is currently known about longitudinal contacts in the actin filament. Multiple crystal structures of actin show similar longitudinal contacts between actin subunits (including both non-crystallographic dimers and symmetry-related dimers), which are thought to mimic inter-subunit contacts in the actin filament (Table 2). However, because of constraints imposed by crystal symmetry, these dimers are unwound, i.e. they lack the natural twist of the actin filament. A detailed analysis of these structures and their implications for our understanding of the actin filament has been carried out by other investigators 32, and will not be repeated here. However, it is important to compare the structure of 3W-Actin to both the actin filament model 3; 4 and the longitudinal dimers observed in crystal structures, with the understanding that the structure of 3W-Actin does not address the conformation of the actin filament per se, but rather the mechanism of recruitment of actin subunits by tandem W domain proteins.
Table 2.
Structures of Longitudinal Actin Dimers.
| PDB | actins per AU |
Description | Symmetry, Resolution (Å) and Cell a, b, c (Å), α, β, γ (°) |
References |
|---|---|---|---|---|
| 3M3N | 2 | Dimer stabilized by tandem W domains |
P6522, 7.0 100.7, 100.7, 458.8, 90.0, 90.0, 120.0 |
This work |
| 2HF3 2HF4 | 1 | non-polymerizable actin mutant (Ala-204Glu/Pro-243Lys) |
C2, 1.8 199.7, 54.1, 39.6, 90.0, 93.2, 90.0 |
42 |
| 2ASM 2ASO | 1 | Complexes with marine macrolides |
C2, 1.6 171.2, 54.7, 40.7, 90.0, 96.0, 90.00 |
45; 46 |
| 2ASP | or | |||
| 2FXU | C2, 1.35 60.1, 56.5, 101.7, 90.0, 94.6, 90.0 |
|||
| 2A5X | 1 | Longitudinally crosslinked actin dimer |
C2, 2.49 207.4, 54.4, 36.2, 90.0, 98.6, 90.0 |
47 |
| 2Q1N 2Q31 | 2 | Longitudinally crosslinked actin dimer |
P21, 2.7 108.1, 71.8, 54.8, 90.0, 104.7, 90.0 |
32 |
| 1Y64 | 1 | Complex with formin homology 2 domain |
C2, 3.05 232.0, 56.2, 100.9, 90.0 107.7, 90.0 |
48 |
| 2HMP | 2 | Non-polymerizable actin, cleaved between Gly42 and Val43 |
P212121, 1.9 64, 198, 69.6, 90.0, 90.0, 90.0 |
49 |
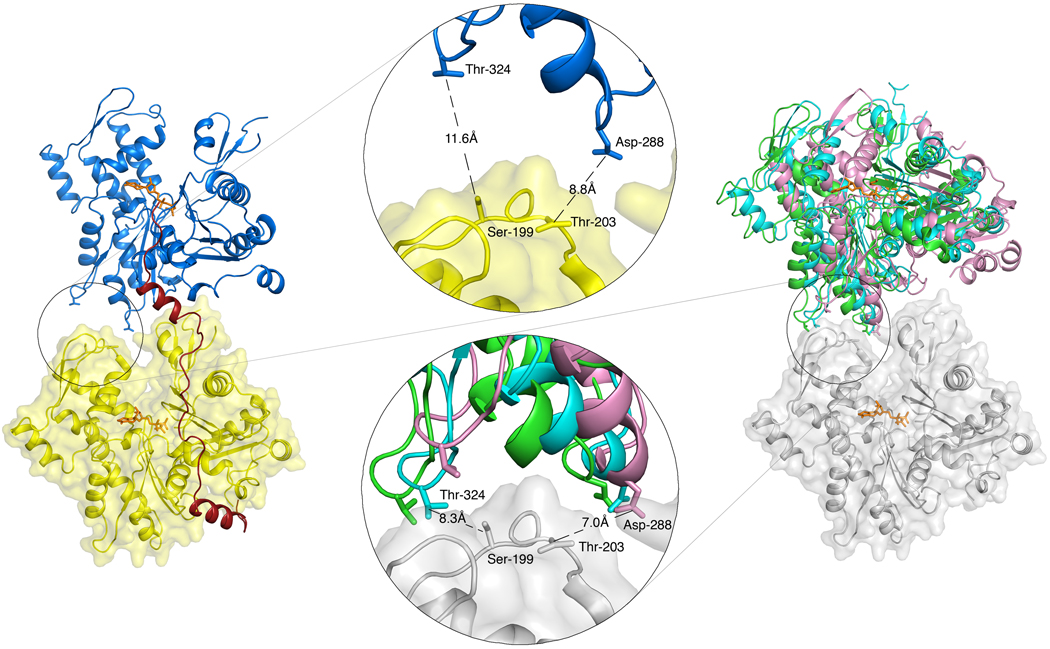
The dimers observed in crystal structures are generally similar and often crystallographically isomorphous. Based on a superimposition of their structures, we have identified three subgroups that diverge more significantly (represented by PDB entries 2FXU, 1Y64 and 2HMP) (Table 2). Compared to a long-pitch dimer of the actin filament model in which consecutive subunits are rotated by ~27° 3; 4, these three subgroups present flat structures, i.e. rotated counterclockwise with respect to the filament dimer by approximately −27° (Fig. 2D) (although the orientation of the axis of rotation is markedly different for entry 2HMP). Remarkably, the longitudinal contacts between subdomains 4 and 3 of neighboring actin subunits are well conserved in the three subgroups (Fig. 3). It thus appears that longitudinal contacts between actin subunits in the filament have a strong tendency to reemerge as crystal contacts in actin structures 32. It is important to note that these structures offer the most accurate view of longitudinal contacts currently available 32, because the resolution of the actin filament model 3; 4 is still insufficient to address specific atomic interactions. Additional longitudinal contacts are thought to involve the D-loop in subdomain 2 33, which is proposed to bind in the hydrophobic cleft between subdomains 1 and 3 of the actin subunit immediately above it 4. However, the D-loop is disordered in all the crystal structures containing longitudinal actin dimers, and its conformation(s) and actual contacts in the filament are unknown.
FIG. 3.
Inter-subunit contacts in the structure of 3W-Actin compared to those of crystallographic actin dimers. Insets show that longitudinal contacts between subdomains 4 and 3 of adjacent actin subunits observed in various crystal structures (right) are mostly broken in the structure of 3W-Actin (left). Representative distances between Cβ atoms are shown for reference. (Two Column Figure)
On the other hand, the rotation between the two actin subunits in the structure of 3W-Actin is approximately −33°, i.e. −60° compared with a longitudinal dimer of the actin filament 3; 4 (Fig. 2D). As a result, the longitudinal contacts observed in other crystal dimers are generally broken in the structure of 3W-Actin (Fig. 3), whereas the contacts involving subdomain 2 are unresolved. Therefore, it appears that the presence of the W domain at the interface between actin subunits breaks the natural tendency that actin has to preserve filament-like longitudinal contacts in crystal structures, and induces a rotation between actin subunits that is of similar magnitude but opposite direction to that of the filament (−33° vs 27°). These results are generally consistent with our previous SAXS studies 22, which revealed an extended (pseudo long-pitch) arrangement of the actin subunits stabilized by tandem W domains. However, the SAXS envelope lacked the resolution to distinguish between the dimer observed here in the crystal structure of 3W-actin and a longitudinal dimer of the actin filament model.
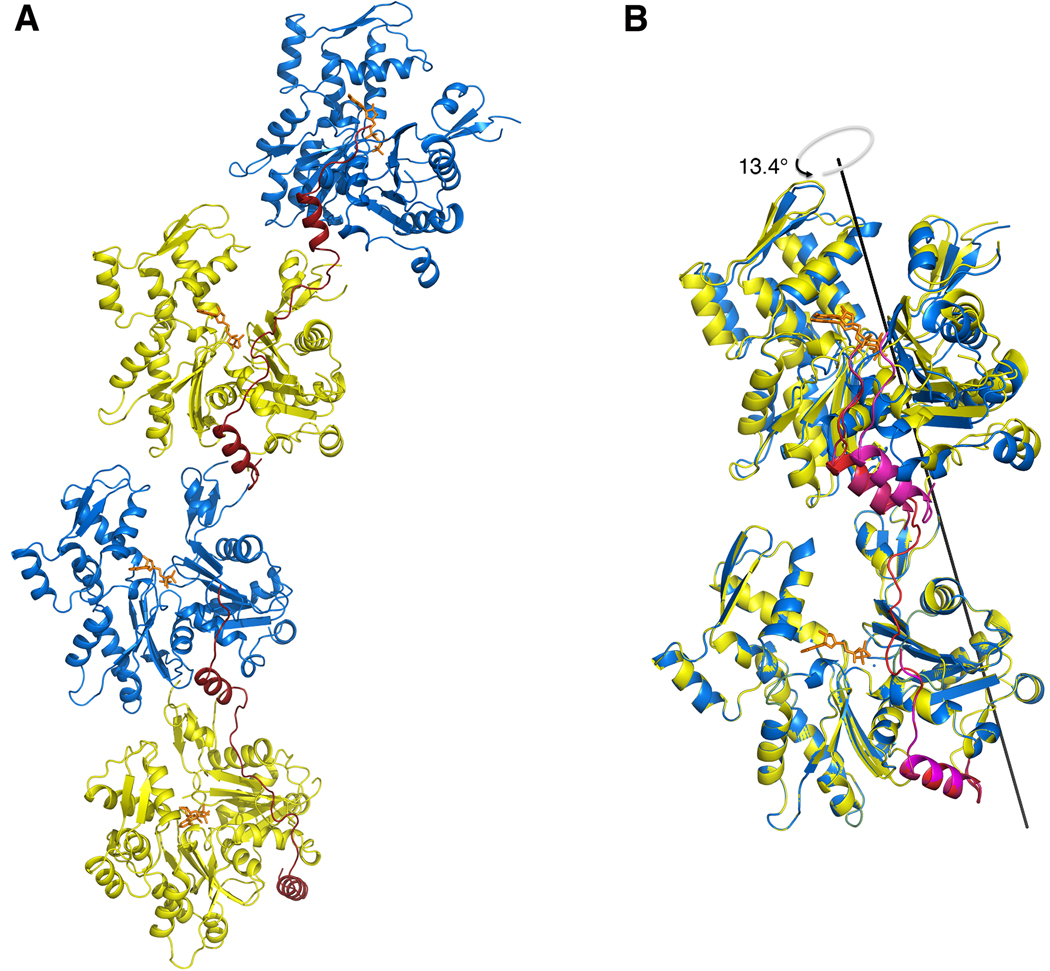
It is interesting to note that there is also a crystal contact in the structure of 3W-Actin (between two adjacent dimers) that resembles the dimer of the asymmetric unit. This so-called ‘crystal’ dimer differs even more significantly from both the actin filament and the other actin dimers described above, due to an overall translation of ~8 Å between actin subunits compared to the dimer of the asymmetric unit (Fig. 4). In the crystal dimer, the crosslink with construct 3W is at the interface between actin subunits, which may explain the added translation. However, it is significant that the actin subunits of both the non-crystallographic and crystal dimers are rotated counterclockwise by about the same amount compared to all the other actin dimers observed in crystal structures, suggesting that this is a general constraint imposed by the W domain at the interface between actin subunits.
FIG. 4.
Comparison of the non-crystallographic and crystallographic dimers in the structure of 3W-Actin. (A) Representation of two consecutive dimers related by crystal symmetry. (B) Superimposition of the non-crystallographic (yellow-blue and red W domains) and crystallographic (blue-yellow and magenta W domains) dimers. Note that despite their general similarity, the crystallographic dimer differs more significantly from other actin dimers observed in crystals structures (Table 2) and the actin filament model. While the actin subunits in the crystallographic dimer are rotated ~13° relative to the non-crystallographic dimer, which undoes part of the initial −60° rotation, there is also a translation of ~8 Å, probably imposed by steric hindrance with the crosslinked W domain. It is nonetheless significant that the actin subunits of both the non-crystallographic and crystallographic dimers are rotated counterclockwise by about the same amount compared to all the other dimers observed in crystal structures, which are generally unwound (see Fig. 2), suggesting that this is a general property of the W domain at the interface between actin subunits. (Two Column Figure)
We conclude that while Spire-like tandem W domains can bring actin subunits into close proximity for nucleation, the conformation of the polymerization nucleus that they form differs significantly from that of the actin filament. This may explain their weak nucleation activity as analyzed next.
Long-pitch nucleation by tandem W domains is suboptimal
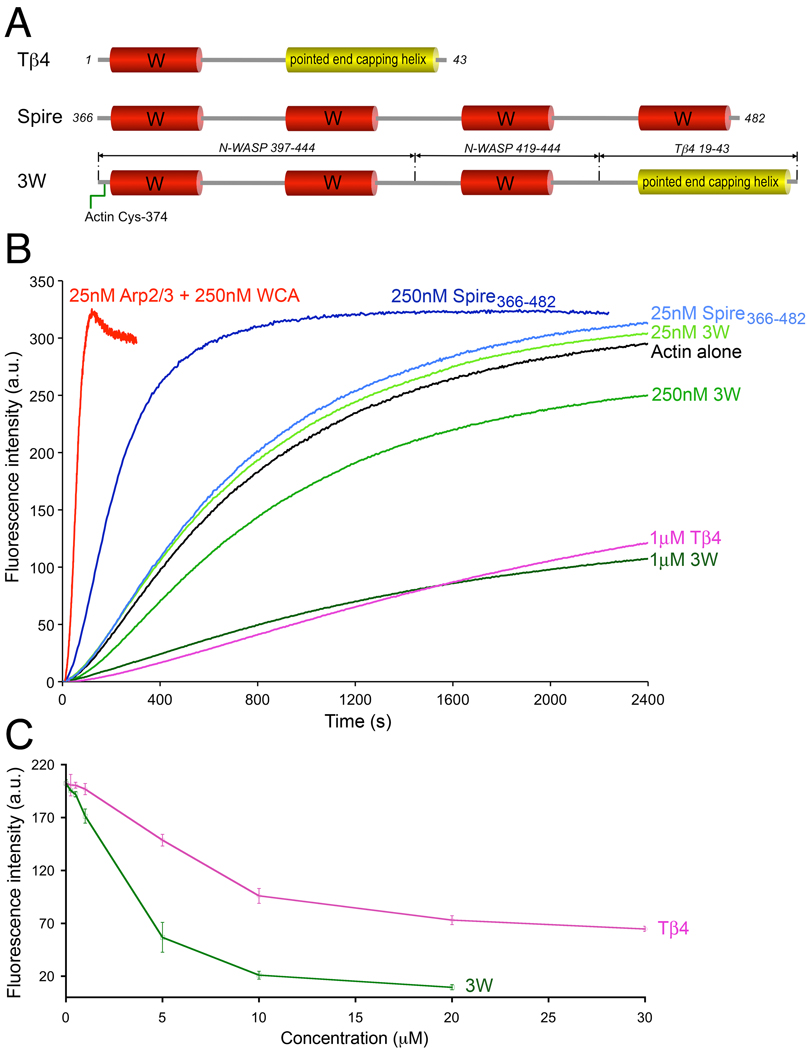
The structural results prompted us to test the polymerization activity of construct 3W as compared to those of the prototypical tandem W domain nucleator Spire, which stabilizes a long-pitch nucleus, and the Arp2/3 complex, which forms a short-pitch nucleus. The nucleation activity of Drosophila Spire 14 has been mapped to the fragment Spire366–482 comprising the four W domains (Fig. 5A), which was used in the current study. We used the pyrene-actin polymerization assay to study the effect of Spire366–482 on the polymerization of 2 µM actin (6% pyrene labeled) by monitoring the fluorescence increase resulting from the incorporation of labeled actin monomers into the filament (Fig. 5B). At the concentration of 25 nM, Spire366–482 had very little effect on actin polymerization (polymerization rate 1.0±0.2 nM/sec as compared to 0.8±0.1 nM/sec for actin alone), whereas the Arp2/3 complex activated by the WCA fragment of mouse N-WASP showed a major increase in polymerization (polymerization rate 31.5±1 nM/sec). However, the nucleation activity of Spire366–482 increased with concentration, becoming a stronger nucleator at 250 nM (polymerization rate 4.8±0.2 nM/sec). The opposite effect was observed with construct 3W, which had no effect on actin polymerization at the concentration of 25 nM, but inhibited polymerization when used at 250 nM. This could be an indication that construct 3W, like Tβ4, sequesters actin monomers (Fig. 5B).
FIG. 5.
Different effects of Spire, 3W, and Tβ4 on actin polymerization. (A) Schematic diagram of Tβ4, the four W domain region of Drosophila Spire, and construct 3W. Note that construct 3W consists of three W domains (occurring naturally in mouse N-WASP) separated by short linkers (as in Spire) and the pointed end capping helix of Tβ4. This construct also contains a Cys residue at the N-terminus that was crosslinked to actin Cys-374 for crystallization, but the crosslink was reduced with DTT to measure the nucleation activity. (B) Time course of polymerization of 2 µM Mg-ATP-actin (6% pyrene-labeled) alone (black) or in the presence of different concentrations of Spire366–482 (different shades of blue), construct 3W (different shades of green), Tβ4 (pink), and 25 nM Arp2/3 complex with 250 nM mouse N-WASP WCA (red). Each experiment was repeated at least three times. The polymerization rates are: Actin (0.8±0.1 nM/sec), Spire (1.0±0.2 nM/sec at 25nM and 4.8±0.2 nM/sec at 250 nM), Arp2/3 complex (31.5±1 nM/sec). (C) Steady-state concentration-dependence of actin monomer sequestration by Tβ4 and 3W. (One Column Figure)
Tβ4 is a short 43-aa polypeptide related to the W domain 8; 9, but it contains an additional helix at the C-terminus that binds atop actin subdomains 2 and 4 34 (Fig. 5A). As a result, and despite the apparent simplicity of its helix-loop-helix design, Tβ4 has the ability to block actin monomer addition to both the pointed and barbed ends of the actin filament, making it an extremely effective monomer sequestering protein 35; 36. Some proteins contain tandem repeats of the Tβ4 fold. Examples include, Acanthamoeba castellanii actobindin 37, Drosophila melanogaster ciboulot 12 and Caenorhabditis elegans tetrathymosin 38, which respectively contain two-and-a-half, three and four copies of the Tβ4 fold. Contrary to tandem repeats of the W domain, that frequently mediate filament nucleation 7, tandem Tβ4 proteins are characterized by their ability to sequester actin monomers 37; 38. Therefore, we asked whether 3W, consisting of a tandem repeat of three W domains followed by the C-terminal helix of Tβ4 (Fig. 5A), would sequester actin monomers. A concentration-dependence analysis of steady-state actin polymerization revealed that construct 3W sequesters actin monomers even more effectively than Tβ4 (Fig. 5C). We had previously shown, using analytical ultracentrifugation, light scattering and native gel electrophoresis, that 3W binds three actin monomers in solution 22, which may explain its stronger sequestering activity compared to Tβ4. Therefore, the effect of 3W on actin polymerization is more closely related to that of tetrathymosin, which binds and sequesters multiple actin monomers 38. Although actobindin and ciboulot also sequester actin monomers, perhaps surprisingly they form 1:1 complexes with actin, indicating that only one of their actin-binding sites is fully functional 12; 37. It thus appears that the simple addition of the pointed end capping helix of Tβ4 to tandem W domains changes their activity from nucleation, as in Spire 14, to monomer sequestration as in Tβ4 35; 36.
CONCLUSIONS
The crystal structure of crosslinked WxActin was found to be nearly undistinguishable from those of non-crosslinked W-Actin complexes. We have used W domain crosslinking in this work, as well as in two previous studies 22; 23. The finding that the structure is not altered in a significant way by the crosslink suggests that this is a structurally sound approach that can be used as a way to stabilize large polymerization complexes, which are intrinsically dynamic, for structural investigation.
Various proteins contain tandem repeats of the W domain 7; 8; 9. While the W domain itself presents well-conserved features (N-terminal helix and LKKT(V) motif), the linkers between W domains are highly variable, and no single structure can be fully representative of this large family of proteins. Irrespective of this variability, the inter-W linkers can be sub-divided into two subgroups: short (as in Spire and N-WASP) and long (as in Cobl linker-2). While 3W is a synthetic construct with no natural counterpart, it is based on the tandem W repeat of N-WASP, and it therefore represents the short inter-W linker subgroup. One general implication of the structure of its complex with actin is that the binding of the W domain is intrinsically incompatible with inter-subunit contacts along the long-pitch helix of the actin filament, which is contrary to what we had anticipated 3; 4. The structure of 3W-Actin further suggests that the actin subunits recruited by tandem W domains with short inter-W linkers are positioned in a way that resembles the long-pitch helix of the actin filament, a conformation that would be expected to favor polymerization. However, due to steric hindrance of the W domain, the contacts between actin subunits in these complexes differ significantly from those of the actin filament. This may explain the weak nucleation activity of Spire as compared to the Arp2/3 complex, formins, Cobl and Lmod, all proteins that are thought to stabilize short-pitch actin nuclei to initiate polymerization. The incompatibility of the W domain with longitudinal inter-subunit contacts in the filament also implies that when the actin nucleus transitions into a filament and begins to elongate, tandem W domain nucleators cannot stay bound to newly formed filaments, and would therefore be unlikely to influence elongation. Steric hindrance of the W domain may also be a contributing factor in the release of NPFs of the Arp2/3 complex from branch junctions once the branch filament begins to elongate (conformational changes within the Arp2/3 complex itself could be another factor). We finally found that the simple addition of the C-terminal pointed end-capping helix of Tβ4 to tandem W domains can change their activity from actin filament nucleation to monomer sequestration.
MATERIALS AND METHODS
Preparation of proteins and protein complexes
Detailed descriptions of the preparation and characterization of the complex of 3W-Actin 22 and the purification of the Arp2/3 complex from bovine brain and preparation of the WCA fragment of mouse N-WASP 23 were reported previously. Actin was purified from rabbit skeletal muscle 39. Tβ4 and the first W domain (amino acids Ser-130 to Ser-160) of Vibrio parahemolyticus VopL (UniProt accession code: Q87GE5) were made as synthetic peptides, and purified by reverse-phase chromatography. During peptide synthesis, an amino acid substitution was made (Val-131->Cys) in the first W domain of VopL, a position chosen based on analysis of the various W-Actin structures 11; 12; 13 as the most favorable for crosslinking to actin Cys-374. The crosslinking reaction was performed by activation of the W domain peptide with DTNB (5,5'-dithiobis(2-nitrobenzoic acid)), before mixing with actin at an actin:W peptide ratio of 1:1.2. The crosslinked fraction was then separated by gel filtration on a S200 column (Pfizer-Pharmacia) in 25 mM Tris-HCl pH 7.5, 100 mM NaCl, 0.2 mM CaCl2 and 0.2 mM ATP.
The fragment 366–482 of Drosophila Spire (Spire366–482), comprising the four W domains, was amplified by PCR from cDNA purchased from Open Biosystems. The PCR product was cloned between the NdeI and EcoRI sites of vector pTYB12 (New England BioLabs). Protein expression was performed in BL21(DE3) cells (Invitrogen) grown in Terrific Broth medium at 37°C until the OD600 was 1.0–1.2. Expression was induced with addition of 0.5 mM isopropylthio-β-D-galactoside for 5 h at 20°C. Cells were resuspended in chitin-column equilibration buffer [20 mM HEPES (pH 7.5), 500 mM NaCl, 1 mM EDTA, and 100 µM PMSF]. After purification on the chitin affinity column and release of the protein by DTT-induced autocleavage of the intein, Spire366–482 was additionally purified on a reverse-phase C18 column (0.1% trifluoroacetic acid and 0–90% acetonitrile), and then dialyzed extensively against 25 mM Tris-HCl pH 7.5, 100 mM NaCl.
Crystallization of the complexes of 3W-Actin and WxActin
The complex of 3W-Actin (consisting of a tandem repeat of three W domains with three actin subunits bound 22) was dialyzed against 20 mM HEPES pH 7.5, 100 mM NaCl, 0.2 mM CaCl2 and 0.2 mM ATP and concentrated to 15 mg/ml using an Amicon centrifugal filter (Millipore). Needle-like crystals grow within hours, or even minutes, using the hanging drop vapor diffusion method at 20°C, and from drops consisting of a 1:1 (v/v) mixture of protein solution and a well solution containing 100 mM CAPS pH 10.0, and 24% polyethylene glycol 3350. However, these crystals did not diffract the X-rays. Crystal quality and diffraction were improved with addition of 10–100 mM RbCl or polyvinylpyrrolidone K15 as additives. The crystals were flash-frozen in liquid nitrogen, with addition of 20% glycerol as cryoprotectant. The crosslinked complex WxActin was concentrated to 5 mg/ml, and crystallized using the hanging drop vapor diffusion method at 20°C from a well solution containing 0.2 M LiNO3 and 20% polyethylene glycol 3350.
The content of the crystals of 3W-Actin was analyzed by non-reducing gel electrophoresis and mass spectrometry, using a Voyager DE Pro MALDI-TOF Mass Spectrometer (Applied Biosystems) and sinapinic acid as a matrix. For this analysis, multiple crystals were collected and washed five times through the crystallization solution by transferring them with a cryo-loop, and then dissolved in water.
Data collection and determination of the structures
X-ray datasets were collected from crystals of WxActin and 3W-Actin at the beamline 17-BM of the IMCA-CAT facility of the Advance Photon Source (Argonne, IL). Data indexation and scaling were carried out with the program HKL2000 (HKL Research, Inc.). The crystals of 3W-Actin diffracted only to 7.0 Å resolution (Table 1). The data in the last resolution shell (7.25 – 7.0 Å) is weak (I/σ 1.1) and only 34.3% complete. Yet, ~70% of the data was obtained between 7.9 – 7.0 Å, with average I/σ of 2.1 and redundancy of 6. This range includes 449 reflections (~20% of the total). Because of the limited resolution, special emphasis was placed on obtaining a highly redundant dataset (the average redundancy is 20.5 for the entire dataset), which should minimize intensity errors.
The structure of WxActin was determined by molecular replacement, using as search model the structure of actin complexed with the W domain of WASP (PDB code: 2A3Z). Molecular replacement and refinement were carried out with the program Phenix 29, and model building was performed with the program Coot 40.
The structure of 3W-Actin was determined by molecular replacement, using the stronger data between 15 and 8 Å resolution, and independently with two different programs; Phaser 30 belonging to the Phenix package 29 and AMoRe 31. The two programs gave the same solution. The likelihood-based scoring function (LLG) of the program Phenix is highly sensitive to the quality of the search model 41. Several search models were tested, including monomeric actin 42, complexes of W-Actin determined as ternary complexes with DNase I 11; 13, the complex of ciboulot-actin with bound latrunculin A 12, the structure of actin with the C-terminal portion of Tβ4 34, and the structure of WxActin determined here. The best-contrasted solution was obtained using the structure of WxActin as search model. Two different models were prepared based on this structure, one consisting of the entire crosslinked complex and one lacking the crosslinked portion (i.e. the last 5 amino acids of actin and the first 3 amino acids of the W domain). These two models were positioned independently using a multibody-body search, and clearly defined the locations of the first (crosslinked) and the second (non-crosslinked) actin subunits of the dimer.
While Phenix was used in the automated mode, a more exhaustive search was performed with the program AMoRe (details in Supplementary Material). AMoRe’s self-rotation function gave a single prominent peak with correlation coefficient 0.62. Thus, while the volume of the unit cell seemed to be compatible with the presence of three W-Actin complexes in the asymmetric unit (corresponding to a Matthews’ coefficient Vm of 2.15 Å3/Da and a solvent content of 43%), only two were found (for a Vm of 3.23 Å3/Da and a solvent content of 62%). We tested many possible configurations in which the orientation of one W-Actin complex was constrained with respect to the other according to the non-crystallographic two-fold axis resulting from the self-rotation function. This gave a clearly contrasted solution for two W-Actin complexes, where the correlation between calculated and observed structure factor amplitudes was 0.66 (0.50 for the next peak that was not contrasted above background). Because of the limited resolution, the only refinement performed after molecular replacement was by rigid body, and using all the diffraction data available.
Actin Polymerization Assay
Pyrene-actin polymerization assays were carried out and analyzed as described 43, using a Cary Eclipse fluorescence spectrophotometer (Varian). All the experiments were performed at 20°C. Prior to data acquisition, 2 µM Mg-ATP-actin (6% pyrene-labeled) was mixed with different concentrations of construct 3W (25 nM, 250 nM or 1 µM), Tβ4 (1 µM), and Spire (25 nM, 250 nM) in F-buffer (10 mM Tris-HCl pH 7.5, 1 mM MgCl2, 50 mM KCl, 1 mM EGTA, 0.02 mg/mL BSA, 0.2 mM ATP, 1 mM DTT, 0.1 mM NaN3). Note that the addition of DTT prevents crosslinking of construct 3W to actin Cys-374 during the polymerization assay. Polymerization rates were measured from the slope of the polymerization curve at 50% polymerization and converted to nM/sec assuming that the total concentration of polymerizable actin monomers is 1.9 µM (2 µM – 0.1 µM, i.e. by subtracting the critical concentration for actin monomer addition to the barbed-end from the total concentration of actin) 43. Steady-state experiments with varying Tβ4 or 3W concentrations were carried out under similar condition by allowing actin to polymerize for 16h.
Miscellaneous
The program DynDom 44 was used to calculate the relative rotation of actin subunits in crystal structures of longitudinal actin dimers. Illustrations of the structures were prepared with the program PyMol (DeLano Scientific LLC).
Supplementary Material
ACKNOWLEDGEMENTS
Supported by National Institutes of Health grant P01 HL086655. Use of IMCA-CAT beamline 17-BM was supported by the Industrial Macromolecular Crystallography Association through a contract with Hauptman-Woodward Medical Research Institute. The Advanced Photon Source was supported by Department of Energy Contract W-31-109-Eng-38.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
PDB ACCESSION NUMBERS
Coordinates and structure factors were deposited under PDB ID 3M1F (WxActin) and 3M3N (3W-Actin).
SUPPLEMENTARY MATERIAL
Supplementary material is available online, including supplementary Material and Methods and Movies S1–S4.
REFERENCES
- 1.Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophys J. 2001;81:667–674. doi: 10.1016/S0006-3495(01)75731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 3.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 4.Oda T, Iwasa M, Aihara T, Maeda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 5.Holmes KC. Structural biology: actin in a twist. Nature. 2009;457:389–390. doi: 10.1038/457389a. [DOI] [PubMed] [Google Scholar]
- 6.Dominguez R. Structural insights into de novo actin polymerization. Curr Opin Struct Biol. 2010;20:217–225. doi: 10.1016/j.sbi.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dominguez R. Actin filament nucleation and elongation factors--structure-function relationships. Crit Rev Biochem Mol Biol. 2009;44:351–366. doi: 10.3109/10409230903277340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paunola E, Mattila PK, Lappalainen P. WH2 domain: a small, versatile adapter for actin monomers. FEBS Lett. 2002;513:92–97. doi: 10.1016/s0014-5793(01)03242-2. [DOI] [PubMed] [Google Scholar]
- 9.Dominguez R. The beta-thymosin/WH2 fold: multifunctionality and structure. Ann N Y Acad Sci. 2007;1112:86–94. doi: 10.1196/annals.1415.011. [DOI] [PubMed] [Google Scholar]
- 10.Dominguez R. Actin-binding proteins--a unifying hypothesis. Trends Biochem Sci. 2004;29:572–578. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Chereau D, Kerff F, Graceffa P, Grabarek Z, Langsetmo K, Dominguez R. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci U S A. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hertzog M, van Heijenoort C, Didry D, Gaudier M, Coutant J, Gigant B, Didelot G, Preat T, Knossow M, Guittet E, Carlier MF. The beta-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell. 2004;117:611–623. doi: 10.1016/s0092-8674(04)00403-9. [DOI] [PubMed] [Google Scholar]
- 13.Lee SH, Kerff F, Chereau D, Ferron F, Klug A, Dominguez R. Structural Basis for the Actin-Binding Function of Missing-in-Metastasis. Structure. 2007;15:145–155. doi: 10.1016/j.str.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Drosophila Spire is an actin nucleation factor. Nature. 2005;433:382–388. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- 15.Ahuja R, Pinyol R, Reichenbach N, Custer L, Klingensmith J, Kessels MM, Qualmann B. Cordon-Bleu Is an Actin Nucleation Factor and Controls Neuronal Morphology. Cell. 2007;131:337–350. doi: 10.1016/j.cell.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liverman AD, Cheng HC, Trosky JE, Leung DW, Yarbrough ML, Burdette DL, Rosen MK, Orth K. Arp2/3-independent assembly of actin by Vibrio type III effector VopL. Proc Natl Acad Sci U S A. 2007;104:17117–17122. doi: 10.1073/pnas.0703196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tam VC, Serruto D, Dziejman M, Brieher W, Mekalanos JJ. A Type III Secretion System in Vibrio cholerae Translocates a Formin/Spire Hybrid-like Actin Nucleator to Promote Intestinal Colonization. Cell Host and Microbe. 2007;1:95–107. doi: 10.1016/j.chom.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 19.Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 20.Zuchero JB, Coutts AS, Quinlan ME, Thangue NB, Mullins RD. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat Cell Biol. 2009;11:451–459. doi: 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chereau D, Boczkowska M, Skwarek-Maruszewska A, Fujiwara I, Hayes DB, Rebowski G, Lappalainen P, Pollard TD, Dominguez R. Leiomodin is an actin filament nucleator in muscle cells. Science. 2008;320:239–243. doi: 10.1126/science.1155313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rebowski G, Boczkowska M, Hayes DB, Guo L, Irving TC, Dominguez R. X-ray scattering study of actin polymerization nuclei assembled by tandem W domains. Proc Natl Acad Sci U S A. 2008;105:10785–10790. doi: 10.1073/pnas.0801650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boczkowska M, Rebowski G, Petoukhov MV, Hayes DB, Svergun DI, Dominguez R. X-ray scattering study of activated Arp2/3 complex with bound actin-WCA. Structure. 2008;16:695–704. doi: 10.1016/j.str.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schutt CE, Myslik JC, Rozycki MD, Goonesekere NC, Lindberg U. The structure of crystalline profilin-beta-actin. Nature. 1993;365:810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- 25.Ferron F, Rebowski G, Lee SH, Dominguez R. Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP. Embo J. 2007;26:4597–4606. doi: 10.1038/sj.emboj.7601874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baek K, Liu X, Ferron F, Shu S, Korn ED, Dominguez R. Modulation of actin structure and function by phosphorylation of Tyr-53 and profilin binding. Proc Natl Acad Sci U S A. 2008;105:11748–11753. doi: 10.1073/pnas.0805852105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aguda AH, Burtnick LD, Robinson RC. The state of the filament. EMBO Rep. 2005;6:220–226. doi: 10.1038/sj.embor.7400363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renault L, Bugyi B, Carlier MF. Spire and Cordon-bleu: multifunctional regulators of actin dynamics. Trends Cell Biol. 2008;18:494–504. doi: 10.1016/j.tcb.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 29.Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC, Adams PD. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 30.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr D Biol Crystallogr. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- 32.Sawaya MR, Kudryashov DS, Pashkov I, Adisetiyo H, Reisler E, Yeates TO. Multiple crystal structures of actin dimers and their implications for interactions in the actin filament. Acta Crystallogr D Biol Crystallogr. 2008;64:454–465. doi: 10.1107/S0907444908003351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reisler E, Egelman EH. Actin structure and function: what we still do not understand. J Biol Chem. 2007;282:36133–36137. doi: 10.1074/jbc.R700030200. [DOI] [PubMed] [Google Scholar]
- 34.Irobi E, Aguda AH, Larsson M, Guerin C, Yin HL, Burtnick LD, Blanchoin L, Robinson RC. Structural basis of actin sequestration by thymosin-beta4: implications for WH2 proteins. Embo J. 2004;23:3599–3608. doi: 10.1038/sj.emboj.7600372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weber A, Nachmias VT, Pennise CR, Pring M, Safer D. Interaction of thymosin beta 4 with muscle and platelet actin: implications for actin sequestration in resting platelets. Biochemistry. 1992;31:6179–6185. doi: 10.1021/bi00142a002. [DOI] [PubMed] [Google Scholar]
- 36.Xue B, Aguda AH, Robinson RC. Models of the actin-bound forms of the beta-thymosins. Ann N Y Acad Sci. 2007;1112:56–66. doi: 10.1196/annals.1415.010. [DOI] [PubMed] [Google Scholar]
- 37.Hertzog M, Yarmola EG, Didry D, Bubb MR, Carlier MF. Control of actin dynamics by proteins made of beta-thymosin repeats: the actobindin family. J Biol Chem. 2002;277:14786–14792. doi: 10.1074/jbc.M112064200. [DOI] [PubMed] [Google Scholar]
- 38.Van Troys M, Ono K, Dewitte D, Jonckheere V, De Ruyck N, Vandekerckhove J, Ono S, Ampe C. TetraThymosinbeta is required for actin dynamics in Caenorhabditis elegans and acts via functionally different actin-binding repeats. Mol Biol Cell. 2004;15:4735–4748. doi: 10.1091/mbc.E04-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85(Pt B):164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 42.Rould MA, Wan Q, Joel PB, Lowey S, Trybus KM. Crystal structures of expressed non-polymerizable monomeric actin in the ADP and ATP states. J Biol Chem. 2006;281:31909–31919. doi: 10.1074/jbc.M601973200. [DOI] [PubMed] [Google Scholar]
- 43.Harris ES, Higgs HN. Biochemical analysis of mammalian formin effects on actin dynamics. Methods Enzymol. 2006;406:190–214. doi: 10.1016/S0076-6879(06)06015-0. [DOI] [PubMed] [Google Scholar]
- 44.Hayward S, Berendsen HJ. Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins. 1998;30:144–154. [PubMed] [Google Scholar]
- 45.Allingham JS, Zampella A, D'Auria MV, Rayment I. Structures of microfilament destabilizing toxins bound to actin provide insight into toxin design and activity. Proc Natl Acad Sci U S A. 2005;102:14527–14532. doi: 10.1073/pnas.0502089102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rizvi SA, Tereshko V, Kossiakoff AA, Kozmin SA. Structure of bistramide A-actin complex at a 1.35 angstroms resolution. J Am Chem Soc. 2006;128:3882–3883. doi: 10.1021/ja058319c. [DOI] [PubMed] [Google Scholar]
- 47.Kudryashov DS, Sawaya MR, Adisetiyo H, Norcross T, Hegyi G, Reisler E, Yeates TO. The crystal structure of a cross-linked actin dimer suggests a detailed molecular interface in F-actin. Proc Natl Acad Sci U S A. 2005;102:13105–13110. doi: 10.1073/pnas.0506429102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- 49.Klenchin VA, Khaitlina SY, Rayment I. Crystal structure of polymerization-competent actin. J Mol Biol. 2006;362:140–150. doi: 10.1016/j.jmb.2006.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.