Abstract
The 21-day experimental gingivitis model, an established noninvasive model of inflammation in response to increasing bacterial accumulation in humans, is designed to enable the study of both the induction and resolution of inflammation. Here, we have analyzed gingival crevicular fluid, an oral fluid comprising a serum transudate and tissue exudates, by LC−MS/MS using Fourier transform ion cyclotron resonance mass spectrometry and iTRAQ isobaric mass tags, to establish meta-proteomic profiles of inflammation-induced changes in proteins in healthy young volunteers. Across the course of experimentally induced gingivitis, we identified 16 bacterial and 186 human proteins. Although abundances of the bacterial proteins identified did not vary temporally, Fusobacterium outer membrane proteins were detected. Fusobacterium species have previously been associated with periodontal health or disease. The human proteins identified spanned a wide range of compartments (both extracellular and intracellular) and functions, including serum proteins, proteins displaying antibacterial properties, and proteins with functions associated with cellular transcription, DNA binding, the cytoskeleton, cell adhesion, and cilia. PolySNAP3 clustering software was used in a multilayered analytical approach. Clusters of proteins that associated with changes to the clinical parameters included neuronal and synapse associated proteins.
Keywords: FTICR−MS, FT-ICR, FT-MS, Fourier transform ion cyclotron mass spectrometry, proteomics, gingivitis, inflammation, iTRAQ, cluster analysis, cilia, ribbon synapses
Short abstract
An established 21-day experimental gingivitis model was adopted to enable the study of the induction and resolution of inflammation. Gingival crevicular fluid, an oral fluid comprising a serum transudate and tissue exudates, was analyzed by LC−MS/MS using Fourier transform ion cyclotron resonance mass spectrometry and iTRAQ isobaric mass tags, in order to establish meta-proteomic profiles of inflammation-induced changes in proteins in healthy young volunteers.
Introduction
The 21-day experimental gingivitis model is an established noninvasive model in humans for investigating the induction and resolution of inflammation in response to increasing bacterial accumulation. From its inception in 1965,1,2 this model of inflammation has been used to assess how drugs,(3) bioactive compounds in dentifrices4−6 and environmental toxins, such as tobacco smoke,7,8 affect the development of inflammation. The model is designed to enable the study of both the induction and resolution of inflammation, which can also be assessed over a relatively short period of time (35 days) and in a controlled manner.
The model involves periodontally and systemically healthy volunteers, who do not brush specified test teeth in one sextant of the dentition, but continue to brush the remaining control teeth over a 21 day period. Accidental brushing of test teeth is prevented by the use of a vinyl shield (mouth guard) to cover them during cleaning. On day 21, the accumulated plaque on all teeth is removed by a trained hygienist and study participants resume their normal oral hygiene practices. Clinical assessment of plaque accumulation and gingival inflammation are made at baseline, day 7, 14, 21 on test and control teeth, following plaque accumulation on test teeth, and normal plaque removal from control teeth. Assessments are repeated again on day 35, following 14 days of resolution of inflammation at test sites, and maintenance of health at control sites. When performed by trained and calibrated experts, these measures show consistent increases in plaque-induced inflammation up to day 21 followed by a return to baseline, preinflammation levels at day 35.9,10 However none of these assessments could potentially form the basis for a predictor of disease as they are all measures of existing inflammation or inducers of inflammation.
A further and more objective assessment that can be made during the experimental gingivitis is that of the volume and composition of gingival crevicular fluid (GCF) and at individual sites around individual teeth. This fluid flows from the crevice between the tooth and the gum and comprises both a serum transdate(11) and tissue exudates.3,10,12−14 During inflammation, the volume and flow of this fluid increases,(1) and this alone can be used to demonstrate induction and/or presence of gingivitis and periodontitis. However, due to its very nature, as a mixed serum transudate and tissue exudate, GCF carries with it proteins from the crevice. Thus its composition also provides a biological and pathological fingerprint of the various physiological and biochemical processes occurring within the gingival tissues. In 1980, Novaes et al.(15) demonstrated that the total protein present in GCF increased with increasing severity of periodontitis, although this was probably due to increases in GCF volume. By 1985, Lamster et al.(16) had measured differences in lactate dehydrogenase, beta-glucuronidase and arylsulfatase activity in GCF collected from experimental gingivitis sites and demonstrated increases in these biomarkers over the course of the 21 day study. Since then, studies have targeted particular cytokines,3,10,12−14,17,18 antibacterial peptides and proteins(19) and many more individual proteins and peptides,9,20−22 revealing increases in pro-inflammatory species during disease. Studies have also examined the effect of diseases, such as type I diabetes mellitus,(23) analysis of many anti-inflammatory or antimicrobial compounds, and environmental conditions, such as smoking,(8) on experimental gingivitis.
Recently, proteomics techniques have greatly increased understanding of the composition of GCF. Surface enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI TOF MS) has been used to examine small proteins and peptides (2.5−30 kDa) from healthy volunteers and gingivitis patients.(24) That study employed a targeted approach to investigate neutrophil defensins. Unfractionated GCF samples were also examined by matrix assisted laser desorption/ionization (MALDI)-TOF MS, for neutrophil defensins.(25) Similarly, Pisano et al(26) used electrospray ionization(27) (ESI)−MS to examine the acid-soluble protein content of GCF from healthy volunteers. They, too, identified the neutrophil defensins, along with cystatin A, statherin and serum albumin. Only in the last year have tandem mass spectrometry (MS/MS) techniques been employed in the examination of GCF. Ngo et al.(28) used both MALDI-TOF/TOF and ESI−MS/MS to identify 66 proteins that had been separated by gel electrophoresis from a single patient, who had a history of periodontal disease but who was in the maintenance phase following treatment. Proteins identified included a number of serum-derived proteins, such as albumin, complement and transferrin. Other proteins that were present included: antimicrobial proteins calgranulin A and B; amylase; cytatin S; and Histone H4. Bostanci et al.(29) used a LC−MSE label-free quantitative technique to identify 154 proteins from either healthy volunteers or patients with aggressive periodontitis. Bostanci et al.(29) identified a wide range of proteins including those mentioned already and a large number of keratins, immunoglobulins, and other intracellular proteins. They also potentially identified a number of bacterial, fungal and viral proteins.
The majority of mass spectrometry-based proteomics is performed using the bottom-up(30) liquid chromatography tandem mass spectrometry (LC−MS/MS) approach. Generally, proteins are digested with trypsin prior to online LC separation. As the peptides elute into the mass spectrometer, they are fragmented typically by collision induced dissociation(31) (CID) resulting in b and y ion series.(32) The MS/MS spectra are searched against protein databases by use of algorithms (e.g., Mascot,(33) SEQUEST,(34) OMSSA(35)) which match the data to theoretical spectra from in-silico digests of proteins. Quantitative proteomics by mass spectrometry can use several methods. Two of the most popular are stable isotope labeled amino acids in cell culture(36) (SILAC) and isobaric tags for relative and absolute quantitation (iTRAQ).(37) In SILAC, cells are grown in media lacking essential amino acids. The media is supplemented with heavy or light amino acids. The labeled and unlabeled peptides elute together and the intensity of the peptide ions can be compared. SILAC is not suitable for clinical samples. A common approach for quantitation of biological fluids(38) and tissue samples(39) is postdigestion labeling with iTRAQ(40) labels. iTRAQ labels consist of a reporter ion with a m/z value of between 113 and 121 and a balance mass (191−183 Da) such that all labeled peptides of the same sequence have the same nominal mass shift (Δ304 Da). Test and control samples are treated with separate iTRAQ labels postdigestion and combined prior to MS analysis. When the peptide ions are fragmented with CID, the reporter ions are cleaved from the peptide and detected in the mass spectrometer. The intensity of the reporter ions can then be quantified. Quantitation with iTRAQ, allows for the analysis up to eight different treatment groups(41) simultaneously. One of the disadvantages of performing CID in ion traps is the instability of ions with m/z values approximately 1/3 that of the parent ion,(42) that is, fragment ions with m/z less than 1/3 of the precursor ions are not detected. iTRAQ reporter ions have m/z 113−121 and therefore ion trap CID of iTRAQ labeled peptides often does not result in quantification. The recent introduction of pulsed-Q dissociation(43) (PQD) has allowed for the analysis of iTRAQ labeled samples in linear ion trap mass analysers. PQD excites ions with a high amplitude Q value; Q is a resonance excitation pulse. When the ions are excited, they collide with neutral gas molecules in a similar manner to CID. The activation profile applied in PQD is different to that of CID meaning that low mass ions are stable and can be detected.
To date, the proteomic profile of GCF during the active induction and subsequent resolution of inflammation under controlled conditions using the 21 day experimental gingivitis model has not been described. Herein we describe the quantitative analysis of GCF, collected from volunteers undergoing experimental gingivitis, by LC−MS/MS using Fourier transform ion cyclotron resonance(44) (FT-ICR) MS and iTRAQ isobaric mass tags, to establish a profile of changes in proteins in healthy young volunteers that may be used to compare to the inflammatory response in other subsets of the population, such as those predisposed to periodontitis.
Materials and Methods
Subjects and Collection
The experimental gingivitis model previously described by Chapple et al.(9) was used, and the study was approved by the South Birmingham Local Research Ethical Committee (LREC 2004/074). Ten nonsmoking volunteers (mean age 21 years; range 19−28, 4 males and 6 females) who had unremarkable medical histories, no periodontal problems (past or present), were not undergoing orthodontic or prosthetic appliance therapy or taking medication that may have affected results were enrolled into the study, and informed consent was obtained. A split mouth design was employed with test sites being the maxillary left 4, 5, and 6 (first and second premolars and first molar on the left-hand side of the upper jaw) in right-handed individuals (n = 9) and the equivalent teeth on the maxillary right side in left-handed individuals (n = 1). Control sites were the corresponding teeth on the contralateral side. Plaque accumulation was assessed using a modified Quigley-Hein index(45) (PI; scores 0−5) and the gingival index (GI) of Löe(46) (scores 0−3) was used to measure gingival inflammation. Bleeding was measured dichotomously as presence or absence following gentle probing and expressed as the percentage of sites that bled upon probing. All volunteers had no evidence of attachment loss (destruction or loss of the connective tissues and bone holding the teeth within their sockets) when admitted to the study. A soft vinyl splint was constructed to cover five teeth, with a 5 mm clearance at each free surface from the marginal tissues. The splint was inserted gently over test teeth before brushing, to ensure against any mechanical or chemical cleaning. The test and control teeth were given a thorough prophylaxis and oral hygiene was closely monitored for 2 weeks prior to the study commencing to ensure pristine gingival health at baseline (see Introduction and Figure 1). GCF samples were collected from both test and control sites on days 0 (baseline), 7, 14, and 21. On day 21, following plaque accumulation at test sites and after GCF sampling and recording of clinical indices, volunteers were given a full mouth prophylaxis and recommenced brushing without using the splint, in order to resolve the experimentally induced inflammation at test sites. Fourteen days later (day 35), the final set of GCF samples was collected and clinical indices recorded.
Figure 1.
Flowchart of study design for test sites. Control sites were treated identically until baseline, but normal plaque removal then continued until day 35.
Gingival crevicular fluid was collected on Periopaper strips (Oraflow, Plainview, NY) over a period of 30 s as previously described.(9) GCF volume was measured using a precalibrated Periotron 8000 (Oraflow, Plainview, NY),(47) from the 3 test teeth and the 3 control teeth. The three test strips were then placed into a cryotube (Appleton Woods, Birmingham, U.K.) containing ammonium bicarbonate (100 mM, 200 μL), sealed and immediately frozen in liquid nitrogen, prior to storage at −80 °C. This procedure was repeated for the 3 control strips, thereby providing one pooled sample from test teeth and one pooled sample from control teeth for each volunteer. Periopaper strips, showed a percentage recovery of 62 ± 5% from spiked plasma (1 μL) (data not shown) as analyzed by the number of proteins identified.
Sample Preparation
Samples were defrosted on ice. From each sample within a time point and test or control site, 200 μL of solution was removed and pooled to give rise to 10 × 1 mL samples: 5 from test sites and 5 from control sites, covering all time points. The pooled samples were vacuum centrifuged and reduced to 200 μL. To each pooled sample dithiothreitol (Fisher Scientific, U.K.) (20 μL, 50 mM) was added and the samples incubated at 60 °C for 40 min. The sample was returned to room temperature before iodoacetamide (Fisher Scientific, U.K.) (100 μL, 22 mM) was added and the samples incubated at room temperature for 25 min. To consume any remaining iodoacetamide, dithiothreitol (2.8 μL, 50 mM) was added. The samples were digested overnight with trypsin Gold (500 ng, Promega, Madison, WI) at 37 °C.
The samples were vacuum centrifuged to dryness, resuspended in trifluoroacetic acid (Fisher Scientific, U.K.) (TFA, 0.5%, 200 μL) and desalted using a Michrom desalting Macrotrap (Michrom, Auburn, CA). The trap was wetted using acetonitrile:water (50:50, 300 μL) and washed with TFA (0.1%, 200 μL). The sample was loaded onto the trap and washed with TFA (0.1%, 200 μL) and eluted in acetonitrile/water (70:30, 200 μL). Samples were vacuum centrifuged to dryness. Samples were resuspended in dissolution buffer (0.5 M triethylammonium bicarbonate, 30 μL). The two day 0 samples were combined and half the sample was taken forward for labeling. The day 7 control was omitted such that 8 samples were labeled in total, as follows. The iTRAQ 8-plex labels (Applied Biosystems, Foster City, CA) were resuspended in isopropanol (50 μL), added to the 8 samples as below, vortexed for 1 min and incubated at room temperature for 2 h. The labels were applied in the following order (sampling time and condition, sample ID): Day 0, 113; Day 7 test, 114; Day 14 control, 115; Day 14 test, 116; Day 21 control, 117; Day 21 test, 118; Day 35 control, 119; and Day 35 test, 121.
The eight labeled samples were combined and vacuum centrifuged dry. The pooled sample was desalted as above and resuspended in mobile phase A (see below, 200 μL). The sample was separated using strong cation exchange high performance liquid chromatography (SCX-HPLC) and fractions collected. The chromatography was performed on an Ettan LC (GE Healthcare Life Science, U.K.) with a Frac-950 fraction collection system. The sample was separated on a polysulfethyl A column (100 mm ×2.1 mm, 5 μm particle size, 200 Åpore size. PolyLC, Columbia, MD) with a javelin guard cartridge (10 mm × 2.1 mm, 5 μm particle size, 200 Åpore size. PolyLC, Columbia, MD). Mobile phase A was potassium dihydrogen orthophosphate (10 mM, pH 3) dissolved in water:acetonitrile (80:20). Mobile phase B was potassium dihydrogen orthophosphate (10 mM), potassium chloride (500 mM, pH 3) dissolved in water/acetonitrile (80:20). The LC gradient ran from 0 to 80% mobile phase B over 73 min. Half of the sample was loaded onto the column and eighteen fractions were collected in eppendorf tubes (1.5 mL). Fractions were combined to give a total of 8 fractions (fractions 2, 18, and 19 were combined; 3, 16, and 17 combined; 4 and 15 combined; 5 and 14 combined; 6 and 13 combined; 7 and 12 combined; 8 and 11 combined; 9 and 10 combined). The combinations gave rise to a minimum number of fractions with similar peptide quantities. The eight fractions were vacuum centrifuged to dryness and desalted as above. The fractions were resuspended in formic acid (0.1%, 50 μL). Each sample was analyzed in triplicate with 5 μL used for mass spectrometric analysis.
Liquid Chromatography−Tandem Mass Spectrometry (LC−MS/MS)
Online liquid chromatography was performed by use of a Micro AS autosampler and Surveyor MS pump (Thermo Fisher Scientific, Bremen, Germany). Peptides were loaded onto a 75 μm (internal diameter) Integrafrit (New Objective, Woburn, MA) C8 resolving column (length 10 cm) and separated over a 40 min gradient from 0% to 40% acetonitrile (Baker, Holland). Peptides were eluted directly (∼350 nL/min) via a Triversa nanospray source (Advion Biosciences, Ithaca, NY) into a 7 T LTQ FT mass spectrometer (Thermo Fisher Scientific), where they were subjected to data-dependent PQD.
Data-Dependent PQD
The mass spectrometer alternated between a full FT-MS scan (m/z 400−2000) and subsequent PQD MS/MS scans of the three most abundant ions above a threshold of 1500. Survey scans were acquired in the ICR cell with a resolution of 100 000 at m/z 400. Precursor ions were subjected to PQD in the linear ion trap. The width of the precursor isolation window was 2 m/z. Only multiply charged precursor ions were selected for MS/MS. PQD was performed with helium gas at a normalized collision energy of 45%, activation Q 0.8 and activation time 200 ms. Automated gain control was used to accumulate sufficient precursor ions (target value 5 × 104, maximum fill time 0.2 s). Dynamic exclusion was used with a repeat count of 1 and exclusion duration of 180 s. Data acquisition was controlled by Xcalibur software V2.1.0 (Thermo Fisher Scientific Inc.).
Data Analysis
The MS/MS spectra were searched against a concatentated forward and reverse IPI human database v3.66 supplemented with bacterial proteins (224 902 entries) using the SEQUEST algorithm in Proteome Discoverer sp 1.0 (Thermo Fisher Scientific). Only the iTRAQ labels were specified as variable modifications, with carboxyamidomethylation of cysteine as a static modification. The data were searched with a precursor mass error of 20 ppm and a fragment mass error of 0.5 Da. The search results were filtered using XCorr vs charge state (peptides reporting XCorr values <2 for 2+ ions, <2.25 for 3+ ions and <2.5 for 4+ or greater ions are rejected) resulting in a false discovery rate of less than 1%. The database used was compiled of human FASTA proteins from the IPI database supplemented with bacterial families identified by Socransky(48) as being associated with periodontal health or periodontal disease. These families were Porphyromonas, Bacterioides, Treponema, Fusobacterium, Prevotella, Campylobacter, Eubacterium, Streptococcus, Capnocytophaga, Eikenella, Actinobaccillus, Actinomyces, Selenomonas and Aggregaterbacter.
Statistical Analysis
Protein database searches resulted in 10 000 protein hits across all the runs. These were filtered for proteins identified in at least three fractions by at least 2 peptides and quantified at all time points. The temporal quantitative profiles of these proteins were analyzed using PolySNAP3 software.(49) PolySNAP3 compares trends using a weighted mean of the Pearson parametric and Spearman nonparametric correlation coefficients employing every measured intensity data point common to all one-dimensional patterns. Cluster numbers are estimated by: principal components analysis using transformed and nontransformed matrices; multidimensional metric scaling (MMDS); gamma statistic, Calinski-Harabasz statistic and C-statistic using either single linkage, group averages, Ward method or complete linkage. The maximum number of clusters was selected as generated by these methods. Data were visualized by 3-dimensional MMDS plots and dendrograms.
Results
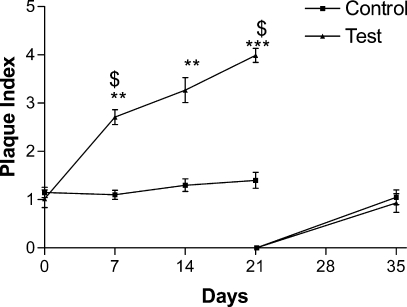
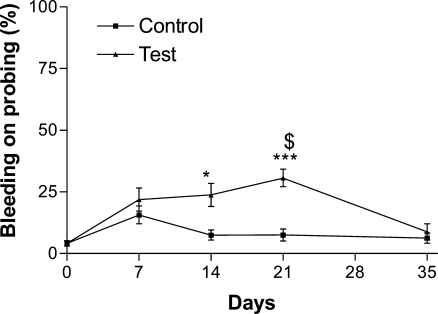
Clinical measurements revealed that the volunteers had complied with the study criteria as shown in Figure 2, where it can be seen that plaque accumulated significantly at the test sites but not the control sites, as assessed by the plaque index. After removal of all plaque from both test and control sites at day 21 microbial recolonization to baseline levels was observed at day 35. Clinical measurements also demonstrated that at test sites inflammation had been induced, as measured by the modified gingival index (Figure 3) and the bleeding index (Figure 4). Both these indices showed significant increases at test sites but not at control sites during plaque accumulation and returned to baseline levels, at day 35, after removal of the inflammatory stimulus at day 21 and resumption of manual brushing (Figure 1). Similarly, GCF volume increased with plaque accumulation at test sites but not at control sites and returned to baseline levels after removal of the inflammatory stimulus (Figure 5).
Figure 2.
Plaque accumulation was assessed using a modified Quigley-Hein index (PI). Data were analyzed by a Kruskal−Wallis test with a Dunn’s post test. *** represents p < 0.001 and ** represents p < 0.01 in comparison to day 0; $ represents p < 0.05 in comparison to control site at the same time point.
Figure 3.
Gingival index (GI) was used to measure gingival inflammation. Data were analyzed by a Kruskal−Wallis test with a Dunn’s post test. ** represents p < 0.01 in comparison to day 0.
Figure 4.
Bleeding on probing was measured dichotomously as presence or absence following gentle probing and expressed as percentage of sites that bled upon probing. Data were analyzed by a Kruskal−Wallis test with a Dunn’s post test. *** represents p < 0.001 and * represents p < 0.05 in comparison to day 0; $ represents p < 0.05 in comparison to control site at the same time point.
Figure 5.
GCF was collected on Periopaper TM strips over 30s and the volume was measured using a precalibrated Periotron 8000TM. Data were analyzed by a Kruskal−Wallis test with a Dunn’s post test. *** represents p < 0.001 and ** represents p < 0.01 in comparison to day 0; $ represents p < 0.05 in comparison to control site at the same time point.
Using a meta-proteomics approach, 16 bacterial and 186 human proteins were identified (Tables 1 and 2). The bacterial proteins showed little temporal variance across the course of the experimental gingivitis, from day 0 to day 35.
Table 1. All Bacterial Proteins Identified from GCF by FTICR−MS/MS, Listing Protein Identity (IPI Accession Number and Description), Mean Coverage, Mean Number of Peptides Used to Identify the Protein, Mean Score, Total Number of Time Identified Across All Runs.
| accession # | description | mean coverage | mean number of peptides | mean score | total times identified |
|---|---|---|---|---|---|
| gi119357403 | helicase domain-containing protein [Chlorobium phaeobacteroides DSM 266] | 2.13 | 2.67 | 5.13 | 3 |
| gi149370586 | type II restriction enzyme, methylase [unidentified eubacterium SCB49] | 1.35 | 3.00 | 2.59 | 3 |
| gi149372617 | hypothetical protein SCB49_12134 [unidentified eubacterium SCB49] | 4.76 | 3.67 | 5.27 | 3 |
| gi149372893 | Glycosyl transferase, group 1 [unidentified eubacterium SCB49] | 3.93 | 2.33 | 3.97 | 3 |
| gi149372926 | ABC transporter ATP-binding protein [unidentified eubacterium SCB49] | 2.89 | 2.40 | 3.26 | 5 |
| gi150008138 | hypothetical protein BDI_1504 [Parabacteroides distasonis ATCC 8503] | 2.29 | 2.20 | 4.23 | 5 |
| gi150010070 | 30S ribosomal protein S2 [Parabacteroides distasonis ATCC 8503] | 6.48 | 3.67 | 3.41 | 3 |
| gi154491686 | hypothetical protein PARMER_01297 [Parabacteroides merdae ATCC 43184] | 2.40 | 2.25 | 4.54 | 4 |
| gi189499472 | chromosome segregation protein SMC [Chlorobium phaeobacteroides BS1] | 1.60 | 2.33 | 1.98 | 3 |
| gi189499611 | cysteine synthase [Chlorobium phaeobacteroides BS1] | 3.27 | 2. 70 | 3.71 | 3 |
| gi212550547 | hypothetical protein CFPG_190 [Candidatus Azobacteroides pseudotrichonymphae genomovar. CFP2] | 3.30 | 5.00 | 9.07 | 3 |
| gi218262131 | hypothetical protein PRABACTJOHN_02373 [Parabacteroides johnsonii DSM 18315] | 4.35 | 2.00 | 4.62 | 4 |
| gi237742543 | fusobacterium outer membrane protein family [Fusobacterium sp. 4_1_13] | 1.06 | 3.17 | 4.15 | 6 |
| gi256841201 | helicase domain-containing protein [Parabacteroides sp. D13] | 1.82 | 2.80 | 3.47 | 5 |
| gi256846448 | fusobacterium outer membrane protein [Fusobacterium sp. 3_1_36A2] | 1.66 | 4.00 | 5.15 | 7 |
| gi260889807 | fusobacterium outer membrane protein [Leptotrichia hofstadii F0254] | 1.41 | 3.25 | 2.95 | 4 |
Table 2. All Human Proteins Identified from GCF by FTICR−MS/MS, Listing Protein Identity (IPI Accession Number and Description), Mean Coverage, Mean Number of Peptides Used to Identify the Protein, Mean Score, Total Number of Time Identified Across All Runs and the Cluster Analysis Groups.
| accession # | description | mean coverage | mean number of peptides | mean score | total times identified |
|---|---|---|---|---|---|
| Cluster 1 | |||||
| Cluster A | Proteins that increase to maximal values at test sites at day 14, before returning to baseline at day 35 | ||||
| IPI:IPI00294242.2 | 28S ribosomal protein S31, mitochondrial | 2.78 | 2 | 2.51 | 3 |
| IPI:IPI00923396.1 | Isoform 2 of Neurofilament heavy polypeptide | 4.11 | 3.6 | 4.98 | 5 |
| IPI:IPI00741005.7 | MAX-interacting protein isoform 2 | 0.82 | 2 | 2.72 | 3 |
| Cluster B1 | Proteins that decrease at test day 35 sites at | ||||
| IPI:IPI00220567.5 | 5-formyltetrahydrofolate cyclo-ligase | 9.85 | 2 | 4.27 | 3 |
| IPI:IPI00023217.2 | cardiac muscle ryanodine receptor | 0.87 | 4.6 | 3.17 | 5 |
| IPI:IPI00927117.1 | CTCL tumor antigen se20−7 | 1.81 | 3.9 | 3.65 | 7 |
| IPI:IPI00448466.3 | Isoform 2 of Ankyrin repeat domain-containing protein 12 | 1.15 | 2.8 | 1.93 | 6 |
| IPI:IPI00942512.1 | keratin 3 | 5.63 | 4.7 | 7.19 | 13 |
| IPI:IPI00008359.1 | Keratin, type II cytoskeletal 2 oral | 6.71 | 5.4 | 9.38 | 11 |
| IPI:IPI00167941.1 | Midasin | 0.7 | 3.6 | 2.97 | 5 |
| IPI:IPI00219168.7 | Spectrin beta chain, brain 4 | 0.95 | 3.5 | 3.71 | 4 |
| Cluster B2i | Proteins largely unchanged across the course of the experimental gingivitis at test sites | ||||
| IPI:IPI00909239.1 | actinin, alpha 1 isoform c | 4.25 | 3.7 | 6.81 | 3 |
| IPI:IPI00946286.1 | alpha 3 type VI collagen isoform 4 precursor | 1.88 | 4.4 | 5.42 | 5 |
| IPI:IPI00218918.5 | Annexin A1 | 8.96 | 3 | 5.27 | 5 |
| IPI:IPI00024095.3 | Annexin A3 | 6.97 | 3 | 5.78 | 4 |
| IPI:IPI00797574.3 | Biorientation of chromosomes in cell division protein 1-like | 1.04 | 4.3 | 3.73 | 4 |
| IPI:IPI00103595.2 | Centrosome-associated protein 350 | 0.67 | 3.3 | 3.53 | 3 |
| IPI:IPI00021828.1 | Cystatin-B | 14.96 | 2.7 | 6.76 | 3 |
| IPI:IPI00032294.1 | Cystatin-S | 7.8 | 2.7 | 4.39 | 3 |
| IPI:IPI00456969.1 | Cytoplasmic dynein 1 heavy chain 1 | 0.68 | 2.3 | 2.41 | 3 |
| IPI:IPI00397930.3 | DBF4-type zinc finger-containing protein 2 | 1.39 | 4.3 | 3.29 | 6 |
| IPI:IPI00335711.5 | Dynein heavy chain 11, axonemal | 0.72 | 2.7 | 2.55 | 3 |
| IPI:IPI00943773.1 | dynein, axonemal, heavy chain 17 | 0.95 | 4.3 | 3.44 | 3 |
| IPI:IPI00642259.2 | Dystonin | 0.8 | 5.7 | 2.36 | 6 |
| IPI:IPI00023711.1 | Envoplakin | 1.51 | 2.7 | 2.01 | 3 |
| IPI:IPI00788953.3 | erythrocyte membrane protein band 4.1-like 2 isoform b | 2.85 | 2.6 | 2.83 | 5 |
| IPI:IPI00007797.3 | Fatty acid-binding protein, epidermal | 10.37 | 2 | 4.36 | 3 |
| IPI:IPI00760833.1 | Isoform 1 of Ankyrin repeat domain-containing protein 26 | 1.29 | 2.7 | 2.95 | 3 |
| IPI:IPI00007834.2 | Isoform 1 of Ankyrin-2 | 0.9 | 4.3 | 2.5 | 4 |
| IPI:IPI00328762.5 | Isoform 1 of ATP-binding cassette subfamily A member 13 | 0.73 | 4.3 | 4.73 | 3 |
| IPI:IPI00152462.1 | Isoform 1 of Dynein heavy chain 3, axonemal | 1.17 | 5 | 2.48 | 3 |
| IPI:IPI00180384.4 | Isoform 1 of Dynein heavy chain 7, axonemal | 0.78 | 3 | 3.7 | 5 |
| IPI:IPI00009866.6 | Isoform 1 of Keratin, type I cytoskeletal 13 | 17.14 | 15.6 | 30.05 | 10 |
| IPI:IPI00175649.3 | Isoform 1 of Leucine-rich repeat serine/threonine-protein kinase 2 | 1.65 | 3.7 | 3.3 | 3 |
| IPI:IPI00186853.6 | Isoform 1 of Pecanex-like protein 2 | 1.16 | 2.3 | 2.78 | 3 |
| IPI:IPI00175151.7 | Isoform 1 of Probable methylcytosine dioxygenase TET2 | 0.95 | 2.3 | 3.09 | 4 |
| IPI:IPI00745872.2 | Isoform 1 of Serum albumin | 6.96 | 6.5 | 10.1 | 11 |
| IPI:IPI00514002.4 | Isoform 1 of StAR-related lipid transfer protein 9 | 0.78 | 3.3 | 4.01 | 3 |
| IPI:IPI00658151.5 | Isoform 1 of Striated muscle preferentially expressed protein kinase | 1.1 | 3.3 | 3.44 | 3 |
| IPI:IPI00939796.1 | Isoform 1 of Zinc finger protein 793 | 4.54 | 3 | 2.44 | 3 |
| IPI:IPI00794668.3 | Isoform 2 of Centrosomal protein of 290 kDa | 1.51 | 3 | 2.79 | 3 |
| IPI:IPI00795958.1 | Isoform 2 of Keratin, type I cuticular Ha6 | 3.98 | 2.4 | 3.66 | 5 |
| IPI:IPI00166205.3 | Isoform 2 of Keratin, type II cytoskeletal 78 | 7.32 | 5 | 6.21 | 3 |
| IPI:IPI00395772.5 | Isoform 2 of Myosin-9 | 1.73 | 2.3 | 2.21 | 3 |
| IPI:IPI00376609.1 | Isoform 2 of Nuclear pore complex protein Nup155 | 1.3 | 2 | 3.3 | 4 |
| IPI:IPI00375454.8 | Isoform 2 of Telomere-associated protein RIF1 | 1.26 | 2.8 | 3.29 | 5 |
| IPI:IPI00023283.3 | Isoform 2 of Titin | 0.88 | 29 | 13.59 | 4 |
| IPI:IPI00783859.2 | Isoform 2 of Vacuolar protein sorting-associated protein 13D | 0.78 | 3 | 1.57 | 4 |
| IPI:IPI00619925.2 | Isoform 3 of Centromere-associated protein E | 1.67 | 5.3 | 4.11 | 4 |
| IPI:IPI00377257.1 | Isoform 3 of Phosphatase and actin regulator 3 | 3.94 | 2.2 | 2.39 | 6 |
| IPI:IPI00843765.1 | Isoform 3 of Spectrin alpha chain, brain | 0.94 | 2.3 | 0.94 | 4 |
| IPI:IPI00220798.1 | Isoform 4 of Bile acid receptor | 4.45 | 2 | 2.86 | 3 |
| IPI:IPI00247295.4 | Isoform 4 of Nesprin-1 | 1.04 | 9 | 5.44 | 6 |
| IPI:IPI00759637.1 | Isoform 4 of Titin | 1.23 | 39.5 | 19.53 | 4 |
| IPI:IPI00401007.2 | Isoform 5 of X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 | 2.02 | 2.3 | 2.63 | 3 |
| IPI:IPI00759542.1 | Isoform 8 of Titin | 0.42 | 11.7 | 2.97 | 3 |
| IPI:IPI00217709.1 | Isoform Beta-1 of DNA topoisomerase 2-beta | 2.84 | 4.3 | 3.37 | 3 |
| IPI:IPI00013933.2 | Isoform DPI of Desmoplakin | 2.49 | 7.9 | 5.69 | 8 |
| IPI:IPI00171196.2 | keratin 13 isoform b | 11.52 | 9.7 | 20.61 | 10 |
| IPI:IPI00384444.6 | Keratin, type I cytoskeletal 14 | 10.82 | 8.8 | 18.68 | 12 |
| IPI:IPI00217963.3 | Keratin, type I cytoskeletal 16 | 12.36 | 9.3 | 18.41 | 17 |
| IPI:IPI00450768.7 | Keratin, type I cytoskeletal 17 | 9.03 | 7.9 | 17.1 | 7 |
| IPI:IPI00479145.2 | Keratin, type I cytoskeletal 19 | 10.4 | 7.4 | 13.74 | 5 |
| IPI:IPI00021298.1 | Keratin, type I cytoskeletal 20 | 6.13 | 5 | 8.5 | 7 |
| IPI:IPI00304458.2 | Keratin, type I cytoskeletal 23 | 4.44 | 3.3 | 4.9 | 4 |
| IPI:IPI00021304.1 | Keratin, type II cytoskeletal 2 epidermal | 5.84 | 5.7 | 11.82 | 6 |
| IPI:IPI00009867.3 | Keratin, type II cytoskeletal 5 | 11.98 | 9.8 | 19 | 12 |
| IPI:IPI00910122.1 | Liprin alpha4 | 3.48 | 4.5 | 4.3 | 4 |
| IPI:IPI00394925.2 | mesoderm posterior 2 homologue | 4.03 | 2 | 3.86 | 4 |
| IPI:IPI00941241.1 | Microtubule-actin cross-linking factor 1 | 0.46 | 2 | 3.06 | 3 |
| IPI:IPI00432363.1 | Microtubule-actin cross-linking factor 1, isoform 4 | 0.88 | 5.8 | 4.17 | 5 |
| IPI:IPI00027883.2 | Microtubule-associated serine/threonine-protein kinase 1 | 1.15 | 2 | 2.4 | 3 |
| IPI:IPI00853536.1 | MOV10-like 1 isoform 2 | 1.63 | 2.6 | 2.42 | 5 |
| IPI:IPI00045914.1 | Msx2-interacting protein | 1.51 | 5.3 | 3.42 | 6 |
| IPI:IPI00103552.4 | Mucin-16 | 0.2 | 3.3 | 2.74 | 3 |
| IPI:IPI00025879.2 | Myosin-1 | 2.05 | 4.8 | 4.82 | 5 |
| IPI:IPI00914847.2 | nebulin isoform 1 | 1.73 | 10 | 4.93 | 4 |
| IPI:IPI00917728.2 | nebulin isoform 2 | 1.92 | 14 | 8.22 | 3 |
| IPI:IPI00742682.2 | Nucleoprotein TPR | 1.09 | 3 | 2.13 | 4 |
| IPI:IPI00298057.2 | Periplakin | 1.97 | 3.4 | 3.97 | 5 |
| IPI:IPI00296909.3 | Poly [ADP-ribose] polymerase 4 | 1.22 | 2.6 | 3.49 | 7 |
| IPI:IPI00005826.1 | Probable E3 ubiquitin-protein ligase HERC2 | 0.56 | 2.3 | 1.49 | 3 |
| IPI:IPI00412541.2 | Probable G-protein coupled receptor 158 | 1.62 | 2 | 2.79 | 3 |
| IPI:IPI00218076.1 | Protein FAM9A | 4.88 | 2.4 | 2.96 | 5 |
| IPI:IPI00007047.1 | Protein S100-A8 | 15.36 | 2.9 | 5.09 | 7 |
| IPI:IPI00939543.1 | Pumilio homologue 1 | 2.38 | 2 | 1.98 | 4 |
| IPI:IPI00013721.2 | Serine/threonine-protein kinase PRP4 homologue | 2.61 | 3 | 2.21 | 4 |
| IPI:IPI00786995.1 | similar to protein kinase, DNA-activated, catalytic polypeptide | 1.51 | 6.3 | 5.66 | 6 |
| IPI:IPI00935432.1 | similar to ubiquitin protein ligase E3 component n-recognin 5 isoform 2 | 0.9 | 2.6 | 3.41 | 7 |
| IPI:IPI00174345.3 | Sperm-associated antigen 17 | 1.2 | 3 | 2.95 | 3 |
| IPI:IPI00303343.7 | Splicing factor, arginine/serine-rich 19 | 2.19 | 2.3 | 2.38 | 4 |
| IPI:IPI00219420.3 | Structural maintenance of chromosomes protein 3 | 1.67 | 2.7 | 2.6 | 3 |
| IPI:IPI00759613.2 | titin isoform N2-A | 0.72 | 23.2 | 11.17 | 11 |
| IPI:IPI00784174.1 | titin isoform novex-3 | 0.56 | 3.8 | 2.01 | 4 |
| IPI:IPI00001159.10 | Translational activator GCN1 | 0.83 | 2.3 | 3.52 | 4 |
| IPI:IPI00289709.3 | Zinc finger imprinted 3 | 3.55 | 2.5 | 2.21 | 4 |
| IPI:IPI00921184.1 | Zinc finger, PHD-type domain containing protein | 3 | 5 | 5.65 | 4 |
| Cluster B2ii | Proteins that increase at test sites at day 21 before returning to baseline at day 35 | ||||
| IPI:IPI00748061.1 | Centrosomal protein 350 kDa | 1.82 | 4.5 | 3.26 | 4 |
| Cluster B2iii | Proteins that increase at test sites at day 14, decrease at day21, returning to baseline at day before 35 | ||||
| IPI:IPI00453473.6 | Histone H4 | 21.9 | 3.9 | 6.76 | 9 |
| IPI:IPI00007729.1 | Isoform 1 of Nucleolar protein 7 | 8.04 | 2 | 1.97 | 3 |
| IPI:IPI00744062.1 | Isoform 2 of Transcription elongation regulator 1 | 2.17 | 3 | 3.3 | 3 |
| IPI:IPI00375294.2 | Laminin subunit alpha-1 | 0.92 | 2.7 | 2.23 | 3 |
| Cluster B2iv | Proteins that increase at test sites at day 35 | ||||
| IPI:IPI00784869.4 | Isoform 1 of Dynein heavy chain 10, axonemal | 1.23 | 5 | 4.97 | 8 |
| IPI:IPI00239405.4 | Isoform 1 of Nesprin-2 | 0.88 | 6.5 | 4.38 | 6 |
| IPI:IPI00892863.1 | Isoform 2 of Collagen alpha-5(VI) chain | 1.61 | 4 | 3.42 | 3 |
| IPI:IPI00855826.1 | Isoform 2 of Ventricular zone-expressed PH domain-containing protein homologue 1 | 2.66 | 2 | 3.8 | 3 |
| IPI:IPI00383970.2 | Mutated melanoma-associated antigen 1-like protein 1 | 3.88 | 2.3 | 2.48 | 3 |
| IPI:IPI00642716.6 | myosin, heavy polypeptide 7B, cardiac muscle, beta | 1.48 | 2.5 | 3.74 | 4 |
| IPI:IPI00001753.2 | Myosin-4 | 1.9 | 5 | 3.48 | 4 |
| IPI:IPI00025880.2 | Myosin-7 | 1.4 | 3 | 3.66 | 7 |
| IPI:IPI00021827.3 | Neutrophil defensin 3 | 13.54 | 2.2 | 4.7 | 11 |
| IPI:IPI00375149.4 | optic atrophy 1 isoform 2 | 2.71 | 2.7 | 4.18 | 3 |
| IPI:IPI00418184.1 | Similar to nonhistone chromosomal protein HMG-1 | 7.33 | 3.3 | 2.18 | 3 |
| IPI:IPI00848233.1 | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homologue isoform 2 precursor | 3.09 | 2.3 | 2.88 | 3 |
| IPI:IPI00307591.5 | Zinc finger protein 609 | 1.25 | 2 | 2.45 | 3 |
| Cluster B2v | Proteins that decrease at test sites at 21 before returning to baseline at day 35 | ||||
| IPI:IPI00009865.3 | Keratin, type I cytoskeletal 10 | 4.22 | 3.7 | 6.95 | 3 |
| Cluster B2vi | Proteins that increase to maximal values at test sites at day 14, before returning to baseline at day 35 | ||||
| IPI:IPI00300786.1 | Alpha-amylase 1 | 3.84 | 2 | 4.8 | 8 |
| IPI:IPI00166331.4 | Ankyrin repeat domain-containing protein 31 | 2.4 | 2.3 | 3.06 | 3 |
| IPI:IPI00022229.1 | Apolipoprotein B-100 | 0.99 | 5 | 5.41 | 5 |
| IPI:IPI00017800.2 | ATP-binding cassette subfamily A member 3 | 1.31 | 2 | 2.47 | 3 |
| IPI:IPI00514867.1 | glutamate receptor, metabotropic 1 isoform beta precursor | 3.86 | 3.7 | 3.33 | 3 |
| IPI:IPI00217465.5 | Histone H1.2 | 11.27 | 2.7 | 1.79 | 3 |
| IPI:IPI00006876.2 | Immediate early response gene 2 protein | 6.28 | 2.7 | 3.97 | 3 |
| IPI:IPI00794880.1 | Isoform 1 of Chromodomain-helicase-DNA-binding protein 7 | 1.33 | 4.5 | 4.1 | 4 |
| IPI:IPI00884996.1 | Isoform 1 of Dynein heavy chain 6, axonemal | 0.69 | 2.7 | 2.66 | 3 |
| IPI:IPI00896403.1 | Isoform 1 of Tetratricopeptide repeat protein GNN | 2.1 | 2.3 | 3.69 | 3 |
| IPI:IPI00376283.1 | Isoform 2 of Retinoblastoma-binding protein 6 | 2.66 | 5.7 | 4.21 | 3 |
| IPI:IPI00218207.1 | Isoform 2 of Spectrin beta chain, brain 2 | 1.49 | 4 | 3.87 | 4 |
| IPI:IPI00412210.1 | Isoform 3 of SLIT-ROBO Rho GTPase-activating protein 3 | 3.9 | 2 | 2.78 | 3 |
| IPI:IPI00218660.3 | Isoform 4 of Inositol 1,4,5-trisphosphate receptor type 1 | 1.2 | 3.7 | 3.98 | 3 |
| IPI:IPI00398776.3 | Isoform 7 of Plectin-1 | 0.41 | 2.3 | 0.54 | 3 |
| IPI:IPI00005859.5 | Keratin, type II cytoskeletal 75 | 8.65 | 7 | 10.93 | 3 |
| IPI:IPI00302329.1 | Myosin-8 | 1.87 | 4 | 2.56 | 4 |
| IPI:IPI00217098.2 | Zinc finger protein 678 | 7.3 | 3 | 3.69 | 3 |
| Cluster B3 | Proteins that decrease at test sites at day 14 before returning to baseline at day 35 | ||||
| IPI:IPI00908515.1 | bromodomain containing 9 isoform 2 | 6.19 | 3.3 | 4.14 | 3 |
| IPI:IPI00174574.1 | Coiled-coil domain-containing protein 147 | 3.47 | 3.3 | 3.74 | 4 |
| IPI:IPI00827754.3 | Ig gamma-3 chain C region | 9.66 | 2.9 | 5.5 | 7 |
| IPI:IPI00878236.2 | Isoform 1 of Fibrous sheath-interacting protein 2 | 0.77 | 5.3 | 4.23 | 4 |
| IPI:IPI00030282.3 | Isoform 1 of Filensin | 3.23 | 3 | 3.5 | 4 |
| IPI:IPI00376383.2 | Isoform 2 of Centriolin | 2.52 | 5 | 3.42 | 3 |
| IPI:IPI00413958.5 | Isoform 2 of Filamin-C | 1.02 | 2.7 | 2.68 | 3 |
| IPI:IPI00889747.1 | Isoform 2 of Intraflagellar transport protein 172 homologue | 1.4 | 2 | 2.24 | 3 |
| IPI:IPI00829833.2 | Isoform 2 of Mucin-19 | 0.72 | 4 | 7.39 | 3 |
| IPI:IPI00384697.2 | Isoform 2 of Serum albumin | 6.39 | 4.4 | 7.56 | 8 |
| IPI:IPI00302008.2 | Isoform 3 of Zinc finger protein 407 | 1.67 | 2 | 2.19 | 4 |
| IPI:IPI00216562.3 | Isoform 4 of Cadherin-23 | 0.81 | 2.7 | 3.6 | 7 |
| IPI:IPI00219941.1 | Isoform A of Oxysterol-binding protein-related protein 1 | 3.89 | 2 | 2.12 | 3 |
| IPI:IPI00306400.9 | Kinesin-like protein KIFC1 | 4.85 | 2.7 | 3.92 | 3 |
| IPI:IPI00218725.3 | laminin alpha 2 subunit isoform b precursor | 1.3 | 3.3 | 4.59 | 3 |
| IPI:IPI00303112.2 | Methylcytosine dioxygenase TET1 | 0.76 | 2.3 | 1.31 | 3 |
| IPI:IPI00152380.3 | Myosin-XV | 1.2 | 4 | 3.17 | 3 |
| IPI:IPI00884926.1 | orosomucoid 1 precursor | 6.72 | 2.3 | 6.55 | 4 |
| IPI:IPI00010471.5 | Plastin-2 | 4.59 | 3 | 6.5 | 4 |
| IPI:IPI00795657.1 | SFRS2IP protein | 1.35 | 2.5 | 2.65 | 4 |
| Cluster B4 | Proteins that increase at test sites at day 14 and remain elevated throughout the experimental gingivitis | ||||
| IPI:IPI00796450.2 | Isoform 1 of Lysosomal-trafficking regulator | 0.67 | 2.2 | 2.07 | 6 |
| IPI:IPI00220327.4 | Keratin, type II cytoskeletal 1 | 5.04 | 3.6 | 7.06 | 9 |
| IPI:IPI00848317.1 | myosin IXB isoform 2 | 0.84 | 3.3 | 2.37 | 4 |
| IPI:IPI00168056.2 | Zinc finger and BTB domain-containing protein 38 | 1.7 | 3.3 | 4.9 | 3 |
| Cluster B5 | Proteins that increase at test sites at day 21 before returning to baseline at day 35 | ||||
| IPI:IPI00014515.3 | Isoform 1 of Serine/threonine-protein phosphatase 4 regulatory subunit 4 | 1.49 | 2.3 | 2.12 | 3 |
| IPI:IPI00426269.2 | Isoform 2 of Leucine-rich repeat-containing protein 7 | 2.41 | 2.7 | 2.77 | 3 |
| Cluster B6 | Proteins that decrease at test sites at day 14 and remain lowered throughout the experimental gingivitis | ||||
| IPI:IPI00301419.2 | COP9 signalosome complex subunit 7a | 5.21 | 2.7 | 3.13 | 3 |
| IPI:IPI00743813.3 | Isoform 1 of Abnormal spindle-like microcephaly associated protein | 1.43 | 4.7 | 4.17 | 7 |
| IPI:IPI00879441.1 | Isoform 3 of Protein strawberry notch homologue 1 | 1.34 | 2.3 | 3.3 | 3 |
| IPI:IPI00299749.4 | Zinc finger protein 16 | 1.96 | 2 | 3.96 | 3 |
| Cluster C | Proteins that decrease at test sites at day 14 before returning to baseline at day 21 | ||||
| IPI:IPI00457261.2 | similar to hCG1645503 | 7.48 | 3.3 | 3.61 | 4 |
| Cluster D | Proteins that increase at day 35 at test sites | ||||
| IPI:IPI00179016.8 | Histone-lysine N-methyltransferase SETD1A | 1.52 | 2.3 | 2.93 | 3 |
| IPI:IPI00006196.3 | Isoform 2 of Nuclear mitotic apparatus protein 1 | 1.76 | 3.3 | 2.68 | 3 |
| Cluster E | Proteins that increase to maximal levels at test sites at day 21 before returning to baseline at day 35 | ||||
| IPI:IPI00747682.1 | Isoform 2 of MAGUK p55 subfamily member 2 | 2.26 | 2.5 | 3.08 | 4 |
| IPI:IPI00298301.4 | Myosin-3 | 2.09 | 4 | 2.59 | 5 |
| IPI:IPI00020153.4 | Protein bassoon | 0.78 | 2.6 | 2.65 | 7 |
| Cluster 2 | Proteins decreased at control sites to minimal levels at day 14 before returning to baseline at day 35 | ||||
| IPI:IPI00942176.1 | A-kinase anchor protein 7 isoform gamma | 5.62 | 2.3 | 3.6 | 3 |
| IPI:IPI00293655.3 | ATP-dependent RNA helicase DDX1 | 1.7 | 3.7 | 2.9 | 3 |
| IPI:IPI00021129.5 | Isoform 1 of AP-3 complex subunit beta-1 | 1.94 | 2 | 2.6 | 4 |
| IPI:IPI00791131.3 | Isoform 1 of Growth hormone-regulated TBC protein 1 | 5.75 | 2 | 3.2 | 3 |
| IPI:IPI00300504.5 | Isoform 1 of Regulator of nonsense transcripts 2 | 1.57 | 2.3 | 3.61 | 3 |
| IPI:IPI00795394.2 | Isoform 2 of Dynein heavy chain 9, axonemal | 0.77 | 3.3 | 2.78 | 9 |
| IPI:IPI00333015.7 | Isoform 2 of Spectrin beta chain, brain 1 | 1.21 | 2.8 | 3.07 | 4 |
| IPI:IPI00884904.1 | Isoform 3 of Protein AHNAK2 | 1.66 | 3.5 | 3.71 | 4 |
| IPI:IPI00219330.2 | Isoform 5 of Interleukin enhancer-binding factor 3 | 3.14 | 2.7 | 2.3 | 3 |
| IPI:IPI00019359.4 | Keratin, type I cytoskeletal 9 | 2.25 | 3.3 | 4.07 | 3 |
| IPI:IPI00300053.4 | Keratin, type II cuticular Hb2 | 3.57 | 2 | 2.29 | 3 |
| IPI:IPI00022463.1 | Serotransferrin | 4.01 | 4.2 | 7.6 | 5 |
| IPI:IPI00787323.2 | Similar to Keratin, type II cytoskeletal 8 | 7.52 | 4.3 | 5.93 | 3 |
| Cluster 3 | Proteins decreased at control sites to minimal levels at day 14 before returning to baseline at day 35 | ||||
| IPI:IPI00294696.4 | Cyclin B1 | 5.06 | 2 | 2.01 | 4 |
| Cluster 4 | Proteins decreased at control sites throughout the experimental gingivitis | ||||
| IPI:IPI00004924.3 | Differentially expressed in FDCP 6 homologue | 3.52 | 2.4 | 3.97 | 5 |
| Cluster 5 | Proteins decreased at control sites at day 14 and before returning to baseline at day 21 and then decreasing again at day 35 | ||||
| IPI:IPI00303335.2 | Nebulin | 1.46 | 8.2 | 3.62 | 6 |
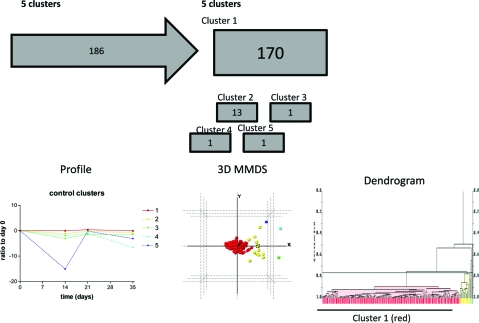
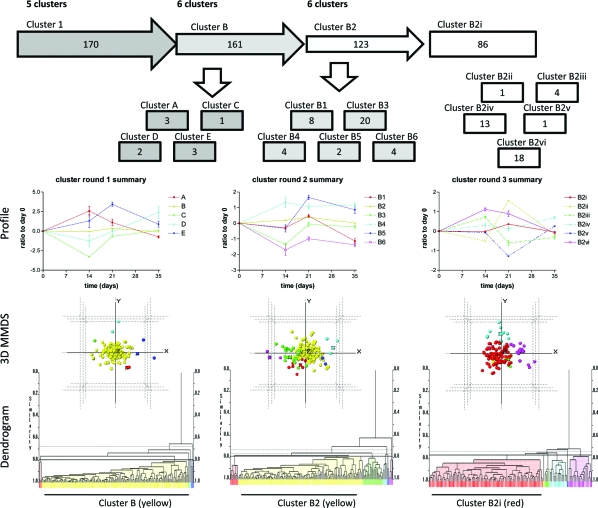
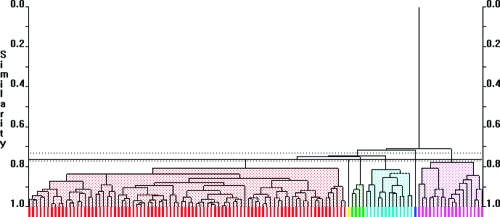
Temporal changes in the abundance of human proteins across the course of the experiment at control sites were analyzed with PolySNAP3 software for patterns different from a stable unchanging profile. 170 proteins showed no change over the course of the experiment (Cluster 1, not deviating from a ratio of 1 from day 0) at control sites, and 16 showed decreases from day 0 to day 35 (Clusters 2−5), at control sites, that did not cluster with the other proteins (maximum number of clusters: 5, range 1−5, median number of clusters 3, Figure 6). These 16 proteins were then removed from subsequent statistical analysis. The proteins in cluster 1 were then used to filter the data from the test sites, which were normalized to control sites. The normalized data for the 170 proteins were analyzed using PolySNAP3 to identify the proteins whose abundances showed the greatest change with development of experimental gingivitis. Five clusters were determined (Clusters A−E, maximum number of clusters: 6, range 2−6, median number of clusters 3, Figure 7). Clusters A and E, containing 3 proteins each, increased to a maximum value at day 14 and day 21 respectively before returning to baseline levels at day 35. Clusters C (1 protein) and D (2 proteins), both decreased at day 14 and then either returned to baseline at days 21 and 35 (Cluster C) or returned to baseline at day 21 and continued to increased to a maximum value at day 35. Cluster B, (161 proteins), appeared to remain constant at baseline levels and throughout the course of the experimental gingivitis.
Figure 6.
Cluster analysis of data set showing the workflow and clusters generated. Control sites across the course of the experimental gingivitis study were clustered using PolySNAP3. Group 1 showed no changes from baseline, whereas groups 2−5 differed from baseline over the course of the experiment (Control clusters). Below the flow diagram are graphs representing the mean (±sem) changes of the proteins in each cluster, which are followed beneath by 3-dimensional metric multidimensional scaling (MMDS) plots and dendrograms showing the relationships of individual proteins with others in the analysis group. Colors of lines on graphs, spheres on 3D plots and bars on dengrograms coordinate to show the same clustering.
Figure 7.
Cluster analysis of data set showing the workflow and clusters generated. Proteins identified from group 1 were used for the next step of the analysis using normalized test data which were clustered using PolySNAP3. Five clusters (clusters A-E) were generated (cluster round 1 summary), with 161 proteins identified in cluster B. The proteins in cluster B were then further analyzed by PolySNAP3 to generate 6 clusters (clusters B1−6; cluster round 2 summary). The largest group from this analysis was group B2 (123 proteins). This group was then further analyzed using PolySNAP3 to generate 6 clusters (clusters B2i-vi, cluster round 3 summary). Below the flow diagram are graphs representing the mean (±sem) changes of the proteins in each cluster, which are followed beneath by 3-dimensional metric multidimensional scaling (MMDS) plots and dendrograms showing the relationships of individual proteins with others in the analysis group. Colors of lines on graphs, spheres on 3D plots and bars on dendrograms coordinate to show the same clustering. It should be noted that the highlighted, largest group on each dendrogram was the group taken forward into the subsequent round of clustering analysis.
Inspection of the dendrogram from this clustering analysis demonstrated that there were still many clusters within group B. Thus further analysis of this group was performed. The second round of clustering revealed a further 6 clusters (clusters B1−6, maximum number of clusters: 6, range 3−6, median number of clusters 4, Figure 7). Clusters B4 (4 proteins) and B5 (2 proteins) demonstrated increases across the course of the experimental gingivitis peaking at days 14 and 21 respectively and remaining elevated at the peak level at day 35. Clusters B1, B3 and B6, containing 8, 20, and 4 proteins respectively, decreased during the course of the study. Cluster B1 showed a small decrease at day 14 and then returned to baseline levels before decreasing greatly at day 35. Cluster B3 decreased at day 14 but returned to baseline at subsequent time points. Cluster B6 decreased at day 14 and remained at this level for the rest of the experimental gingivitis. Cluster B2 (123 proteins) did not fluctuate at this level of detail across the course of the experimental gingivitis and these proteins were again taken for further clustering analysis. That analysis gave rise to another 6 clusters (clusters B2i-vi, maximum number of clusters: 6, range 3−6, median number of clusters 4, Figure 7). At this point two observations were made: (1) the largest group (B2i) showed a slight increase at day 21, i.e., no longer showed little variation across the course of the experimental gingivitis and (2) the variation in the ratio to day 0 for all the clusters was now between 0−1 unit at its maximum. Therefore, further rounds of clustering analysis were not pursued, even though there were still a large number of proteins in the largest cluster (cluster B2i, n = 86). Inspection of the patterns generated from this third and final clustering showed that cluster B2ii (1 protein) showed a peak at day 21; cluster B2iii (4 proteins) peaked at day 14 and then decreased at days 21 and 35; cluster B2iv (13 proteins) showed little change over 14 and 21 days but an increase at day 35; cluster B2v (1 protein) showed a decrease at day 21; and cluster B2vi (18 proteins) showed an increase at day 14 that was sustained to day 21 before returning to baseline at day 35. Fold changes for all proteins in Cluster 1 can be found in the supplemental Table 1.
Gene ontology analysis for location revealed the majority of proteins identified to cover the entire range of nuclear, plasma membrane, cytoplasmic and extracellular compartments (Figure 8). Gene ontology analysis for function identified a wide range of possibilities with the greatest numbers of proteins for cellular transcription, DNA binding, cytoskeletal functions, cell adhesion, defense against bacteria and cilia (Figure 8). Particular clusters did not preferentially fall into one of these categories and the proteins functions were diverse throughout the individual clusters. It should be noted that between 35 and 40% proteins did not have gene ontology terms that could be used.
Figure 8.
Analysis of the proteomic data showing component and function gene ontology. Gene Otology pie charts were generated using Swiss-Prot database with Inforsense, embedded in Proteome Discoverer sp1.0 from Thermo Fisher Scientific Inc.
Discussion
Across the course of experimentally induced gingivitis, we identified 16 bacterial and 186 human proteins by combining iTRAQ and LC−MS/MS on an FT-ICR mass spectrometer. The abundances of the bacterial proteins identified did not vary temporally. This observation may be used as confirmation that the GCF samples were not significantly contaminated with plaque during sampling. Furthermore, Fusobacterium species (Fusobacterium outer membrane proteins) were detected and although this family, particularly Fusobacterium nucleatum which is part of the orange complex identified by Socransky et al,(48) has being associated with increased periodontal pocket depth and worsening clinical parameters, it is also associated with nonperiodontitis sites.(48) Other bacterial proteins identified were not immediately obvious candidates from oral pathogens.
The human proteins identified spanned a wide range of compartments and functions. As GCF comprises both a serum transudate and tissue exudates, it is unsurprising to find both extracellular and intracellular components within samples. With respect to the extracellular components, serum proteins, such as albumin, IgG3 and apolipoprotein B-100; as well as proteins displaying antibacterial properties, such as neutrophil defensin, cystatins B and S and Annexins A1 and A3, were found. The serum components (albumin and IgG3) clustered together (cluster B3) and showed a decrease at day 14 at test sites. At this time point, the GCF volume from test sites was already significantly increased above control sites. On the other hand apolipoprotein B-100 falls into cluster B2vi, which follows the profile of the increase in GCF volume and bleeding index at test sites. Apolipoprotein B-100 is the protein component of low density lipoprotein, which has a diameter of approximately 26 nm. This particle is unlikely to be able to pass across the permeable sulcular membrane and could potentially be used as a marker of increased microulceration of the sulcular epithelium. Cystatins B and S and Annexins A1 and A3 were found in cluster B2i, which was largely unchanged across the course of the experimental gingivitis. Cystatin B and Annexin A1 were found by Bostanci et al.(29) only in healthy volunteers and not in developing inflammatory disease. That they seem not to change across the course of our experiment would support the idea that they are not associated with the development of moderate gingival inflammation. Neutophil defensin was found in cluster B2iv that showed the greatest increases at day 35 for the test sites. This may indicate a role in maintaining a healthy state upon resolution of inflammation. Bostanci et al.(29) also detected this protein at healthy sites, although the GCF volume at these sites is equivalent to day 7 in this experimental gingivitis study, perhaps highlighting a potential role of neutrophil defensin in population based health screening.
Intracellular proteins identified had functions such as cellular transcription, DNA binding, cytoskeletal functions, cell adhesion, and cilia proteins. Starting with cytoskeletal proteins, a large number of keratins were identified, which was expected from sampling adjacent to epithelial tissues. The oral epithelium has one of the highest turnover rates(50) and this will have contributed to the large number of keratins seen. The majority of keratins were clustered together in cluster B2i, which showed the least variation across the course of the experimental gingivitis, perhaps indicating that the differentiation or proliferation of the epithelium was not affected during the course of the induced inflammation, again consistent with a sulcular rather than a pocket lining epithelium that characterizes periodontitis. However, it is acknowledged that some keratin contamination could have occurred during sample handling. Other cytoskeletal and associated proteins, such as titin and actin- and microtubule- associated proteins, also fell into cluster B2i. These proteins could be components of desquamated epithelial cells released during the turnover of the epithelium. There were also several axonemal dynein proteins grouped to cluster B2i. To our knowledge, these cilia-associated proteins have not been identified in GCF before. However, a number of studies51−53 have used histology to investigate changes in the gingivae in periodontal disease. Saglie et al.(53) used scanning electron microscopy to investigate the pocket epithelium from patients with advanced periodontal disease. At high resolution they observed individual epithelial cells, bacteria and erythrocytes; however, large amounts of cilia were not seen, such as might be expected with lung epithelium. Primary cilia, however, are present on nearly all cells, including internal stratified oral epithelium,(54) and play a chemo- or mechano-sensory role.(55) Although they are not changed in this experimental gingivitis model, it will be interesting to determine how cilia proteins change with chronic periodontal diseases as it has been proposed that ciliopathies may impair wound healing.(55)
Among the cellular transcription and DNA binding proteins that were identified were two histone proteins: Histone H4 and Histone H1.2. They did not cluster together; however, they were both in clusters (B2iii and B2vi, respectively) that showed an increase at test sites at day 14. These proteins are associated with chromatin and may indicate the presence of neutrophil extracellular traps (NETs).(56) In addition to phagocytosis, neutrophils can discharge decondensed chromatin and associated proteins to trap and immobilize bacteria. These NETs have been shown to be present in the gingival crevice of periodontitis patients.(57) Neutrophils are the most important leukocytes in the periodontal tissues and have been implicated in periodontitis patients as being hyper-active and hyper-reactive.58,59 Further evidence for neutrophils may be taken from the presence of Plastin-2 (or l-plastin), which was also identified by Bostanci et al.(29) Plastin-2 is an actin bundling protein that plays a role in signaling following Fcγ receptor stimulation in adherent neutrophils. Here we found Plastin-2 clusters within cluster B3, which demonstrated a decrease at day 14 at test sites, which might indicate a less adherent phenotype in neutrophils at this stage of the inflammatory response. Bostanci et al.(29) reported this protein in aggressive periodontitis patients only, perhaps showing a further change in the behavior of neutrophils in diseased rather than healthy volunteers.
In this study, we have used PolySNAP3 clustering software in a multilayered analytical approach. For ease of comprehension, an analogy may be used of examining the clusters of proteins in a manner similar to “peeling away the layers of an onion” or “zooming in to different levels of detail in the clustering”. The first layer isolated those proteins that did not maintain a broadly steady flat baseline in the control sites during the course of the experimental gingivitis. The greatest change was seen with Nebulin (cluster 5); however, other isoforms of this protein (nebulin isoform 1 and 2) were clustered to cluster B2i which showed the least variation across the course of the experimental gingivitis. Other proteins in clusters 2−4 showed decreases from baseline. Cluster analysis of the entire data set (not shown) always showed these particular proteins as lying outside the central core (Figure 6 control cluster 3D MMDS plot), thus they may be viewed as outliers. Following the exclusion of these proteins the normalized test data was clustered revealing the clusters that changed the most across the course of the experimental gingivitis and then subsequent clustering of the largest group showed the next level of changes, and so on in the third round of clustering. Two groups (clusters A and E) in the first round of clustering and one (cluster B2vi) in the third round of clustering showed increases at days 14, 21, and 14 respectively, which were similar to increases in the clinical parameters. Cluster A contained disparate proteins involved in neuronal axons, transcription and protein production; cluster E contained proteins involved in signal transduction, actin-myosin based movement and synaptic transmission; and cluster B2vi contained many proteins including actin-myosin based movement and synaptic transmission again. A common theme from these clusters might be the presence of neuronal and synapse associated proteins. The most abundant is protein bassoon which is a constituent of ribbon synapses. Sterling and Matthews (2005)(60) suggest that ribbon synapses “occur wherever synaptic exocytosis is evoked by graded depolarisation and where signalling requires a high rate of sustained release” such as in the terminals of photoreceptors and auditory and vestibular hair cells. These synapses use glutamate as their primary neurotransmitter and this would fit with the identification of metabotropic glutamate receptors in cluster B2vi. Histological investigations of gingival biopsies will be required to verify the presence of ribbon synapses within the gingivae as their presence has not been seen before. The role of their destruction during experimental gingivitis remains to be fully investigated, particularly with the knowledge that substance P, a neuropeptide, released from sensory neurones and an inducer of inflammation and pain, is increased in gingivitis and periodontitis,(61) and that in a recent transcriptomics investigation Offenbacher et al (2009)(62) found that after changes to immune response pathways, neural process pathways where the second most abundant pathways activated during a similar experimental gingivitis model.
Lastly, cytokines have been investigated extensively in GCF as biomarkers of gingival inflammation. However, in this study we have not been able to detect any cytokines, consistent with Bostanci et al.(29) This is probably due to the cytokines being at low, subattomol, concentrations.
To conclude, here we have shown for the first time the quantitative analysis of temporal changes of proteins in gingival crevicular fluid, using a nonpresumptive approach. Proteins identified included proteins already known to the dental community and verification of newly identified proteins in other mass spectral studies. We have also identified new proteins that highlight structural components of the gingivae that have not been seen before and that warrant further investigation.
Acknowledgments
We gratefully acknowledge Unilever Oral Care UK (Unilever Oral Care, Bebington, UK) (MMG, HRG, JBM and ILCC) and the Wellcome Trust (074131) (HJC) for funding.
Supporting Information Available
Supplemental table. This material is available free of charge via the Internet at http://pubs.acs.org.
Supplementary Material
References
- Loe H.; Holm-Pederson P. Absence and Presence of Fluid from Normal and Inflamed Gingivae. Periodontics 1965, 3 (4), 171–177. [PubMed] [Google Scholar]
- Loe H.; Theilade E.; Jensen S. B. Experimental Gingivitis in Man. J. Periodontol. 1965, 36 (3), 177–187. [DOI] [PubMed] [Google Scholar]
- Heasman P. A.; Offenbacher S.; Collins J. G.; Edwards G.; Seymour R. A. Flurbiprofen in the Prevention and Treatment of Experimental Gingivitis. J. Clin. Periodontol. 1993, 20 (10), 732–738. [DOI] [PubMed] [Google Scholar]
- Putt M. S.; Vanderweijden G. A.; Kleber C. J.; Saxton C. A. Validation of a 21-Day, Partial-Mouth Gingivitis Model for Evaluating Chemotherapeutic Dentifrices. J. Periodontal Res. 1993, 28 (4), 301–307. [DOI] [PubMed] [Google Scholar]
- Saxton C. A.; Huntington E.; Cummings D. The effect of dentifrices containing Triclosan on the development of gingivitis in a 21-day experimental gingivitis study. Int. Dent. J. 1993, 43 (4), 423–429. [PubMed] [Google Scholar]
- Lang N. P.; Anton E.; Gabriel Y.; Salvi G. E.; Pjetursson B. E.; Winston J. L.; He T. An experimental gingivitis study to evaulate the clinical effects of stannous fluoride dentrifice. Oral Health Prev. Dent. 2004, 2 (4), 369–376. [PubMed] [Google Scholar]
- Bergstrom J.; Preber H. The Influence of Cigarette-Smoking on the Development of Experimental Gingivitis. J. Periodontal Res. 1986, 21 (6), 668–676. [DOI] [PubMed] [Google Scholar]
- Giannopoulou C.; Cappuyns I.; Mombelli A. Effect of smoking on gingival crevicular fluid cytokine profile during experimental gingivitis. J. Clin. Periodontol. 2003, 30 (11), 996–1002. [DOI] [PubMed] [Google Scholar]
- Chapple I. L. C.; Socransky S. S.; Dibart S.; Glenwright D. H.; Matthews J. B. Chemiluminescent assay of alkaline phosphatase in human gingival crevicular fluid: Investigations with an experimental gingivitis model and studies on the source of the enzyme within crevicular fluid. J. Clin. Periodontol. 1996, 23 (6), 587–594. [DOI] [PubMed] [Google Scholar]
- Wright H. J.; Chapple I. L. C.; Matthews J. B. Levels of TGF beta 1 in gingival crevicular fluid during a 21-day experimental model of gingivitis. Oral Dis. 2003, 9 (2), 88–94. [DOI] [PubMed] [Google Scholar]
- Brill N.The gingival pocket fluid. Studies of its occurrence, composition and effect. PhD Thesis from Royal Dental College, Copenhagen, Denmark, 1962.
- Kinane D. F.; Adonogianaki E.; Moughal N.; Winstanley F. P.; Mooney J.; Thornhill M. Immunocytochemical Characterization of Cellular Infiltrate, Related Endothelial Changes and Determination of Gcf Acute-Phase Proteins during Human Experimental Gingivitis. J. Periodontal Res. 1991, 26 (3), 286–288. [DOI] [PubMed] [Google Scholar]
- Adonogianaki E.; Moughal N. A.; Mooney J.; Stirrups D. R.; Kinane D. F. Acute-Phase Proteins in Gingival Crevicular Fluid during Experimentally-Induced Gingivitis. J. Periodontal Res. 1994, 29 (3), 196–202. [DOI] [PubMed] [Google Scholar]
- Gonzales J. R.; Herrmann J. M.; Boedeker R. H.; Francz P. I.; Biesalski H.; Meyle J. Concentration of interleukin-l beta and neutrophil elastase activity in gingival crevicular fluid during experimental gingivitis. J. Clin. Periodontol. 2001, 28 (6), 544–549. [DOI] [PubMed] [Google Scholar]
- Novaes A. B. Jr.; Shapiro L.; Fillios L. C.; Wood N. Gingival fluid fucose to protein ratios as indicators of the severity of periodontal disease. J. Periodontol. 1980, 51 (2), 88–94. [DOI] [PubMed] [Google Scholar]
- Lamster I. B.; Hartley L. J.; Oshrain R. L.; Gordon J. M. Evaluation and Modification of Spectrophotometric Procedures for Analysis of Lactate-Dehydrogenase, Beta-Glucuronidase and Arylsulfatase in Human Gingival Crevicular Fluid Collected with Filter-Paper Strips. Arch. Oral Biol. 1985, 30 (3), 235–242. [DOI] [PubMed] [Google Scholar]
- Offenbacher S.; Beck J.; Obste M. O. T. R. Effects of Periodontal Therapy on Rate of Preterm Delivery: A Randomized Controlled Trial Reply. Obstet. Gynecol. 2010, 115 (2), 386–386. [DOI] [PubMed] [Google Scholar]
- Offenbacher S.; Barros S.; Mendoza L.; Mauriello S.; Priesser J.; Moss K.; de Jager M.; Apsiras M. Changes in gingival crevicular fluid inflammatory mediator levels during the induction and resolution of experimental gingivitis in humans. J. Clin. Periodontol. 2010, 37 (4), 324–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannopoulou C.; Andersen E.; Demeurisse C.; Cimasoni G. Neutrophil Elastase and Its Inhibitors in Human Gingival Crevicular Fluid during Experimental Gingivitis. J. Dent. Res. 1992, 71 (2), 359–363. [DOI] [PubMed] [Google Scholar]
- Moughal N. A.; Adonogianaki E.; Thornhill M. H.; Kinane D. F. Endothelial-Cell Leukocyte Adhesion Molecule-1 (Elam-1) and Intercellular-Adhesion Molecule-1 (Icam-1) Expression in Gingival Tissue during Health and Experimentally-Induced Gingivitis. J. Periodontal Res. 1992, 27 (6), 623–630. [DOI] [PubMed] [Google Scholar]
- Chapple I. L. C.; Garner I.; Saxby M. S.; Moscrop H.; Matthews J. B. Prediction and diagnosis of attachment loss by enhanced chemiluminescent assay of crevicular fluid alkaline phosphatase levels. J. Clin. Periodontol. 1999, 26 (3), 190–198. [DOI] [PubMed] [Google Scholar]
- Que M. L.; Andersen E.; Mombelli A. Myeloid-related protein (MRP)8/14 (calprotectin) and its subunits MRP8 and MRP14 in plaque-induced early gingival inflammation. J. Clin. Periodontol. 2004, 31 (11), 978–984. [DOI] [PubMed] [Google Scholar]
- Salvi G. E.; Franco L. M.; Braun T. M.; Lee A.; Persson G. R.; Lang N. P.; Giannobile W. V. Pro-inflammatory biomarkers during experimental gingivitis in patients with type 1 diabetes mellitus: a proof-of-concept study. J. Clin. Periodontol. 2010, 37 (1), 9–16. [DOI] [PubMed] [Google Scholar]
- Dommisch H.; Vorderwulbecke S.; Eberhard J.; Steglich M.; Jepsen S. SELDI-TOF-MS of gingival crevicular fluid-A methodological approach. Arch. Oral Biol. 2009, 54 (9), 803–809. [DOI] [PubMed] [Google Scholar]
- Lundy F. T.; Orr D. F.; Shaw C.; Lamey P.; Linden G. J. Detection of individual human neutrophil alpha-defensins (human neutrophil peptides 1, 2 and 3) in unfractionated gingival crevicular fluid - A MALDI-MS approach. Mol. Immunol. 2005, 42 (5), 575–579. [DOI] [PubMed] [Google Scholar]
- Pisano E.; Cabras T.; Montaldo C.; Piras V.; Inzitari R.; Olmi C.; Castagnola M.; Messana I. Peptides of human gingival crevicular fluid determined by HPLC-ESI-MS. Eur. J. Oral Sci. 2005, 113 (6), 462–468. [DOI] [PubMed] [Google Scholar]
- Fenn J. B.; Mann M.; Meng C. K.; Wong S. F.; Whitehouse C. M. Electrospray Ionization for Mass-Spectrometry of Large Biomolecules. Science 1989, 246 (4926), 64–71. [DOI] [PubMed] [Google Scholar]
- Ngo L. H.; Veith P. D.; Chen Y. Y.; Chen D.; Darby I. B.; Reynolds E. C. Mass spectrometric analyses of peptides and proteins in human gingival crevicular fluid. J. Proteome Res. 2010, 9 (4), 1683–1693. [DOI] [PubMed] [Google Scholar]
- Bostanci N.; Heywood W.; Mills K.; Parkar M.; Nibali L.; Donos N. Application of Label-Free Absolute Quantitative Proteomics in Human Gingival Crevicular Fluid by LC/MSE (Gingival Exudatome). J. Proteome Res. 2010, 9, 2191–2199. [DOI] [PubMed] [Google Scholar]
- Armirotti A. Bottom-Up Proteomics. Curr. Anal. Chem. 2009, 5 (2), 116–130. [Google Scholar]
- Wells J. M.; McLuckey S. A. Collision-induced dissociation (CID) of peptides and proteins. Biol. Mass Spectrom. 2005, 402, 148–185. [DOI] [PubMed] [Google Scholar]
- Roepstorff P.; Fohlman J. Proposal for a Common Nomenclature for Sequence Ions in Mass-Spectra of Peptides. Biomed. Mass Spectrom. 1984, 11 (11), 601–601. [DOI] [PubMed] [Google Scholar]
- Perkins D. N.; Pappin D. J. C.; Creasy D. M.; Cottrell J. S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20 (18), 3551–3567. [DOI] [PubMed] [Google Scholar]
- Eng J. K.; Mccormack A. L.; Yates J. R. An Approach to Correlate Tandem Mass-Spectral Data of Peptides with Amino-Acid-Sequences in a Protein Database. J. Am. Soc. Mass Spectrom. 1994, 5 (11), 976–989. [DOI] [PubMed] [Google Scholar]
- Geer L. Y.; Markey S. P.; Kowalak J. A.; Wagner L.; Xu M.; Maynard D. M.; Yang X. Y.; Shi W. Y.; Bryant S. H. Open mass spectrometry search algorithm. J. Proteome Res. 2004, 3 (5), 958–964. [DOI] [PubMed] [Google Scholar]
- Mann M. Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell Biol. 2006, 7 (12), 952–958. [DOI] [PubMed] [Google Scholar]
- DeSouza L.; Diehl G.; Rodrigues M. J.; Guo J.; Romaschin A. D.; Colgan T. J.; Siu K. W. M. Search for cancer markers from endometrial tissues using differentially labeled tags iTRAQ and clCAT with multidimensional liquid chromatography and tandem mass spectrometry. J. Proteome Res. 2005, 4 (2), 377–386. [DOI] [PubMed] [Google Scholar]
- Kaspar H.; Dettmer K.; Chan Q.; Daniels S.; Nimkar S.; Daviglus M. L.; Stamler J.; Elliott P.; Oefner P. J. Urinary amino acid analysis: A comparison of iTRAQ (R)-LC-MS/MS, GC-MS, and amino acid analyzer. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2009, 877 (20−21), 1838–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralhan R.; DeSouza L. V.; Matta A.; Tripathi S. C.; Ghanny S.; DattaGupta S.; Thakar A.; Chauhan S. S.; Siu K. W. M. iTRAQ-Multidimensional Liquid Chromatography and Tandem Mass Spectrometry-Based Identification of Potential Biomarkers of Oral Epithelial Dysplasia and Novel Networks between Inflammation and Premalignancy. J. Proteome Res. 2009, 8 (1), 300–309. [DOI] [PubMed] [Google Scholar]
- DeSouza L.; Diehl G.; Rodrigues M. J.; Guo J. Z.; Romaschin A. D.; Colgan T. J.; Siu K. W. M. Search for cancer markers from endometrial tissues using differentially labeled tags iTRAQ and clCAT with multidimensional liquid chromatography and tandem mass spectrometry. J. Proteome Res. 2005, 4 (2), 377–386. [DOI] [PubMed] [Google Scholar]
- Phanstiel D.; Unwin R.; McAlister G. C.; Coon J. J. Peptide Quantification Using 8-Plex Isobaric Tags and Electron Transfer Dissociation Tandem Mass Spectrometry. Anal. Chem. 2009, 81 (4), 1693–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham C.; Glish G. L.; Burinsky D. J. High amplitude short time excitation: A method to form and detect low mass product ions in a quadrupole ion trap mass spectrometer. J. Am. Soc. Mass Spectrom. 2006, 17 (1), 81–84. [DOI] [PubMed] [Google Scholar]
- Guo T.; Gan C. S.; Zhang H.; Zhu Y.; Kon O. L.; Sze S. K. Hybridization of Pulsed-Q Dissociation and Collision-Activated Dissociation in Linear Ion Trap Mass Spectrometer for iTRAQ Quantitation. J. Proteome Res. 2008, 7 (11), 4831–4840. [DOI] [PubMed] [Google Scholar]
- Comisaro M. B.; Marshall A. G. Fourier-Transform Ion-Cyclotron Resonance Spectroscopy. Chem. Phys. Lett. 1974, 25 (2), 282–283. [Google Scholar]
- Lobene R. R.; Soparkar P. M.; Newman M. B. Use of dental floss. Effect on plaque and gingivitis. Clin. Prev. Dent. 1982, 4 (1), 5–8. [PubMed] [Google Scholar]
- Loe H. Gingival Index Plaque Index and Retention Index Systems. J. Periodontol. 1967, 38 (6p2), 610–. [DOI] [PubMed] [Google Scholar]
- Chapple I. L. C.; Landini G.; Griffiths G. S.; Patel N. C.; Ward R. S. N. Calibration of the Periotron 8000 (R) and 6000 (R) by polynomial regression. J. Periodontal Res. 1999, 34 (2), 79–86. [DOI] [PubMed] [Google Scholar]
- Socransky S. S.; Haffajee A. D.; Cugini M. A.; Smith C.; Kent R. L. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998, 25 (2), 134–144. [DOI] [PubMed] [Google Scholar]
- Barr G.; Dong W.; Gilmore C. J. PolySNAP3: a computer program for analysing and visualizing high-throughput data from diffraction and spectroscopic sources. J. Appl. Crystallogr. 2009, 42, 965–974. [Google Scholar]
- Rowat J. S.; Squier C. A. Rates of Epithelial-Cell Proliferation in the Oral-Mucosa and Skin of the Tamarin Monkey (Saguinus-Fuscicollis). J. Dent. Res. 1986, 65 (11), 1326–1331. [DOI] [PubMed] [Google Scholar]
- Brecx M. C.; Schlegel K.; Gehr P.; Lang N. P. Comparison between Histological and Clinical-Parameters during Human Experimental Gingivitis. J. Dent. Res. 1986, 65, 182–182. [DOI] [PubMed] [Google Scholar]
- Seymour G. J.; Powell R. N.; Cole K. L.; Aitken J. F.; Brooks D.; Beckman I.; Zola H.; Bradley J.; Burns G. F. Experimental Gingivitis in Humans - a Histochemical and Immunological Characterization of the Lymphoid-Cell Subpopulations. J. Periodontal Res. 1983, 18 (4), 375–385. [DOI] [PubMed] [Google Scholar]
- Saglie F. R.; Pertuiset J. H.; Rezende M. T.; Sabet M. S.; Raoufi D.; Carranza F. A. Bacterial invasion in experimental gingivitis in man. J. Periodontol. 1981, 58, 837–846. [DOI] [PubMed] [Google Scholar]
- Warfvinge K.; Elofsson R. Single Modified Cilia Displayed by Cells of Human Internal Stratified Epithelia (Oral Cavity, Vagina). Cell Tissue Res. 1988, 251 (2), 237–241. [DOI] [PubMed] [Google Scholar]
- Veland I. R.; Awan A.; Pedersen L. B.; Yoder B. K.; Christensen S. T. Primary Cilia and Signaling Pathways in Mammalian Development, Health and Disease. Nephron Physiol. 2009, 111 (3), 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs T. A.; Abed U.; Goosmann C.; Hurwitz R.; Schulze I.; Wahn V.; Weinrauch Y.; Brinkmann V.; Zychlinsky A. Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 2007, 176 (2), 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitkov L.; Klappacher M.; Hannig M.; Krautgartner W. D. Extracellular neutrophil traps in periodontitis. J. Periodontal Res. 2009, 44 (5), 664–672. [DOI] [PubMed] [Google Scholar]
- Matthews J. B.; Wright H. J.; Roberts A.; Ling-Mountford N.; Cooper P. R.; Chapple I. L. C. Neutrophil hyper-responsiveness in periodontitis. J. Dental Res. 2007, 86 (8), 718–722. [DOI] [PubMed] [Google Scholar]
- Matthews J. B.; Wright H. J.; Roberts A.; Cooper P. R.; Chapple I. L. C. Hyperactivity and reactivity of peripheral blood neutrophils in chronic periodontitis. Clin. Exp. Immunol. 2007, 147 (2), 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling P.; Matthews G. Structure and function of ribbon synapses. Trends Neurosci. 2005, 28 (1), 20–29. [DOI] [PubMed] [Google Scholar]
- Linden G. J.; McKinnell J.; Shaw C.; Lundy F. T. Substance P and neurokinin A in gingival crevicular fluid in periodontal health and disease. J. Clin. Periodontol. 1997, 24 (11), 799–803. [DOI] [PubMed] [Google Scholar]
- Offenbacher S.; Barros S. P.; Paquette D. W.; Winston J. L.; Biesbrock A. R.; Thomason R. G.; Gibb R. D.; Fulmer A. W.; Tiesman J. P.; Juhlin K. D.; Wang S. L.; Reichling T. D.; Chen K. S.; Ho B. Gingival Transcriptome Patterns During Induction and Resolution of Experimental Gingivitis in Humans. J. Periodontol. 2009, 80 (12), 1963–1982. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.