Abstract
Tyrosine nitration in proteins is an important post-translational modification (PTM) linked to various pathological conditions. When multiple potential sites of nitration exist, tandem mass spectrometry (MS/MS) methods provide unique tools to locate the nitro-tyrosine(s) precisely. Electron capture dissociation (ECD) is a powerful MS/MS method, different in its mechanisms to the “slow-heating” threshold fragmentation methods, such as collision-induced dissociation (CID) and infrared multiphoton dissociation (IRMPD). Generally, ECD provides more homogeneous cleavage of the protein backbone and preserves labile PTMs. However recent studies in our laboratory demonstrated that ECD of doubly charged nitrated peptides is inhibited by the large electron affinity of the nitro group, while CID efficiency remains unaffected by nitration. Here, we have investigated the efficiency of ECD versus CID and IRMPD for top-down MS/MS analysis of multiply charged intact nitrated protein ions of myoglobin, lysozyme, and cytochrome c in a commercial Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer. CID and IRMPD produced more cleavages in the vicinity of the sites of nitration than ECD. However the total number of ECD fragments was greater than those from CID or IRMPD, and many ECD fragments contained the site(s) of nitration. We conclude that ECD can be used in the top-down analysis of nitrated proteins, but precise localization of the sites of nitration may require either of the “slow-heating” methods.
The significance of tyrosine nitration in proteins has been recognized in a growing number of publications over the past decade.(1) This post-translational modification (PTM) is one of several occurring during oxidative stress caused by radical species.2,3 It has been linked to such pathological conditions as Alzheimer’s disease,(4) cardiovascular disease,(5) and atherothrombotic diseases.(6)In vivo, the main nitrating agent for this modification is thought to be the peroxynitrite anion (OONO−), formed in the reaction of the superoxide anion (O2−•) with nitric oxide (NO•). The product of tyrosine nitration is 3-nitrotyrosine with the NO2 group in the ortho-position to the phenol. Other nitrating agents can also react with tyrosine and its nitration is regarded as a marker for general nitrative stress.(1) Importantly, tyrosine nitration is a selective process occurring only at specific tyrosine residues.1,7,8 Therefore, a full description of the biological processes involving nitrotyrosine requires precise knowledge of the nitration site(s) in the protein(s).
Mass spectrometry (MS) provides reliable tools for both identification of nitrated proteins and high-resolution identification of PTM sites in the protein. A combination of ESI with online separation of proteins by liquid chromatography (LC−MS) has been used to identify 3-nitrotyrosine residues in complex protein mixtures.8−12 While mass shifts of the peptide fingerprints can be used to identify the nitration in protein segments, only tandem MS/MS experiments are able to provide precise localization of PTMs sites on the protein backbone. The mainstream approach for locating the sites of nitration is to obtain mass fingerprints of the peptides from a proteolytic digest of a complex protein mixture or an individual nitrated protein and further analyze the peptides by fragmentation MS/MS methods, such as collision induced dissociation (CID).11,12 However, MS/MS analysis of protein digests (the “bottom-up” approach) allows the possibility that peptides with PTMs may be missed due to poor or insufficient separation on the HPLC column or poor ionization efficiency. The alternative approach is “top-down” protein characterization, in which intact proteins are ionized and subsequently analyzed in a mass spectrometer.13−17 The complexity of the resulting mass spectra requires a high-resolution technique, such as Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometry. The key advantage of the top-down approach is that the measured mass of an intact protein may be compared with the mass calculated from the known protein sequence, thus providing information about all the modifications present in the protein and retaining connectivity between the modifications. In the case of multiple modifications, several mass peaks related to protein isoforms may be present in the mass spectra, and their relative intensities provide information on the extent of each of the modifications. MS/MS fragments of intact proteins can be analyzed to localize the sites of modifications in a similar way to that for peptides from protein digests. The top-down method has been demonstrated on proteins up to ∼200 kDa.(17) The downside of this method is that the time of MS/MS analysis is longer than in the bottom-up approach, due to a much larger number of fragmentation channels.
Several MS/MS fragmentation techniques are available for both bottom-up and top-down approaches. “Slow-heating” methods,(18) such as CID or infrared multiphoton dissociation (IRMPD),19,20 utilize different techniques to excite and dissociate molecular ions thermally. CID and IRMPD produce heterolytic cleavages of the amide bonds in the polypeptide chain giving rise to b and y fragment ions that contain the N- or C-terminus, respectively.(21) As thermal methods, they cleave the weakest bonds first. Since its introduction in 1998, electron capture dissociation(22) (ECD) and its sister method, electron transfer dissociation (ETD),(23) have also become popular in MS/MS studies. ECD is a fast fragmentation technique whereby cleavage of the peptide backbone occurs following low-energy (<0.2 eV) electron capture and the subsequent cascade of intramolecular radical-driven reactions, the precise pathways of which are still of some discussion.24−27 ECD cleavage of the N−Cα bonds occurs producing mainly N-terminus c′ and C-terminus z• (or c• and z′) fragment ions. One of the advantages of ECD over the thermal methods is that it provides a more uniform pattern of cleavages along the backbone, with the only exception being cleavage of the N-terminal to proline(28) and thus leads to greater peptide sequence coverage.29,30 Unlike the thermal methods, disulfide bridges are efficiently cleaved by ECD of peptides,(24) and following capture of a second electron, fragments from the peptide segment inside the “disulfide loop” can be produced. Furthermore, ECD fragments retain labile post-translational modifications,(31) while CID and IRMPD tend to cleave them. Examples of the efficient use of ECD for localizing PTM sites include phosphorylation,32,33 N- and O-glycosylation,34,35 ubiquitination,(36) sumoylation,(37) and others. Nevertheless, there have been observations that ECD is not universally efficient for all possible peptide modifications. We have recently demonstrated that addition of nitration to tyrosine severely inhibits the production of ECD sequence fragments in peptides.(38) A similar effect was reported by the Beauchamp group for benzyl modifications of cysteine which have an electron affinity (EA) of ≥1.00 eV.(39) Specifically, 3-nitrobenzylcysteine (EA = 1.00 eV) and 3,5-dinitrobenzylcysteine (EA = 1.65 eV), termed “electron predators”, inhibit peptide backbone cleavage by ECD and the related electron transfer dissociation (ETD) completely.(39) Apparently 3-nitrotyrosine, structurally similar to nitrobenzylcysteine, was also acting as an “electron predator” in our ECD experiments. However, we demonstrated that ECD of the triply charged nitrated peptides resulted in some singly charged sequence fragments, which may be the products of secondary electron capture.(38) That result indicated that top-down ECD of intact nitrated proteins may be efficient, as multiple electron capture by multiply charged protein ions usually occurs,24,30 the hypothesis which we put to test in this work.
In this study we optimize and compare top-down ECD, CID, and IRMPD of nitrated proteins: myoglobin, cytochrome c, and hen egg white lysozyme (HEWL). Our choice of proteins was due to the different behaviors of their un-nitrated forms under ECD. Previously, we have shown that z > 14+ cations of unmodified myoglobin fragment extensively under ECD in our instrument.(40) ECD of unmodified cytochrome c does not produce fragments from the vicinity of Cys14 and Cys17, where the heme group is covalently attached to the protein.40,41 Native lysozyme has four disulfide bonds, which have to be cleaved by ECD first in order to produce backbone fragments from the interior of the molecule, i.e., multiple electron capture is required. Thus nitrated myoglobin represents a system where ECD can be affected only by the presence of the nitrated tyrosine, while native lysozyme and cytochrome c represent systems where ECD efficiency can be affected by other modifications in addition to nitration. For comparison, ECD, CID, and IRMPD of reduced and alkylated nitrated lysozyme were also carried out. Finally, we compare our results for top-down ECD and IRMPD of these nitrated proteins with our recent results on ECD of peptides containing 3-nitrotyrosine.(38)
Experimental Section
Materials
In this work we used methanol (Fisher Scientific, Leicestershire, U.K.), water (J. T. Baker, Deventer, The Netherlands), formic acid (Fisher Scientific), nitric acid, disodium tetraborate, boric acid, sodium nitrite, sodium phosphate, sodium chloride, ammonium acetate, and ammonium bicarbonate (Fisher Scientific), dithiothreitol (DTT), and iodoacetamide (IAA) (Sigma-Aldrich, Poole, Dorset, U.K.). Equine skeletal myoglobin, chicken egg lysozyme, and cytochrome c (horse heart) were purchased from Sigma-Aldrich and used without further purification.
Preparation of Nitrated Proteins for MS/MS Analysis
Our aim was to prepare nitro-proteins in the state(s) of nitration found in vivo. Electrosynthetic modification of proteins has been shown, under various conditions, to oxidize specific amino acid residues including tyrosine.42−45 Although in vitro chemical nitration may perhaps provide larger total yields of modified proteins, it has been observed to be less selective and specific than electrochemical nitration. For example, in vitro nitration of lysozyme and cytochrome c using peroxynitrite46,47 results in combinations of mono-, bis-, and tris-nitration, among other PTM modifications, and complex separation of products is necessary. Furthermore, analysis of tryptic hydrolysates of lysozyme nitrated in vitro by myeloperoxidase revealed that Tyr23/Trp28 were modified to a higher extent than Tyr20 (specific site of nitration) and additionally that Trp62/Trp63 were oxidized and nitrated.(46) The same residues were also the main targets of peroxynitrite, which also hydroxylated Trp108/Trp111.
Detailed information on our method of electrochemical nitration of myoglobin and lysozyme and the protocol for reduction/alkylation of disulfide bonds in lysozyme have been described previously.38,42−44 Briefly, a water-cooled electrochemical cell with platinum electrode (lysozyme and cytochrome c) or boron-doped diamond electrode (lysozyme and myoglobin) was used. After the reaction was stopped, myoglobin samples were extensively dialyzed.(38) We estimate that protein loss during dialysis is <10%. No further LC separation of nitrated proteins from the unmodified myoglobin was carried out. Samples were subsequently freeze-dried and stored at −20 °C for later use. The reaction products from the electrooxidative nitration of lysozyme were separated by fast protein liquid chromatography (LC), and reduction/alkylation of the disulfide bonds was carried out after the LC separation as described previously.43,44 The electrochemical nitration of cytochrome c was carried out following the same procedure as for lysozyme, but the protein solution was ∼2.5 less concentrated than those of myoglobin and lysozyme. The reaction products from cytochrome c nitration were LC separated using the same mobile phases and gradients as in the case of lysozyme, but the column used was a Hitrap SP sepharose Fast Flow, 5 mL × 5 mL, from GE Heathcare. After LC separation, the fraction containing proteins was extensively dialyzed against 10 mM ammonium acetate (pH = 5.8) and finally concentrated, as described previously for lysozyme.43,44
Mass Spectrometry
All tandem mass spectrometry analysis was performed on a Thermo Finnigan LTQ FT mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Protein samples were buffer exchanged in 49.5/49.5% of H2O/CH3OH and 1% of formic acid using an Amicon centrifugal filter device, 3 or 10 kDa cutoff (Millipore). The final concentration was 1−3 μM for myoglobin and lysozyme, for cytochrome c it was below 1 μM. Samples were directly injected into the LTQ FT by use of an Advion Biosciences Triversa Nanomate electrospray source (Advion Biosciences, Ithaca, New York) at a flow rate of 200 nL min−1. All MS and MS/MS spectra were acquired in the ICR cell with a resolution of 100 000 at m/z 400. In total, 150−250 microscans (transients) were averaged for each fragmentation spectrum. Precursor ions (single charge state) were isolated in the linear ion trap (LTQ). The automatic gain control (AGC) target was 2 −10 × 106 ion counts with maximum fill time of 2 s. The isolation width for the ions from the samples of unmodified proteins and fractions of mono- and bis-nitrated lysozyme was 20−50 Th (single step isolation). Isolation of the ions of nitrated myoglobin and cytochrome c was achieved in two or three steps in order to exclude the ions of unmodified protein present in the sample. The final isolation width was 10−15 Th. Narrowing the isolation width below these values lead to a serious depletion of the isolated ions and, thus, a much smaller number of fragments detected. However, we used narrower five-step isolation with a final width of 5−10 Th for investigation of neutral losses from nitrated protein ions upon ECD. CID was carried out in the LTQ with a normalized collision energy of 9−20%, duration of 30 ms, and Q = 0.25. Electrons for ECD were produced by an indirectly heated barium−tungsten cylindrical dispenser cathode (5.1 mm diameter, 154 mm from the cell, 1 mm off axis) (Heat-Wave Laboratories, Watsonville, California). The current across the electrode was ∼1.1 A. The ECD duration was 5−10 ms, and the ECD energy was in the range of 2.5−4% (corresponding to a cathode potential of −0.275 to −1.775 V). IRMPD of the protein ions was carried out in the ICR cell using a 75 W in-built CO2 laser (Synrad, Mikilteco, Washington) for 100 ms. IRMPD energy was measured as a percent of the maximum (i.e., 75 W). Raw MS data were analyzed by use of Xcalibur 2.05 software (Thermo Fisher Scientific), where the Xtract program was used for calculating monoisotopic masses (44% fit factor, 25% remainder). ProSight PTM (https://prosightptm.scs.uiuc.edu) was used to search for b, y and c, z protein fragment ions in the single protein mode. The mass accuracy for the search was set at 10 ppm.
Results
Nitrated Myoglobin
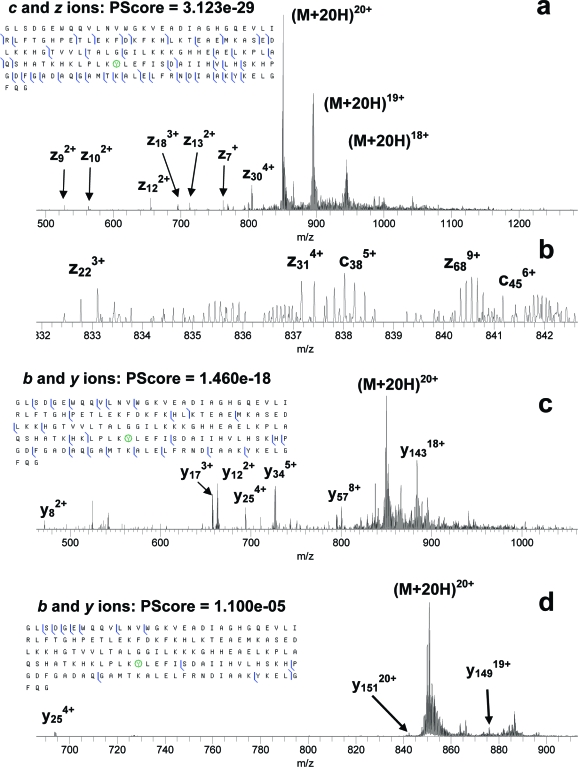
Electrospray mass spectrometry of the nitrated myoglobin indicated that both unmodified and mononitrated proteins with relative ratio of 1:1.25 were present in the sample. Nitration of myoglobin was expected to take place at Tyr103.(42) MS/MS of nitrated myoglobin ions was carried out for five charge states: 14+, 16+, 18+, 20+, and 22+, Table 1. For each charge state the total number of fragments from ECD was greater than that from IRMPD or CID. For each MS/MS method, the number of fragments varied with the charge state but not monotonically. MS/MS spectra and fragmentation diagrams for the 20+ ions of nitrated myoglobin are given in Figure 1. MS/MS fragmentation diagrams for the 20+ ions of unmodified myoglobin are given in Figure S-1 in the Supporting Information. ECD of the unmodified myoglobin produced extensive cleavages, while IRMPD and CID resulted in smaller numbers of fragments. CID of unmodified myoglobin produced more fragments than IRMPD (35 versus 26). Nitration resulted in a reduction of the total number of ECD fragments (from 84 to 45), and ECD cleavage sites were located at least 5 residues C-terminal and 11 N-terminal to the site of nitration, Tyr103, versus two and five residues, respectively, in the unmodified protein, Figure 1a. A decrease in the number of ECD cleavages around Tyr103 upon nitration was observed for all charge states. IRMPD of the nitrated myoglobin produced cleavages of the protein backbone closer to the site of nitration than ECD, Figure 1c. IRMPD produces more fragments than CID from the nitrated protein (25 vs 12), Figure 1c,d. Nitration does not seem to have any serious effect on the efficiency of IRMPD (26 and 25 IRMPD fragments produced from unmodified and nitrated myoglobin, respectively). The number of CID fragments reduced significantly after nitration (35 and 12 fragments before and after nitration, respectively). The reason for that appears to be of the instrumental nature (see the discussion below). Additionally, our ECD, CID, and IRMPD results all confirm unequivocally that Tyr103, not Tyr146, is the site of nitration. However, fewer fragments confirming nitration on Tyr103 were produced by CID of the 20+ ions than by ECD or IRMPD. The same was true for the other charge states.
Table 1. Numbers of MS/MS Fragments from ECD, IRMPD, and CID of Nitrated Myoglobina.
| ECD |
IRMPD |
CID |
||||
|---|---|---|---|---|---|---|
| charge state z | total no. of fragments | no. of fragments containing nY103 | total no. of fragments | no. of fragments containing nY103 | total no. of fragments | no. of fragments containing nY103 |
| 22+ | 33 | 6 | 17 | 9 | 18 | 6 |
| 20+ | 45 | 10 | 25 | 12 | 12 | 8 |
| 18+ | 36 | 5 | 16 | 7 | 12 | 4 |
| 16+ | 33 | 10 | 23 | 7 | 8 | 5 |
| 14+ | 47 | 16 | 22 | 8 | 19 | 11 |
nY103 indicates nitrotyrosine.
Figure 1.
ECD (a, b), IRMPD (c), and CID (d) MS/MS of mononitrated myoglobin. Nitrated tyrosine residues are circled in the fragment summaries.
Nitrated Lysozyme
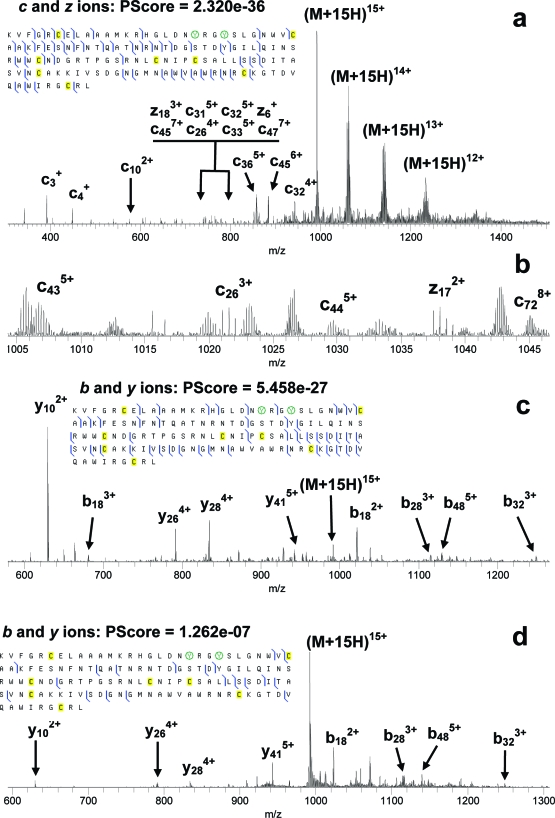
Electrospray mass spectrometry of the two LC fractions from lysozyme nitration indicated that one of them contained mostly mononitrated protein and the other mostly bis-nitrated protein. MS/MS of unmodified, mono, and bis-nitrated lysozyme was carried out for both nonreduced and reduced protein samples. Initial nitration of lysozyme was expected to take place at Tyr23 followed by nitration of Tyr20.43,44 MS/MS spectra and fragmentation diagrams for the most abundant charge state, 15+, of reduced bis-nitrated lysozyme are presented in Figure 2. Summaries of fragments from ECD MS/MS of the 10+ ions of nonreduced lysozyme are presented in Figure S-2 in the Supporting Information. Higher charge states of nonreduced lysozyme could not be obtained with abundances sufficient for MS/MS investigation. MS/MS spectra and fragmentation diagrams for the 15+ charge state of reduced mononitrated lysozyme are presented in Figures S-3−S-5 in the Supporting Information.
Figure 2.
ECD (a, b), IRMPD (c), and CID (d) MS/MS of bis-nitrated lysozyme. Nitrated tyrosine residues are circled.
Only small numbers of fragments could be obtained from ECD of nonreduced lysozyme, Figure S-2 in the Supporting Information. As the cleavage of a disulfide bond by ECD results in one of the two cysteines reduced by the recombined proton,(24) the search for ECD fragments from the nonreduced lysozyme was carried out assuming a −1 Da mass shift on one of the cysteines from each of the four S−S bonds, no mass shift on the other cysteine, and vice versa. However, the fragments found indicate that only the Cys6−Cys127 disulfide bond was cleaved by ECD, with Cys6 being reduced. The other disulfide bonds appear not to be cleaved by ECD. A total of 13 c′ fragments between amino acid residues 3 and 20 and only two z• fragments close to the C-terminus were produced from unmodified native lysozyme, Figure S-2a in the Supporting Information. ECD of mono- or bis-nitrated nonreduced lysozyme produces fewer fragments than ECD of unmodified nonreduced lysozyme, Figure S-2b,c in the Supporting Information. Furthermore, ECD does not cleave beyond amino acid residue 13. Thus ECD cleavages do not reach the site(s) of nitration, and none of the ECD fragments contains the site(s) of nitration. Use of activated ion (AI) ECD, as described in our recent publication,(40) did not lead to the appearance of new ECD fragments, although depletion of the charge-reduced ions was observed, and the intensities of the ECD fragments increased in comparison with the standard ECD. CID and IRMPD of nonreduced nitrated lysozyme produced only a few fragments, as they could not cleave the disulfide bonds (data not shown). Lower charge states, z < 10, of nonreduced lysozyme produced even fewer MS/MS fragments than the 10+ state.
As expected, reduction/alkylation of the disulfide bonds provided a remarkable improvement in the sequence coverage by MS/MS. For bis-nitrated lysozyme, the total number of fragment ions was 56, 47, and 21 from ECD, IRMPD, and CID, respectively, Figure 2. For mononitrated lysozyme, the total number of fragments was 59, 49, and 25 from ECD, IRMPD, and CID, respectively, Figures S-3−S-5 in the Supporting Information. In the summary of MS/MS fragments from mononitrated lysozyme (Figures S-3−S-5 in the Supporting Information), the nitration site is assumed to be at Tyr23. However neither ECD, nor IRMPD, nor CID data exclude the possibility that mono-nitration occurs at Tyr20. The number of MS/MS fragments containing the site of nitration in mononitrated lysozyme is 26, 14, and 9 for ECD, IRMPD, and CID, respectively. The number of fragments containing the sites of nitration in bis-nitrated lysozyme is 23, 13, and 8 for ECD, IRMPD, and CID, respectively. Thus, ECD produced the largest number of nTyr-containing fragments in both cases. ECD cleaved the protein backbone two and three positions C-terminal of Tyr23 in mono- and bis-nitrated lysozyme, respectively. However no ECD cleavages closer than eight positions N-terminal of Tyr23 were produced. In contrast to that, IRMPD cleaves much closer to the nitration site(s) and, furthermore, cleaves between Tyr20 and Tyr23 in bis-nitrated lysozyme, Figure 2c. CID of the nitrated lysozyme is the least efficient of the three MS/MS techniques, as discussed below.
Nitrated Cytochrome c
Unmodified cytochrome c ions with a charge state z ≥ 9+ produce fragments extensively under ECD in our instrument with the exception of the region around the covalent heme group attachment to the protein backbone.(40) Previously we also pointed out the importance of searching for c• and z′ ion fragments in ECD spectra of cytochrome c, as for some charge states of cytochrome c their number exceeds that of c′ and z• fragments.(40)
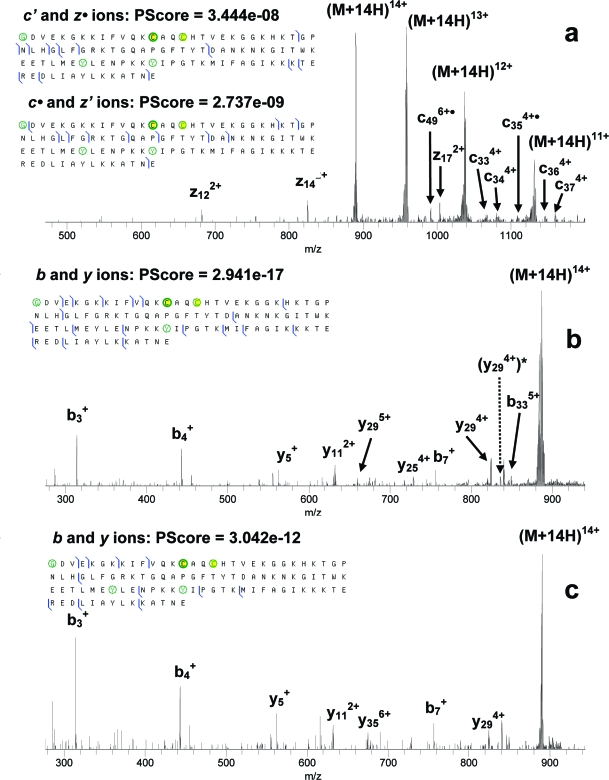
Nitration of cytochrome c was not as efficient as in the cases of myoglobin and lysozyme. Electrospray mass spectra of the nitrated cytochrome c samples were dominated by the unmodified protein ions with small peaks corresponding to mono- and bis-nitrated protein (∼20 and 15% of the abundance of the native protein ions, respectively). LC separation of the sample containing nitrated cytochrome c using a strong cation exchange column was not successful. Thus a small isolation window of 10 Th was used in order to isolate the ions of mononitrated protein from the ions of both unmodified and bis-nitrated protein for MS/MS investigation. Such a narrow isolation led to a reduction in the number of trapped ions and adversely affected the MS/MS analysis, i.e., the majority of ECD fragments from the mononitrated protein were not observed with intensities detectable above the noise level. ECD of bis-nitrated cytochrome c was more efficient, as a larger isolation m/z width could be used. Additionally, in all the MS/MS experiments on cytochrome c the most abundant charge state, 14+, was used in order to maximize the number of fragments. MS/MS spectra and summaries of fragments from nitrated cytochrome c are given in Figure 3. Although several ECD fragments, including c• and z′-type ions, containing the possible sites of nitration were found for bis-nitrated cytochrome c, the total sequence coverage was still poor in comparison with myoglobin and reduced lysozyme ECD data. CID of mono- and bis-nitrated cytochrome c was not efficient and produced only a few fragments at N-terminus (data not shown). IRMPD of mononitrated cytochrome c was more efficient than ECD and produced several backbone cleavages around the possible sites of nitration, Figure 3b. Both the ECD and IRMPD data allowed localization of the site(s) of nitration in the protein, and a comparison could be made with other methods of nitration. Cytochrome c has four tyrosine residues: Tyr48, Tyr67, Tyr74, and Tyr97. Tyr74 and Tyr97 are solvent-exposed, and the mononitration was expected to take place at either of them, as previously observed with peroxynitrite as a nitrating agent.45,46 Tyr48 and Tyr67 are less exposed, but Tyr67 is located close to Tyr 74 and can be a target for secondary nitration. Batthyany et al.(48) observed nitration in bis-nitrated cytochrome c at Tyr74 and Tyr67 or Tyr97 and Tyr67. Interestingly, the authors did not report simultaneous nitration at Tyr74 and Tyr97.
Figure 3.
ECD (a) and IRMPD (b, c) of mono- (b) and bis-nitrated (a, c) cytochrome c. N-acetylated glycine, nitrated tyrosines, and cysteines bound to the heme group are circled.
In our MS/MS experiments, IRMPD produced six and four C-terminal fragments between Tyr74 and Tyr97 in mono- and bis-nitrated protein, respectively, which did not contain the nitro group. Four fragments, again without the nitro group, were produced from the same region by ECD of bis-nitrated cytochrome c. Only one C-terminal IRMPD fragment containing nTyr97 was observed from mononitrated protein ((y294+)* in Figure 3b). Furthermore, no fragments containing nTyr48 were observed for ECD or IRMPD, and three unmodified N-terminal ECD fragments between Tyr48 and Tyr67 indicated the absence of nitration at Tyr48. Thus, while there is some evidence for initial nitration of Tyr97, the majority of the data suggests that initial nitration occurs at Tyr74. As Tyr48 and Tyr97 appear to be less probable sites of nitration, the site of the secondary nitration is suggested to be Tyr67.
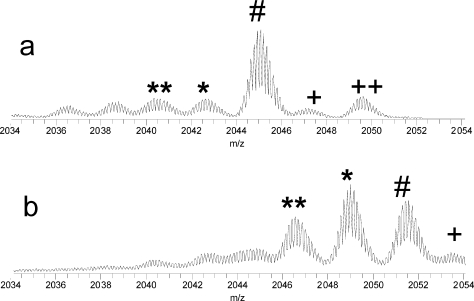
Recently we reported abundant losses of neutral species, including hydroxyl radicals, water, and ammonia, from nitrated peptides following ECD.(38) We have also recorded abundant losses of neutral species from ECD of the intact nitrated proteins. However, significant loss of neutrals even from the unmodified proteins was present under ECD, as in the case of lysozyme, Figure 4a. A significant increase in the relative abundances of the fragment ions resulting from neutral losses was observed upon nitration, Figure 4b, but the neutral losses due to the nitration could not be easily separated from the original neutral losses from the unmodified protein.
Figure 4.
Loss of neutrals from lysozyme ions under ECD. (a) Unmodified and (b) singly nitrated lysozyme. The # symbols label (M + 10H)7+ reduced ions, ∗ and ∗∗ label losses of 17 and 34 Da, respectively, and + and ++ label peaks of naturally oxidized protein.
Discussion
ECD was not completely inhibited in the vicinity of the site(s) of nitration: it produced cleavages at a distance of two and three residues from nTyr23 in mono- and bis-nitrated lysozyme, respectively, Figure 2. However, the total number of cleavages in the vicinity of the nitration site(s) is larger in the case of IRMPD for all three proteins investigated. Table 2 summarizes the numbers of cleavages produced by ECD or IRMPD within six residues N- or C-terminal to the site(s) of nitration. IRMPD not only produced more cleavages in that important region but also cleaved between the two nitro-tyrosines in bis-nitrated lysozyme and cytochrome c, while ECD did not, Figures 2 and 3. That result is consistent with the previous reports on suppression of ECD by nitration in doubly charged peptides and higher efficiency of the “slow-heating” MS/MS methods in such cases.38,39 However, in contrast to MS/MS of peptides, ECD provided the largest total number of fragments from intact mononitrated myoglobin, mono- and bis-nitrated reduced lysozyme, and bis-nitrated cytochrome c (including c• and z′ fragments).
Table 2. Number of Cleavages within 6 Amino Acid Residues Both N- and C-Terminal to the Site(s) of Nitration.
| ECD | IRMPD | |
|---|---|---|
| mononitrated myoglobin | 1 | 2 |
| mononitrated lysozyme | 2 | 4 |
| bis-nitrated lysozyme | 2 | 6 |
| mononitrated cytochrome c | n/a | 3 |
| bis-nitrated cytochrome c | 0 | 3 |
Sequence coverage by ECD was greater for nitrated myoglobin and reduced nitrated lysozyme than for nonreduced nitrated lysozyme and nitrated cytochrome c, where other internal modifications were present: disulfide bonds (lysozyme) and a heme group (cytochrome c). In the case of ECD of nonreduced nitrated lysozyme, the poor sequence coverage was due to the limited cleavage of the disulfide bonds by ECD. Cleavage of all four S−S bonds in lysozyme would require the capture of at least four electrons. We did record up to four consecutive electron capture events taking place for the 10+ charge state of nitrated lysozyme, which was indicated by the presence of (M + 10H)6+ charge-reduced ions in the ECD mass spectra. There is a possibility that both the disulfide bond and backbone cleavages occurred, but the resulting ECD fragments were held together by noncovalent interactions in the protein. However post-ECD IR activation, which we had previously used to release ECD fragments from the charge-reduced ions,(40) did not produce new fragments. Therefore we conclude that multiple disulfide bond cleavages were either not efficient or were not proceeded by efficient backbone cleavages.
ECD in cytochrome c was hindered by the presence of two nitro groups and one heme group. While nitro groups act as electron predators,38,39 the heme group is known to interfere with the normal course of events during ECD.(41) We also note that the amount of sample of nitrated cytochrome c was much smaller than that of nitrated myoglobin or lysozyme, because the reaction of nitration of cytochrome c was less efficient than for the other two proteins. Consequently, all three MS/MS methods were less efficient for cytochrome c than for myoglobin and reduced lysozyme.
CID produced more fragments than IRMPD from unmodified myoglobin (35 vs 26, Figure S-1 in the Supporting Information). For all the nitrated proteins studied, CID produced fewer fragments than IRMPD. The reasons for such behavior are not completely clear. Both MS/MS technique are “slow-heating” threshold fragmentation methods. IRMPD is often regarded as a more efficient fragmentation technique, as it acts upon not only the isolated precursor ions but also upon their primary fragments, thus producing additional smaller fragment ions. Contrary to that, the CID waveform is tuned to affect mostly the isolated precursor ions. Additionally, in our instrument CID is carried out in the linear ion trap and the fragments transported to and analyzed in the ICR cell, while IRMPD is carried out directly in the ICR cell. Transmission of CID fragments to the ICR cell could induce additional ion losses. Furthermore, in order to provide sufficient abundance of the fragment ions from top-down MS/MS analysis of intact proteins, one has to isolate and fragment a much larger number of precursor ions than in the case of bottom-up MS/MS analysis of peptides. This is due to a much larger number of dissociation channels in MS/MS of intact proteins. The number of precursor ions we typically isolate in the linear ion trap corresponds to an AGC value of 106−107 for intact proteins, while for peptide ions AGC values are 1−2 orders of magnitude smaller. The larger number of intact protein ions in the LTQ should cause stronger space-charge effects during CID. All CID measurements presented in this work were taken at the CID threshold energy. We observed that above the threshold to CID the precursor protein ions were quickly depleted but that did not lead to the production of new fragments. This may be an indication that many of the precursor ions were expelled from the LTQ during CID, rather than fragmented, or lost during transportation from the LTQ to the ICR cell for mass analysis. The effect was greater for CID of nitrated myoglobin and cytochrome c, where a two or three-step narrow isolation was used in order to isolate the nitrated protein ions from the remaining unmodified proteins. A plausible explanation for this CID behavior is that the space charge effects in the LTQ were aggravated by the narrow isolation and led to destabilization of the fragment ion trajectories during collisional excitation. Both ECD and IRMPD should suffer less from this effect, as they produce fragments inside the ICR cell and losses of the fragments there should be minimal.
Conclusions
Our previous studies demonstrated that the high electron affinity of the nitro group inhibits normal ECD process in nitrotyrosine-containing peptides.(38) Results from top-down MS/MS on tyrosine nitration in myoglobin, lysozyme, and cytochrome c presented in this work indicate that ECD can be used to study intact nitrated proteins. ECD provided the largest total number of fragments from the intact nitrated proteins among the three MS/MS methods tried: ECD, IRMPD, and CID. Although ECD produced fewer cleavages in the vicinity of the nitration site than IRMPD, there were sufficient numbers of ECD fragments which contained the site of nitration. However, if other modifications, such as multiple disulfide bonds or a heme group, are present, ECD of the intact protein could be further inhibited. The most appropriate approach for the top-down MS/MS analysis of intact nitrated proteins seems to be a combination of different methods of fragmentation following the reduction and alkylation of any disulfide bonds present.
Acknowledgments
The Wellcome Trust (Grants 074131 and 080998) (H.J.C. and V.A.M., respectively), the Ramon y Cajal Programme (Grant CTQ2006-14959), and the Ministerio de Ciencia y Tecnologia (Grant CTQ2007-62345), Spain (J.I.), are acknowledged for funding.
Supporting Information Available
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
Supplementary Material
References
- Abello N.; Kerstjens H. A. M.; Postma D. S.; Bischoff R. J. Proteome Res. 2009, 8, 3222–3238. [DOI] [PubMed] [Google Scholar]
- Ischiropoulos H. Arch. Biochem. Biophys. 1998, 356, 1–11. [DOI] [PubMed] [Google Scholar]
- Greenacre S. A.; Ischiropoulos H. Free Radical Res. 2001, 34, 541–581. [DOI] [PubMed] [Google Scholar]
- Good P. F.; Werner P.; Hsu A.; Olanow C. W.; Perl D. P. Am. J. Pathol. 1996, 149, 21–28. [PMC free article] [PubMed] [Google Scholar]
- Shishehbor M. H.; Aviles R. J.; Brennan M. L.; Fu X. M.; Goormastic M.; Pearce G. L.; Gokce N.; Keaney J. F.; Penn M. S.; Sprecher D. L.; Vita J. A.; Hazen S. L. JAMA, J. Am. Med. Assoc. 2003, 289, 1675–1680. [DOI] [PubMed] [Google Scholar]
- Parastatidis I.; Thomson L.; Burke A.; Chernysh I.; Nagaswami C.; Visser J.; Stamer S.; Liebler D. C.; Koliakos G.; Heijnen H. F. G.; Fitzgerald G. A.; Weisel J. W.; Ischiropoulos H. J. Biol. Chem. 2008, 283, 33846–33853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischiropoulos H. Biochem. Biophys. Res. Commun. 2003, 305, 776–83. [DOI] [PubMed] [Google Scholar]
- Jiao K.; Mandapati S.; Skipper P. L.; Tannenbaum S. R.; Wishnok J. S. Anal. Biochem. 2001, 293, 43–52. [DOI] [PubMed] [Google Scholar]
- Wong P. S.; van der Vliet A. Methods Enzymol. 2002, 359, 399–410. [DOI] [PubMed] [Google Scholar]
- Ghesquière B.; Colaert1 N.; Helsens K.; Dejager L.; Vanhaute C.; Verleysen K.; Kas K.; Timmerman E.; Goethals M.; Libert C.; Vandekerckhove J.; Gevaert K. Mol. Cell. Proteomics 2009, 8, 2642–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappetta G.; Corbo C.; Palmese A.; Marino G.; Amoresano A. Proteomics 2009, 9, 1524–1537. [DOI] [PubMed] [Google Scholar]
- Amoresano A.; Chiappetta G.; Pucci P.; D’Ischia M.; Marino G. Anal. Chem. 2007, 79, 2109–2117. [DOI] [PubMed] [Google Scholar]
- Kelleher N. L.; Lin H. Y.; Valaskovic G. A.; Aaserud D. J.; Fridriksson E. K.; McLafferty F. W. J. Am. Chem. Soc. 1999, 121, 806–812. [Google Scholar]
- Kelleher N. L. Anal. Chem. 2004, 76, 196A–203A. [PubMed] [Google Scholar]
- Bogdanov B.; Smith R. D. Mass Spectrom. Rev. 2005, 24, 168–200. [DOI] [PubMed] [Google Scholar]
- Parks B. A.; Jiang L.; Thomas P. M.; Wenger C. D.; Roth M. J.; Boyne M. T.; Burke P. V.; Kwast K. E.; Kelleher N. L. Anal. Chem. 2007, 79, 7984–7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X. M.; Jin M.; Breuker K.; McLafferty F. W. Science 2006, 314, 109–112. [DOI] [PubMed] [Google Scholar]
- McLuckey S. A.; Goeringer D. E. J. Mass Spectrom. 1997, 32, 461–474. [DOI] [PubMed] [Google Scholar]
- Little D. P.; Speir J. P.; Senko M. W.; O’Connor P. B.; McLafferty F. W. Anal. Chem. 1994, 66, 2809–2815. [DOI] [PubMed] [Google Scholar]
- Woodin R. L.; Bomse D. S.; Beauchamp J. L. J. Am. Chem. Soc. 1978, 100, 3248–3250. [Google Scholar]
- Roepstorff P.; Fohlman J. Biomed. Mass Spectrom. 1984, 11, 601–601. [DOI] [PubMed] [Google Scholar]
- Zubarev R. A.; Kelleher N. L.; McLafferty F. W. J. Am. Chem. Soc. 1998, 120, 3265–3266. [Google Scholar]
- Syka J. E. P.; Coon J. J.; Schroeder M. J.; Shabanowitz J.; Hunt D. F. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubarev R. A.; Kruger N. A.; Fridriksson E. K.; Lewis M. A.; Horn D. M.; Carpenter B. K.; McLafferty F. W. J. Am. Chem. Soc. 1999, 121, 2857–2862. [Google Scholar]
- Leymarie N.; Costello C. E.; O’Connor P. B. J. Am. Chem. Soc. 2003, 125, 8949–8958. [DOI] [PubMed] [Google Scholar]
- Syrstad E. A.; Turecek F. J. Am. Soc. Mass Spectrom. 2005, 16, 208–224. [DOI] [PubMed] [Google Scholar]
- Simons J. Chem. Phys. Let. 2010, 484, 81–95. [Google Scholar]
- Cooper H. J.; Hudgins R. R.; Hakansson K.; Marshall A. G. Int. J. Mass Spectrom. 2003, 228, 723–728. [Google Scholar]
- Axelsson J.; Palmblad M.; Hakansson K.; Hakansson P. Rapid Commun. Mass Spectrom. 1999, 13, 474–477. [DOI] [PubMed] [Google Scholar]
- Zubarev R. A.; Horn D. M.; Fridriksson E. K.; Kelleher N. L.; Kruger N. A.; Lewis M. A.; Carpenter B. K.; McLafferty F. W. Anal. Chem. 2000, 72, 563–573. [DOI] [PubMed] [Google Scholar]
- Kelleher R. L.; Zubarev R. A.; Bush K.; Furie B.; Furie B. C.; McLafferty F. W.; Walsh C. T. Anal. Chem. 1999, 71, 4250–4253. [DOI] [PubMed] [Google Scholar]
- Shi S. D. H.; Hemling M. E.; Carr S. A.; Horn D. M.; Lindh I.; McLafferty F. W. Anal. Chem. 2001, 73, 19–22. [DOI] [PubMed] [Google Scholar]
- Stensballe A.; Jensen O. N.; Olsen J. V.; Haselmann K. F.; Zubarev R. A. Rapid Commun. Mass Spectrom. 2000, 14, 1793–1800. [DOI] [PubMed] [Google Scholar]
- Hakansson K.; Cooper H. J.; Emmett M. R.; Costello C. E.; Marshall A. G.; Nilsson C. L. Anal. Chem. 2001, 73, 4530–4536. [DOI] [PubMed] [Google Scholar]
- Mirgorodskaya E.; Roepstorff P.; Zubarev R. A. Anal. Chem. 1999, 71, 4431–4436. [DOI] [PubMed] [Google Scholar]
- Cooper H. J.; Heath J. K.; Jaffray E.; Hay R. T.; Lam T. T.; Marshall A. G. Anal. Chem. 2004, 76, 6982–6988. [DOI] [PubMed] [Google Scholar]
- Cooper H. J.; Tatham M. H.; Jaffray E.; Heath J. K.; Lam T. T.; Marshall A. G.; Hay R. T. Anal. Chem. 2005, 77, 6310–6319. [DOI] [PubMed] [Google Scholar]
- Jones A. W.; Mikhailov V. A.; Iniesta J.; Cooper H. J. J. Am. Soc. Mass Spectrom. 2010, 21, 268–277. [DOI] [PubMed] [Google Scholar]
- Sohn C. H.; Chung C. K.; Yin S.; Ramachandran P.; Loo J. A.; Beauchamp J. L. J. Am. Chem. Soc. 2009, 131, 5444–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailov V. A.; Cooper H. J. J. Am. Soc. Mass Spectrom. 2009, 20, 763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn D. M.; Breuker K.; Frank A. J.; McLafferty F. W. J. Am. Chem. Soc. 2001, 123, 9792–9799. [DOI] [PubMed] [Google Scholar]
- Kendall G.; Cooper H. J.; Heptinstall J.; Derrick P. J.; Walton D. J.; Peterson I. R. Arch. Biochem. Biophys. 2001, 392, 169–179. [DOI] [PubMed] [Google Scholar]
- Matters D.; Cooper H. J.; McDonnell L.; Iniesta J.; Heptinstall J.; Derrick P.; Walton D.; Peterson I. Anal. Biochem. 2006, 356, 171–181. [DOI] [PubMed] [Google Scholar]
- Iniesta J.; Esclapez-Vicente M. D.; Heptinstall J.; Walton D. J.; Peterson I. R.; Mikhailov V. A.; Cooper H. J. Enzyme Microb. Technol. 2010, 46, 472–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniesta J.; Cooper H. J.; Marshall A. G.; Heptinstall J.; Walton D. J.; Peterson I. R. Arch. Biochem. Biophys. 2008, 474, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz S. M.; Prado F. M.; Di Mascio P.; Augusto O. Arch. Biochem. Biophys. 2009, 484, 127–133. [DOI] [PubMed] [Google Scholar]
- Abriata L. A.; Cassina A.; Tórtora V.; Marín M.; Souza J. M.; Castro L.; Vila A. J.; Radi R. J. Biol. Chem. 2009, 284, 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batthyany C.; Souza J. M.; Duran R.; Cassina A.; Cervenansky C.; Radi R. Biochemistry 2005, 44, 8038–8046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.