Abstract
Ethanol metabolism in the liver induces oxidative stress and altered cytokine production preceding myofibroblast activation and fibrogenic responses. The purpose of this study was to determine how ethanol affects the fibrogenic response in precision-cut liver slices (PCLS). PCLS were obtained from chow-fed male Wistar rats (200–300 g) and were cultured up to 96 h in medium, 25 mM ethanol, or 25 mM ethanol and 0.5 mM 4-methylpyrazole (4-MP), an inhibitor of ethanol metabolism. Slices from every time point (24, 48, 72, and 96 h) were examined for glutathione (GSH) levels, lipid peroxidation [thiobarbituric acid-reactive substance (TBARS) assay], cytokine production (ELISA and RT-PCR), and myofibroblast activation [immunoblotting and immunohistochemistry for smooth muscle actin (SMA) and collagen]. Treatment of PCLS with 25 mM ethanol induced significant oxidative stress within 24 h, including depletion of cellular GSH and increased lipid peroxidation compared with controls (P < 0.05). Ethanol treatment also elicited a significant and sustained increase in interleukin-6 (IL-6) production (P < 0.05). Importantly, ethanol treatment accelerates a fibrogenic response after 48 h, represented by significant increases in SMA and collagen 1α(I) production (P < 0.05). These ethanol-induced effects were prevented by the addition of 4-MP. Ethanol metabolism induces oxidative stress (GSH depletion and increased lipid peroxidation) and sustained IL-6 expression in rat PCLS. These phenomena precede and coincide with myofibroblast activation, which occurs within 48 h of treatment. These results indicate the PCLS can be used as in vitro model for studying multicellular interactions during the early stages of ethanol-induced liver injury and fibrogenesis.
Keywords: smooth muscle actin, collagen, IL-6, oxidative stress
ethanol-induced liver fibrosis is a pronounced wound-healing response due to the chronic assault by multiple factors. These factors may include, but are not limited to elevated lipopolysaccharide (LPS) plasma levels (ethanol-induced leaky gut), generation of reactive oxygen species (ROS) with concomitant antioxidant depletion and pericentral hypoxia, formation of toxic and profibrogenic ethanol metabolites (acetaldehyde, hydroxyethyl radicals, lipid peroxidation products), and a complex inflammatory response. Exposure to these factors results in increased hepatocellular damage and activation of hepatic stellate cells (HSC) and myofibroblasts (14).
Currently many animal and in vitro models are used to examine the mechanism(s) and potential treatment strategies associated with alcoholic liver injury and disease. In vivo animal models that induce liver damage and fibrosis include voluntary and involuntary ethanol feeding, chemical or surgical treatment, and diet modification in conjunction with ethanol feeding (18, 35, 36). In vitro models primarily consist of cell lines, which represent specific cell types within the liver. These include hepatoma cell lines; hepatocyte-like cell lines; various stellate, macrophage/Kupffer, or endothelial cell lines; and isolated primary cells (4, 5a, 6, 13, 26, 32). All of these models have limitations for conducting research in alcoholic liver disease. In vivo models do not develop overt liver fibrosis (voluntary feeding) with just alcohol alone, and chemical or surgical treatments induce fibrosis that is not consistent with or representative of human alcoholic liver injury. Additional drawbacks include difficulties in examining the early events that occur after alcohol treatment. The in vitro models are limited in that they represent the effects of ethanol on single cell types. These models do not take into account the potential cell-cell and cell-matrix interactions that occur in three-dimensional liver architecture.
To address some of these concerns with the current models of liver injury and fibrosis, an ex vivo/in vitro model representative of the whole organ has been developed that shows significant ethanol-induced damage in as little as 24 h. This model uses precision-cut liver slices (PCLS) originating from Wistar rats, cultured in the presence or absence of ethanol in a roller system (20). Over a 96-h time period, this model efficiently metabolizes ethanol, produces acetaldehyde, develops a reduced redox state and steatosis, and exhibits impaired albumin secretion (20). All of these phenomena are characteristics of early liver injury. Interestingly, in the presence of 4-methylpyrazole (4-MP), an inhibitor of ethanol metabolism, all of the ethanol-mediated effects are ameliorated or significantly reduced, indicating the metabolites of ethanol are responsible for the damage.
Previous studies using PCLS cultured in media only have shown that a moderate wound-healing response (fibrogenesis) occurs by 72 to 96 h as assessed by collagen deposition (34). Addition of CCl4, a known inducer of liver injury and fibrosis, to PCLS in culture induces a more robust fibrogenic response, which occurs by 48 h (31). Therefore, the purpose of this study was to determine whether PCLS treated with ethanol develop overt fibrogenesis within the 96-h culture period, consistent with previous results using this model, indicating that ethanol metabolism induces liver injury similar to what occurs in vivo (20). Expression of smooth muscle actin (SMA) and collagen 1α(I) were examined as markers of fibrogenesis. Markers of oxidative stress and cytokine profiles were also examined as possible predictors and/or causes of ethanol-induced fibrogenesis in this model.
MATERIALS AND METHODS
Male Wistar rats were purchased from Charles River Laboratories (Wilmington, MA) and maintained on a Purina rat chow diet. All animals were allowed free access to their food and water up to 1 h prior to euthanasia. All procedures were approved by the Animal Subcommittee of the Omaha Veterans Affairs Medical Center and are in accordance with the National Institutes of Health Guidelines on the Use of Laboratory Animals.
PCLS
PCLS were prepared as previously published (20). Rats weighing 200–300 g were anesthetized with isoflurane. The liver was excised and placed into oxygenated V-7 cold preservation buffer (Vitron, Tucson, AZ). Cylindrical tissue cores (8 mm) were cut using a hand-held coring tool, loaded into the Vitron tissue slicer. Slices (250-μm thickness) were cut using a 45-mm rotary blade and floated into ice-cold oxygenated V-7 preservation buffer. Slices were equilibrated by gentle shaking in serum-free Williams E medium (Sigma Chemical, St. Louis, MO) containing d-glucose and gentamicin (WEGG) under 95% O2-5% CO2 (carbogen) at 37°C for 30 min. Some slices were processed at this point and were designated as time 0 (t0). The remaining slices were floated onto a titanium screen containing rollers from Vitron. These rollers were inserted into sterile 20-ml glass vials containing 1.7 ml of serum-free WEGG medium, WEGG medium containing 25 mM ethanol, or WEGG medium containing 25 mM ethanol and 0.5 mM 4-MP. The vials were capped with lids containing a 1-mm hole for oxygen transfer. Vials were placed into the dynamic organ culture incubator from Vitron and were incubated at 37°C with carbogen (1.5 l/min). The media was replenished every 24 h up to 96 h.
Ethanol Metabolism and Viability Measurements
Ethanol metabolism and acetaldehyde (AA) production was measured in media at each time point by headspace gas chromatography as described previously (30). Viability was determined by measuring lactate dehydrogenase activity in the slices and corresponding media from each time point as described previously (20).
GSH Assay
PCLS were incubated as described above. At each time point, PCLS were rinsed with PBS, pH 7.2, and assayed for total cellular glutathione by use of the total glutathione detection system from Assay Designs (Ann Arbor, MI). Briefly, slices were sonicated on ice in PBS, pH 7.2 (150 μl PBS/slice). Protein concentrations were determined by using a BCA protein assay kit from Pierce (Rockford, IL). Ice-cold 10% (wt/vol) metaphosphoric acid was added to each sonicate (5% final), and after mixing the samples were centrifuged at 14,000 g 10 min at 4°C. The clarified supernatant was diluted 1:10, and 25 μl was serially diluted in duplicate on microtiter plates. The reaction mix containing glutathione reductase was added to each well, and the absorbance at 410 nm was measured at 1-min intervals for 5 min by using a MRXII plate reader with Revelation Software (Dynex Technologies, Chantilly, VA). Cellular glutathione (GSH) concentrations were calculated against a GSH standard and were normalized to the total protein detected in the samples. To determine the amount of oxidized glutathione (GSSG) in the samples, aliquots of the supernatants and GSSG standards were incubated with 4-vinylpyridine (40 mM final) 1 h at room temperature (RT) according to the manufacturer's instructions. These samples were serially diluted and assayed as above.
Lipid Peroxidation
PCLS were analyzed for lipid peroxidation by use of the thiobarbituric acid-reactive substance (TBARS) assay system from Cayman Chemical (Ann Arbor, MI). Briefly, PCLS were lysed with PBS-RIPA buffer [PBS, pH 7.4, 0.5% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 1 mM Na-EDTA, and 5 μl/ml protease inhibitor cocktail (Sigma Chemical)] as described previously (25). The supernatant was assayed for protein concentration and TBARS. Samples (30 μl) and malondialdehyde (MDA) standards were incubated with 20 μl of SDS solution and 1 ml of the color reagent containing thiobarbituric acid for 1 h in a boiling water bath according to the manufacturer's instructions. The samples were placed on ice for 10 min and centrifuged at 1,600 g for 10 min at 4°C. Samples and standards were placed in a microtiter plate, and the absorbance at 540 nm was measured. The amount of MDA was calculated against the standard curve and was normalized to the amount of protein in each sample.
Analysis of SMA and Collagen 1α(I)
RIPA lysates from PCLS equivalent to 50 μg were resolved under reducing conditions by 10% SDS-PAGE for the detection of SMA. For collagen 1α(I) forms, lysates equivalent to 100 μg were resolved under reducing conditions by 8% SDS-PAGE. Proteins were transferred to Immun-Blot polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA). Membranes were blocked 30 min in Odyssey blocking buffer (LI-COR, Lincoln, NE) at 37°C. Blots were incubated with either a 1:2,000 dilution of anti-SMA mouse monoclonal antibody (Sigma Chemical) or a 1:1,000 anti-collagen 1α(I) rabbit polyclonal antibody (Rockland Immunochemicals, Gilbertsville, PA) followed by 1:15,000 dilutions of IRDye conjugated anti-mouse or anti-rabbit secondary antibodies (LI-COR). Blots were scanned with an Odyssey IR Scanner (LI-COR). Data for each time point and condition were expressed as the densitometric volume of SMA or collagen 1α(I) relative to the densitometric volume of the t0 sample. Tubulin was used (1:4,000 anti-tubulin mouse monoclonal antibody; Sigma Chemical) as the loading control for each lane.
Immunohistochemical Analysis
At each time point, PCLS were rinsed with PBS, pH 7.4, and fixed overnight in 10% buffered formalin. Paraffin embedding, section cutting, and picrosirius red staining of the fixed PCLS were performed by the Histology Core facility at the University of Nebraska Center, Omaha, NE. Slides were analyzed by using a Nikon Eclipse 80i at 10× power and Nis-Elements 3.0 software (Nikon, Melville, NY). Digital photographs were then subjected to histomorphometry to quantitate the amount of Sirius red staining present (24). Briefly, images were subjected to color deconvolution and threshold analysis of 10 random fields per slide. MBF-ImageJ software was used to quantify the amount of intensity for each field (7).
ELISA Analysis for IL-6
Media for each condition were collected after 24, 48, 72 and 96 h of incubation. Detection of media IL-6 was accomplished using OptEIA ELISA (Pharmingen, Palo Alto, CA) according to the manufacturer's instructions. Briefly, ELISA plates (Immunlon IV; Dynatech, Chantilly, VA) were coated with 100 μl/well of capture antibody diluted in coating buffer (0.1 M carbonate, pH 9.5). The plates were incubated overnight at RT. Plates were washed and blocked for 1 h at RT with 200 μl/well assay diluent. The IL-6 standard and samples (100 μl) were pipetted into appropriate wells. Plates were sealed and incubated at RT for 2 h. After washing, 100 μl of working detector was added to each well, sealed, and incubated for 1 h at RT. After washing, 100 μl of substrate solution (1:1 substrate A/substrate B) was added to each well and incubated for 30 min at RT in the dark. Stop solution (2 N H2SO4, 50 μl/well) was added and plates were read at 450 nm (570 nm correction) on a MR7000 MicroPlate Reader (Dynatech).
Real-Time PCR Analysis for Cytokines
RNA extraction.
RNA was extracted from rat PCLS by using an RNeasy Mini Kit per the Qiagen protocol (Qiagen, Valencia, CA). RNA was quantified and its integrity was verified by electrophoresis of 1 μg RNA on a 1% agarose gel stained with ethidium bromide. Gel bands were detected by exposure on a Fluor-S multiimager and analyzed by Quantity One software (Bio-Rad).
RNA expression assay.
RNA samples from PCLS were converted into cDNA by using a High Capacity cDNA Archive system from Applied Biosystems (Foster City, CA) according to the manufacturer's instructions. Briefly, a master mix containing reverse transcription buffer, dNTPs, random primers, MultiScribe reverse transcriptase, and nuclease-free water was incubated with 50 ng of RNA for 10 min at RT and 120 min at 37°C. The cDNA was subjected to RT-PCR using TaqMan Gene Expression Assay primers for IL-6 from Applied Biosystems. Using a 7500 Real-Time PCR System from Applied Biosystems, we amplified cDNA by using amplification steps set up by the manufacturer designed to work with the TaqMan primers as per Applied Biosystems protocol for RNA expression. Message levels were analyzed via the 7500 software, and data were expressed in fold change in mRNA expression from t0.
Statistical Analysis
The results were expressed as means ± SE. Comparisons between PCLS treated with ethanol and untreated controls at each time point were made via ANOVA followed by a post hoc t-test; P < 0.05 was considered significant.
RESULTS
Ethanol Treatment Accelerates a Fibrogenic Response in Rat PCLS
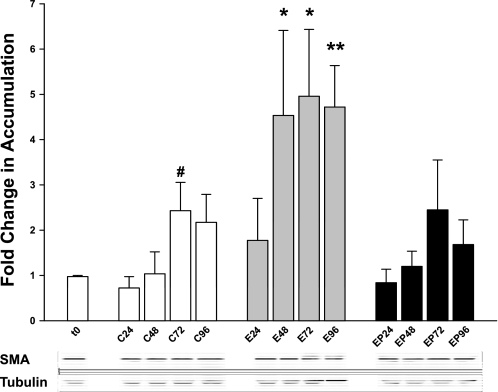
Rat PCLS were previously determined to be a good model for studying alcohol-induced liver injury (20). The next step was to determine whether ethanol treatment could induce or accelerate a fibrogenic response in the PCLS. SMA is a well-used indicator of fibrogenesis and is expressed in myofibroblast-type cells, which in the liver consist primarily of activated HSC and portal fibroblasts. Initially, the ability of rat PCLS to metabolize ethanol and the efficacy of 4-MP to inhibit ethanol metabolism were confirmed by headspace gas chromatography (Table 1) on media from each time point. These results are consistent with those reported previously (20), indicating that rat PCLS efficiently metabolize ethanol and produce AA throughout 96 h. Also, addition of 0.5 mM 4-MP concurrently with 25 mM ethanol effectively inhibits ethanol metabolism and production of AA. Slices from each time point and condition were lysed and used for immunoblot analysis. Blots were probed for SMA and tubulin (reference protein). Densitometric analysis revealed that ethanol treatment (E) induced the accumulation of SMA within 48 h, a 4.5-fold increase over t0 and a 4.4-fold increase over the corresponding control (C) (Fig. 1, t0 = 1.01 ± 0.34; C48 = 1.04 ± 0.53; E48 = 4.53 ± 2.06 relative units). The ethanol-induced increase in SMA accumulation was sustained over the 96-h time period, maintaining almost a fivefold increase over t0 (Fig. 1, E72 = 4.96 ± 1.62; E96 = 4.72 ± 1.0 relative units) and an approximate twofold increase over corresponding controls (C72 = 2.48 ± 0.68; C96 = 2.17 ± 0.68 relative units). Addition of 4-MP concurrently with ethanol (EP) prevented the ethanol-induced increase in SMA accumulation at all time points (Fig. 1), indicating that ethanol metabolism and metabolites are responsible for SMA upregulation.
Table 1.
Ethanol metabolism and acetaldehyde production
| Condition | Ethanol Metabolized, μmol•mg protein−1•24 h−1 | Acetaldehyde Produced, μM |
|---|---|---|
| 24 h | ||
| E | 13.64 ± 1.99 | 8.0 ± 2.0 |
| EP | 4.23 ± 0.56 | 6.0 ± 3.0 |
| 48 h | ||
| E | 12.96 ± 3.19 | 52.0 ± 24.8 |
| EP | 2.42 ± 0.52 | 0.0 ± 0.0 |
| 72 h | ||
| E | 9.61 ± 2.68 | 102.0 ± 33.6 |
| EP | 4.37 ± 1.44 | 2.0 +/−2.2 |
| 96 h | ||
| E | 9.02 ± 1.01 | 122.0 ± 44.3 |
| EP | 3.45 ± 0.75 | 4.0 ± 2.7 |
Rat precision-cut liver slices (PCLS) were cultured ≤96 h in the absence or presence of 25 mM ethanol and/or 0.5 mM 4-methylpyrazole (4-MP). E, presence of 25 mM ethanol; EP, ethanol + 0.5 mM 4-MP. Every 24 h, samples were analyzed by headspace gas chromatography for ethanol and acetaldehyde concentrations as described in materials and methods. Note that 4-MP effectively inhibited both ethanol metabolism and acetaldehyde production as observed previously (20); n = 5 different rat experiments.
Fig. 1.
Smooth muscle actin (SMA) accumulation in rat precision-cut liver slices (PCLS) treated with 25 mM ethanol and/or 4-methylpyrazole (4-MP). Rat PCLS from all treatment groups and times were lysed with RIPA as described in materials and methods. Lysates were analyzed by SDS-PAGE and immunoblotting as described. Accumulation of SMA at each time point was normalized to time 0 (t0). Tubulin was used as a loading control. Representative bands are shown below the graph. C, control; E, ethanol; EP, ethanol + 4-MP; numbers represent hours of treatment. #P < 0.05 compared with t0; *P < 0.05 compared with corresponding controls; **P < 0.001 compared with corresponding control; n = 6 different rat experiments.
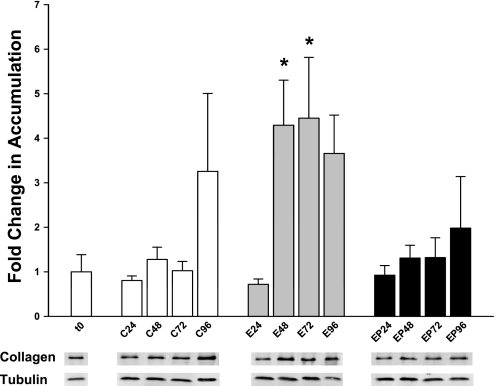
An additional marker of fibrogenesis is the appearance of collagen 1α(I), produced by activated myofibroblasts. Since ethanol treatment amplified and accelerated SMA accumulation, rat PCLS were examined for collagen 1α(I) deposition. To examine collagen 1α(I) accumulation, slices from each time point and condition were lysed and used for immunoblotting analysis. Blots were probed for collagen 1α(I) and tubulin. Densitometric analysis showed that collagen 1α(I) accumulation was unchanged after 24 h of culture, regardless of treatment (Fig. 2). At 48 h, ethanol treatment induced a 4.3-fold increase in collagen accumulation over t0 (t0 = 1.0 ± 0.42; E48 = 4.29 ± 1.01 relative units). This increase was sustained over 96 h (E72 = 4.45 ± 1.36; E96 = 3.66 ± 0.86). In contrast, collagen 1α(I) accumulation in control PCLS wasn't increased until 96 h, and even then it was not significantly different from t0 (C96 = 3.26 ± 1.76 relative units). Addition of 4-MP simultaneously with ethanol prevented the ethanol-induced increase in collagen 1α(I). These results are similar to those found for SMA accumulation and indicate that myofibroblast-like cells are activated by ethanol metabolism.
Fig. 2.
Collagen accumulation in PCLS treated with 25 mM ethanol and/or 4-MP up to 96 h. Rat PCLS were treated as described in materials and methods. Every 24 h, slices were lysed with RIPA as described and subjected to SDS-PAGE and immunoblotting. Collagen accumulation for each time point was normalized to t0. Tubulin was used as a loading control. Representative bands corresponding to collagen and tubulin for each time point and condition are shown below the graph. *P < 0.05; n = 5 different rat experiments.
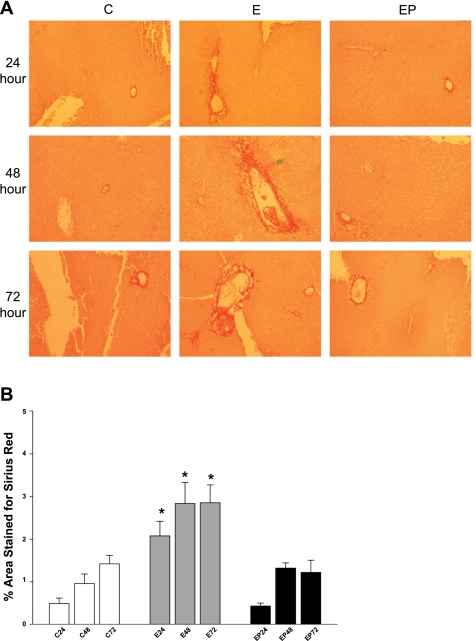
PCLS were also analyzed by immunohistochemistry for collagen deposition. Slices incubated in the presence or absence of 25 mM ethanol or 4-MP were fixed and paraffin embedded. Slides were cut and stained with picrosirius red, which stains fibrillar collagen. Analysis of staining revealed that collagen deposition in the tissue became pronounced at 24 h of ethanol exposure and became more intense and widespread at 48 and 72 h (Fig. 3A). In contrast, staining of control slices revealed moderate collagen deposition after 48 h that continued through 72 h. Treatment of slices with ethanol and 4-MP prevented the collagen deposition observed with ethanol alone. Sirius red staining was confirmed quantitatively by using MBF-ImageJ Software (Fig. 3B). These results indicate that ethanol metabolism induces myofibroblast activation and a fibrogenic response, consistent with the SMA and collagen 1α(I) immunoblot data described above (Figs. 1 and 2).
Fig. 3.
A: Sirius red stain of PCLS. PCLS were incubated in the absence (C) or presence of 25 mM ethanol (E) or ethanol + 0.5 mM 4-MP (EP) up to 72 h. At each point, PLCS were rinsed, fixed with buffered formalin, and embedded in paraffin. Sections (4 μm thick) were stained with Sirius red. Slides were analyzed with a Nikon Eclipse 80i at 10× power and Nis-Elements 3.0 software (Nikon, Melville, NY). Photographs are representative of 4 separate animal experiments. B: quantification of Sirius red staining of PCLS. PCLS were incubated in the absence or presence of 25 mM ethanol or ethanol + 0.5 mM 4-MP up to 72 h. Samples were stained for Sirius red and photographed. Pictures were deconvoluted, subjected to threshold analysis, and quantified by use of MBF-ImageJ software. PCLS slides from 4 separate animals using 10 random fields per slide were analyzed. *P < 0.01 compared with control or ethanol + 4-MP.
Ethanol Induces Oxidative Stress in Rat PCLS That Precedes Fibrogenesis
Since ethanol metabolism and metabolites amplified and accelerated the fibrogenic response in rat PCLS, the next goal was to examine possible causes of the response. Previous studies in other models have associated ethanol-induced oxidative stress with a fibrogenic response (1, 8, 15, 39). Some common indicators for oxidative stress include depletion of GSH and increased lipid peroxidation.
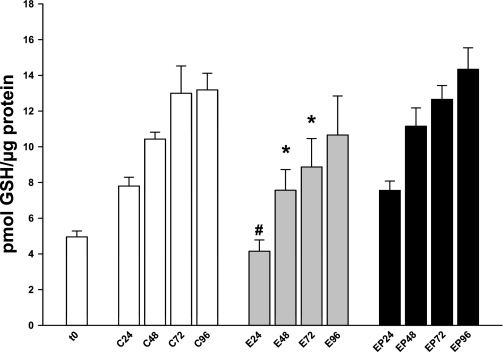
Initial studies examined GSH levels (Fig. 4, Table 2). At 24 h, control PCLS expressed 7.8 ± 0.55 pmol GSH/μg protein, whereas the ethanol-treated slices had only 53% of control levels (4.15 ± 0.71 pmol/μg). This trend continued through 72 h. The ethanol treated slices had consistently and significantly lower GSH levels than controls [48 h, C = 10.43 ± 0.43 pmol/μg vs. E = 7.56 ± 1.29 pmol/μg (73% of C); 72 h, C = 13.0 ± 1.71 vs. E = 8.87 ± 1.78 (68% of C)]. By 96 h, the ethanol-treated slices, although still lower than the corresponding controls, had more than doubled the total GSH observed at 24 h. GSH in the controls also increased over the time period, consistent with a previous study using PCLS (34). Addition of 4-MP concurrently with ethanol treatment prevented the ethanol-induced GSH depletion, indicating ethanol metabolism is responsible for the lower GSH levels (Fig. 4, Table 2). Further experiments to determine the amount of GSSG showed that almost no GSSG was present, suggesting the depletion may occur due an ethanol-induced efflux of GSH from the PCLS into the media as shown previously (5).
Fig. 4.
Total cellular GSH levels. Rat PCLS were treated with ethanol in the absence or presence of 4-MP as described in materials and methods. Every 24 h, slices were analyzed for total cellular GSH by using a kit according to manufacturer's instructions. #P < 0.001 compared with corresponding control; *P < 0.05 compared with corresponding control; n = 5 different rat experiments.
Table 2.
Comparison of oxidative stress and viability
| Condition | GSH Level, pmol GSH/μg protein | Lipid Peroxidation, pmol TBARS/μg protein | % Cytotoxicity |
|---|---|---|---|
| t0 | 4.95 ± 0.37 | 1.08 ± 0.16 | 0.0 |
| 24 h | |||
| C | 7.80 ± 0.55 | 1.59 ± 0.50 | 6.93 ± 1.31 |
| E | 4.15 ± 0.71c | 3.24 ± 0.61b | 6.45 ± 1.35 |
| EP | 7.55 ± 0.59 | 1.26 ± 0.49 | 1.81 ± 0.97e |
| 48 h | |||
| C | 10.43 ± .43 | 1.73 ± 0.25 | 9.89 ± 1.37 |
| E | 7.56 ± 1.29a | 3.28 ± 0.45b | 13.07 ± 3.78 |
| EP | 11.15 ± 1.14 | 1.41 ± 0.34 | 2.74 ± 1.44e |
| 72 h | |||
| C | 13.00 ± 1.71 | 1.69 ± 0.32 | 12.56 ± 2.26 |
| E | 8.87 ± 1.78a | 2.89 ± 0.44b | 20.86 ± 5.95 |
| EP | 12.65 ± 0.87 | 1.33 ± 0.33 | 5.51 ± 2.87f |
| 96 h | |||
| C | 13.18 ± 1.04 | 2.01 ± 0.54d | 23.46 ± 3.36 |
| E | 10.66 ± 2.45 | 4.23 ± 0.76a | 23.07 ± 0.97 |
| EP | 14.33 ± 1.35 | 2.31 + 1−0.37d | 12.80 ± 7.81 |
Rat PCLS were cultured ≤96 h in the absence or presence of 25 mM ethanol and/or 0.5 mM 4-MP. Every 24 h, samples were analyzed for GSH levels, lipid peroxidation [thiobarbituric acid-reactive substance (TBARS)], and cytotoxicity (LDH leakage into media) as described in materials and methods. C, absence of ethanol; t0, time 0.
P < 0.05, compared with the corresponding control;
P < 0.005 compared with the corresponding control;
P < 0.001, compared with the corresponding control;
P < 0.05 compared with t0;
P < 0.01 compared with the corresponding control and ethanol;
P < 0.05 compared with the corresponding ethanol; n = 6 different rat experiments.
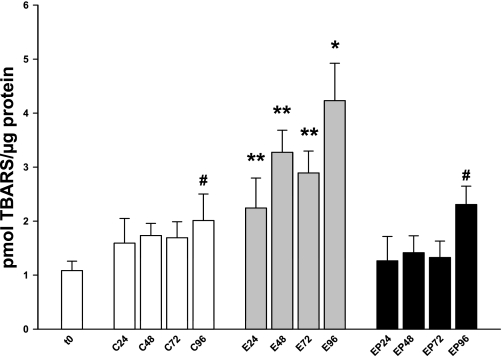
Depletion of GSH has been shown to correlate with increased lipid peroxidation in other models of ethanol-induced liver injury (1, 15). To determine whether ethanol metabolism induced lipid peroxidation in rat PCLS, slices were processed every 24 h and analyzed for lipid peroxidation products (mainly MDA and 4-hydroxynonenal) via a TBARS assay. By 24 h, ethanol-treated PCLS exhibited a twofold increase in TBARS over t0 (Fig. 5, Table 2) and a 1.4-fold increase over the corresponding control (t0 = 1.08 ± 0.15 pmol/μg protein, C = 1.59 ± 0.5 pmol/μg protein, E = 2.24 ± 0.61 pmol/μg protein). At all subsequent times, ethanol treatment induced ∼1.7–2.1-fold increases in lipid peroxidation over the corresponding controls (48 h, C = 1.73 ± 0.25 vs. E = 3.28 ± 0.45; 72 h, C = 1.69 ± 0.32 vs. E = 2.89 ± 0.44; 96 h, C = 2.01 ± 0.54 vs. E = 4.23 ± 0.76 pmol/μg protein). Notably, lipid peroxidation in control slices remained relatively constant and was not significantly elevated over t0 until 96 h (Fig. 5, Table 2). This could be due to the length of culture and diminishing viability (Table 2, % cytotoxicity), or it may be related to the onset of fibrogenesis observed previously for control PCLS (34). Addition of 4-MP prevented ethanol-induced lipid peroxidation (Fig. 5, Table 2). In fact, the TBARS profile for the ethanol + 4-MP group over 96 h mimicked that of the control slices and indicates ethanol metabolism is responsible for lipid peroxidation.
Fig. 5.
Thiobarbituric acid-reactive substance (TBARS) analysis of rat PCLS after ethanol treatment. Rat PCLS were treated with ethanol in the absence or presence of 4-MP as described in materials and methods. Every 24 h, slices were analyzed for TBARS by using a kit according to manufacturer's instructions. **P < 0.05 compared with corresponding controls; *P < 0.005 compared with corresponding control; #P < 0.05 compared with t0; n = 6 different rat experiments.
Ethanol Induces Sustained IL-6 Response in Rat PCLS
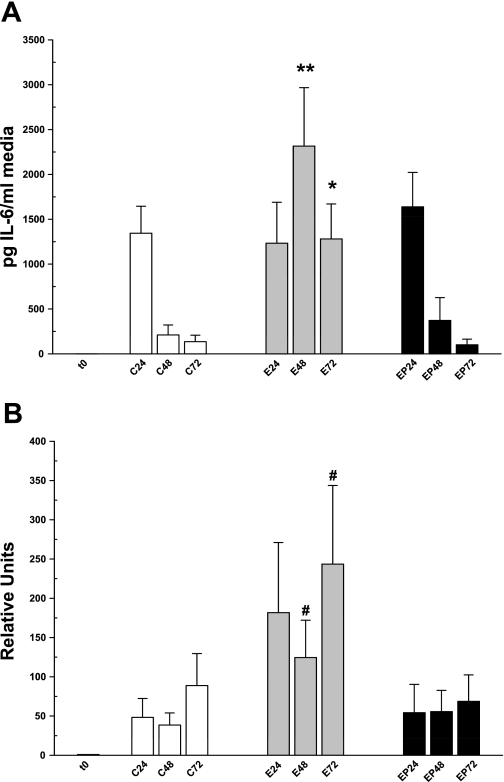
Many studies have noted an association between oxidative stress and cytokine expression in ethanol-induced liver injury (2, 23). Therefore, the goal of these experiments was to determine whether ethanol affected rat PCLS in a similar manner. Rat PCLS were incubated as before and were processed for RT-PCR and the media were collected for cytokine ELISA assays. Figure 6 shows both IL-6 secretion (A) and message profiles (B) for the different treatment groups over time. At 24 h, IL-6 was secreted robustly by all treatment groups (Fig. 6A; C = 758.4 ± 184.4; E = 713.3 ± 229.4; EP = 769.1 ± 192.4 pg IL-6/ml media). Consistent with this, IL-6 message was not significantly different at 24 h among the groups (Fig. 6B; C = 48.3 ± 26.4, E = 181.7 ± 97.8, EP = 54.2 ± 32.3). Within 48 h, IL-6 secretion in control slices dropped eightfold to 93.6 ± 53.6 pg IL-6/ml media and stayed at this level at 72 h of culture (Fig. 6A; 60.2 ± 34.7 pg IL-6/ml media). In contrast, IL-6 secretion was sustained at a high level (almost 14-fold higher than controls) over 72 h in slices treated with ethanol (48 h, 1,285.1 ± 275.1; 72 h, 831.8 ± 206.3 pg IL-6/ml media). Analysis of IL-6 message shows a corresponding significant increase at both 48 and 72 h in ethanol-treated PCLS (Fig. 6B; C48 = 38.5 ± 17, E48 = 124.5 ± 52.1, C72 = 88.8 ± 44.7, E72 = 243.6 ± 109.6). Simultaneous treatment of PCLS with ethanol and 4-MP prevented both the sustained IL-6 secretion and increased message levels found with ethanol treatment alone (Fig. 6, A and B), indicating that increased IL-6 expression was due to ethanol metabolism and metabolites. It is important to note that addition of 4-MP to control slices had no effect on oxidative stress, cytokine production, or induction of fibrogenesis (data not shown).
Fig. 6.
IL-6 secretion in media (A) and mRNA levels (B) from rat PCLS treated with 25 mM ethanol and/or 4-MP. A: media from all treatment groups and times were analyzed by using an ELISA to IL-6 as described in materials and methods. B: slices from all treatment groups and times were used for RT-PCR by using primers to IL-6 as described in materials and methods. #P < 0.05 compared with corresponding controls; *P < 0.005 compared with corresponding controls; **P < 0.0005 compared with corresponding control; n = 6 different rat experiments.
DISCUSSION
Previously, this laboratory has shown that PCLS from Wistar rats are a good model for ethanol-induced liver injury (20). This model exhibits many ethanol-induced features present in vivo, including a reduced redox state, fatty liver, and impaired protein trafficking, while maintaining viability and differentiated liver functions over 96 h of culture. The present study was undertaken to determine whether ethanol treatment could induce a fibrogenic response in PCLS. PCLS have already been shown to exhibit a moderate fibrogenic response (including increased collagen and SMA expression) in the absence of any treatment, but this occurred late in the culture period (72–96 h) and is thought to be a wound-healing response (34). Results from this study demonstrate that PCLS develop a fibrogenic state within 48 h of ethanol treatment, as evidenced by increased SMA and collagen 1α(I) and collagen deposition. This ethanol-induced response is more robust and precedes the response observed in controls. Induction of a robust fibrogenic response within 48 h makes this model uniquely suited to examine different treatment strategies to resolve the fibrogenic response. Theoretically, this means that early fibrotic events could be induced and resolved within a week, which hasn't been done in any animal models of ethanol-induced liver injury. This ethanol-induced fibrogenic response also provides an opportunity to examine how myofibroblast activation and propagation affects the other cell types within the context of liver architecture, as opposed to studies involving isolated liver cells. In addition, events leading to myofibroblast activation can be examined within the first 48 h. Studying how this model reacts to ethanol could lead to insights concerning the progression of the human disease and the development of effective treatment and/or preventative regimens for alcoholic liver disease.
Previous studies have indicated that oxidative stress is involved in liver injury and fibrosis (1, 8, 15, 39). Consistent with these studies, this manuscript shows that ethanol metabolism by rat PCLS induces oxidative stress within 24 h (GSH depletion and increased lipid peroxidation), which precedes fibrogenesis. Notably, neither the control nor the ethanol + 4-MP group exhibited significant increases in either GSH depletion or lipid peroxidation, supporting the idea that ethanol-induced oxidative stress contributes to fibrogenesis. However, it will be interesting to examine the role of oxidative stress in fibrogenesis in the PCLS by using GSH depleters and H2O2, for example, to determine whether these would accelerate the fibrogenic response. Also, the role of antioxidants as potential treatments or preventers of fibrogenesis will also be examined to determine the role played by oxidative stress.
Lipid peroxidation induced by oxidative stress generates aldehydes like MDA and 4-hydroxynonenal (8). These aldehydes rapidly form protein adducts, leading to cellular damage and a fibrogenic response. This laboratory has previously shown that two aldehydes produced by ethanol metabolism (AA and MDA) interact to form MDA-AA adducts on hepatic proteins, called MAA (29, 37, 38). MAA-modified proteins have been shown to generate proinflammatory and profibrogenic responses in nonparenchymal cells (9, 28). Since PCLS treated with ethanol can produce both of these aldehydes, the possibility exists that MAA-adducts could be formed in this system. Future studies will examine ethanol-induced generation of MAA-adducted proteins in PCLS to help understand the effects of these adducts on liver function and fibrosis.
Ethanol can also induce an inflammatory response in the liver. Previous studies have shown that chronic ethanol ingestion induces hepatocellular damage that promotes recruitment and activation of inflammatory cells (Kupffer cells, neutrophils, CD4+ T-helper cells) (10, 22). The activated inflammatory cells produce ROS and secrete cytokines including TNF-α, IL-1β, and IL-6 as part of the acute-phase response, which can induce and perpetuate myofibroblast activation. In the present study, only a limited acute-phase cytokine response, represented by sustained IL-6 production, was induced by ethanol treatment. Evaluation of TNF-α and IL-1β secretion (data not shown) did not reveal the presence of either cytokine due to ethanol treatment. This could be due to many factors, including 1) a limited amount of either cytokine being produced that is taken up continuously in a paracrine fashion, and 2) peak expression of the cytokines occurring earlier than 24 h, which could be suppressed by sustained IL-6 production (12). Future studies to deplete IL-6 and/or enhance TNF-α will be helpful in determining the importance of cytokines in this model of accelerated fibrogenesis.
Many studies indicate that IL-6 is a multifunctional cytokine, having antifibrogenic and profibrogenic effects (11, 17). The length of time liver tissue is exposed to IL-6 appears to determine its response to the cytokine. Short-term exposure has been shown to enhance liver survival and regeneration, whereas more chronic exposure to IL-6 can sensitize liver to injury and death (19). In fact, patients with alcoholic liver disease have been shown to have elevated levels of IL-6 (16, 21), and multiple studies in different models of liver injury have implicated sustained exposure to IL-6 enhances liver injury and fibrosis (12, 19). In contrast to controls, PCLS treated with ethanol exhibit sustained secretion of high levels of IL-6, which may contribute to the decreased viability and induction of fibrogenesis in this model. These results support the idea that acute vs. sustained IL-6 exposure have very different effects on liver function. Future studies will examine the pathway(s) underlying the sustained production of IL-6 and will examine whether IL-6 secretion is essential for induction of fibrogenesis. Also, previous studies indicated that IL-6 expression induced the downregulation of CYP2E1 expression (3, 27), which could explain the low expression of CYP2E1 in control PCLS after 24 h of culture previously reported by this laboratory (20).
The results from this study showing ethanol-induced increases in SMA and collagen 1α(I) accumulation in PCLS are similar to previous studies in which PCLS were chemically treated (CCl4) to induce fibrogenesis (31, 33). However, the results from this study are difficult to compare with the CCl4 studies since the majority of their data involves examination of mRNA levels of various fibrogenic markers and the time frame for examination of fibrogenesis only goes to 48 h, whereas the present study demonstrates induction of fibrogenesis by examining protein levels of SMA and collagen through 96 h. It will be interesting in future studies to compare CCl4 and ethanol treatments relating to the extent of and mechanism(s) leading to fibrogenesis in PCLS.
In summary, ethanol metabolism by PCLS induces oxidative stress and a sustained IL-6 response. These ethanol-induced effects result in a rapid fibrogenic response (within 48 h) that mimics the in vivo situation but does not necessitate the introduction of other cofactors (i.e., LPS, CCl4, iron, etc.). Importantly, this model system will allow for the examination of multiple pathways and the contribution of environmental factors that may lead to fibrogenesis, all under tightly controlled conditions. This system may be useful in refining and developing current and novel treatment strategies for eventual clinical use, making it an extremely good preclinical model for screening these potential agents.
GRANTS
This work was supported by National Institute on Alcohol Abuse and Alcoholism Grants R01 AA10435, R37 AA07818, and R21 AA15505-01A2. Additional support was provided by the Department of Veterans Affairs National Merit Review Program and the Department of Internal Medicine at University of Nebraska Medical Center.
DISCLOSURES
The authors have no financial conflicts of interest.
REFERENCES
- 1.Albano E. Alcohol, oxidative stress and free radical damage. Proc Nutr Soc 65: 278– 290, 2006 [DOI] [PubMed] [Google Scholar]
- 2.Albano E. Oxidative mechanisms in the pathogenesis of alcoholic liver disease. Mol Aspects Med 29: 9– 16, 2008 [DOI] [PubMed] [Google Scholar]
- 3.Carlson TJ, Billings RE. Role of nitric oxide in the cytokine-mediated regulation of cytochrome P-450. Mol Pharmacol 49: 796– 801, 1996 [PubMed] [Google Scholar]
- 4.Cederbaum AI, Wu D, Mari M, Bai J. CYP2E1-dependent toxicity and oxidative stress in HepG2 cells. Free Radic Biol Med 31: 1539– 1543, 2001 [DOI] [PubMed] [Google Scholar]
- 5.Choi DW, Kim SY, Kim SK, Kim YC. Factors involved in hepatic glutathione depletion induced by acute ethanol administration. J Toxicol Environ Health A 60: 459– 469, 2000 [DOI] [PubMed] [Google Scholar]
- 5a.Clemens DL, Calisto LE, Sorrell MF, Tuma DJ. Ethanol metabolism results in a G2/M cell-cycle arrest in recombinant Hep G2 cells. Hepatology 38: 385– 393, 2003 [DOI] [PubMed] [Google Scholar]
- 6.Clemens DL, Halgard CM, Miles RR, Sorrell MF, Tuma DJ. Establishment of a recombinant hepatic cell line stably expressing alcohol dehydrogenase. Arch Biochem Biophys 321: 311– 318, 1995 [DOI] [PubMed] [Google Scholar]
- 7.Collins TJ. ImageJ for microscopy. Biotechniques 43: 25– 30, 2007 [DOI] [PubMed] [Google Scholar]
- 8.De Minicis S, Brenner DA. Oxidative stress in alcoholic liver disease: role of NADPH oxidase complex. J Gastroenterol Hepatol 23, Suppl 1: S98– S103, 2008 [DOI] [PubMed] [Google Scholar]
- 9.Duryee MJ, Klassen LW, Freeman TL, Willis MS, Tuma DJ, Thiele GM. Lipopolysaccharide is a cofactor for malondialdehyde-acetaldehyde adduct-mediated cytokine/chemokine release by rat sinusoidal liver endothelial and Kupffer cells. Alcohol Clin Exp Res 28: 1931– 1938, 2004 [DOI] [PubMed] [Google Scholar]
- 10.Duryee MJ, Klassen LW, Thiele GM. Immunological response in alcoholic liver disease. World J Gastroenterol 13: 4938– 4946, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao B. Cytokines, STATs and liver disease. Cell Mol Immunol 2: 92– 100, 2005 [PubMed] [Google Scholar]
- 12.Greenwel P, Iraburu MJ, Reyes-Romero M, Meraz-Cruz N, Casado E, Solis-Herruzo JA, Rojkind M. Induction of an acute phase response in rats stimulates the expression of alpha 1(I) procollagen messenger ribonucleic acid in their livers. Possible role of interleukin-6. Lab Invest 72: 83– 91, 1995 [PubMed] [Google Scholar]
- 13.Groneberg DA, Grosse-Siestrup C, Fischer A. In vitro models to study hepatotoxicity. Toxicol Pathol 30: 394– 399, 2002 [DOI] [PubMed] [Google Scholar]
- 14.Guo J, Friedman SL. Hepatic fibrogenesis. Semin Liver Dis 27: 413– 426, 2007 [DOI] [PubMed] [Google Scholar]
- 15.Han D, Hanawa N, Saberi B, Kaplowitz N. Mechanisms of liver injury. III. Role of glutathione redox status in liver injury. Am J Physiol Gastrointest Liver Physiol 291: G1– G7, 2006 [DOI] [PubMed] [Google Scholar]
- 16.Hill DB, Marsano L, Cohen D, Allen J, Shedlofsky S, McClain CJ. Increased plasma interleukin-6 concentrations in alcoholic hepatitis. J Lab Clin Med 119: 547– 552, 1992 [PubMed] [Google Scholar]
- 17.Horiguchi N, Wang L, Mukhopadhyay P, Park O, Jeong WI, Lafdil F, Osei-Hyiaman D, Moh A, Fu XY, Pacher P, Kunos G, Gao B. Cell type-dependent pro- and anti-inflammatory role of signal transducer and activator of transcription 3 in alcoholic liver injury. Gastroenterology 134: 1148– 1158, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iredale JP. Models of liver fibrosis: exploring the dynamic nature of inflammation and repair in a solid organ. J Clin Invest 117: 539– 548, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin X, Zimmers TA, Perez EA, Pierce RH, Zhang Z, Koniaris LG. Paradoxical effects of short- and long-term interleukin-6 exposure on liver injury and repair. Hepatology 43: 474– 484, 2006 [DOI] [PubMed] [Google Scholar]
- 20.Klassen LW, Thiele GM, Duryee MJ, Schaffert CS, DeVeney AL, Hunter CD, Olinga P, Tuma DJ. An in vitro method of alcoholic liver injury using precision-cut liver slices from rats. Biochem Pharmacol 76: 426– 436, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latvala J, Hietala J, Koivisto H, Jarvi K, Anttila P, Niemela O. Immune responses to ethanol metabolites and cytokine profiles differentiate alcoholics with or without liver disease. Am J Gastroenterol 100: 1303– 1310, 2005 [DOI] [PubMed] [Google Scholar]
- 22.Li JT, Liao ZX, Ping J, Xu D, Wang H. Molecular mechanism of hepatic stellate cell activation and antifibrotic therapeutic strategies. J Gastroenterol 43: 419– 428, 2008 [DOI] [PubMed] [Google Scholar]
- 23.Nagata K, Suzuki H, Sakaguchi S. Common pathogenic mechanism in development progression of liver injury caused by non-alcoholic or alcoholic steatohepatitis. J Toxicol Sci 32: 453– 468, 2007 [DOI] [PubMed] [Google Scholar]
- 24.Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol 23: 291– 299, 2001 [PubMed] [Google Scholar]
- 25.Schaffert CS, Sorrell MF, Tuma DJ. Expression and cytoskeletal association of integrin subunits is selectively increased in rat perivenous hepatocytes after chronic ethanol administration. Alcohol Clin Exp Res 25: 1749– 1757, 2001 [PubMed] [Google Scholar]
- 26.Schaffert CS, Todero SL, McVicker BL, Tuma PL, Sorrell MF, Tuma DJ. WIF-B cells as a model for alcohol-induced hepatocyte injury. Biochem Pharmacol 67: 2167– 2174, 2004 [DOI] [PubMed] [Google Scholar]
- 27.Siewert E, Bort R, Kluge R, Heinrich PC, Castell J, Jover R. Hepatic cytochrome P450 down-regulation during aseptic inflammation in the mouse is interleukin 6 dependent. Hepatology 32: 49– 55, 2000 [DOI] [PubMed] [Google Scholar]
- 28.Thiele GM, Duryee MJ, Freeman TL, Sorrell MF, Willis MS, Tuma DJ, Klassen LW. Rat sinusoidal liver endothelial cells (SECs) produce pro-fibrotic factors in response to adducts formed from the metabolites of ethanol. Biochem Pharmacol 70: 1593– 1600, 2005 [DOI] [PubMed] [Google Scholar]
- 29.Tuma DJ, Thiele GM, Xu D, Klassen LW, Sorrell MF. Acetaldehyde and malondialdehyde react together to generate distinct protein adducts in the liver during long-term ethanol administration. Hepatology 23: 872– 880, 1996 [DOI] [PubMed] [Google Scholar]
- 30.Tuma DJ, Zetterman RK, Sorrell MF. Inhibition of glycoprotein secretion by ethanol and acetaldehyde in rat liver slices. Biochem Pharmacol 29: 35– 38, 1980 [DOI] [PubMed] [Google Scholar]
- 31.Van de Bovenkamp M, Groothuis GM, Draaisma AL, Merema MT, Bezuijen JI, van Gils MJ, Meijer DK, Friedman SL, Olinga P. Precision-cut liver slices as a new model to study toxicity-induced hepatic stellate cell activation in a physiologic milieu. Toxicol Sci 85: 632– 638, 2005 [DOI] [PubMed] [Google Scholar]
- 32.Van de Bovenkamp M, Groothuis GM, Meijer DK, Olinga P. Liver fibrosis in vitro: cell culture models and precision-cut liver slices. Toxicol In Vitro 21: 545– 557, 2007 [DOI] [PubMed] [Google Scholar]
- 33.Van de Bovenkamp M, Groothuis GM, Meijer DK, Olinga P. Liver slices as a model to study fibrogenesis and test the effects of anti-fibrotic drugs on fibrogenic cells in human liver. Toxicol In Vitro 22: 771– 778, 2008 [DOI] [PubMed] [Google Scholar]
- 34.Vickers AE, Saulnier M, Cruz E, Merema MT, Rose K, Bentley P, Olinga P. Organ slice viability extended for pathway characterization: an in vitro model to investigate fibrosis. Toxicol Sci 82: 534– 544, 2004 [DOI] [PubMed] [Google Scholar]
- 35.Wasser S, Tan CE. Experimental models of hepatic fibrosis in the rat. Ann Acad Med Singapore 28: 109– 111, 1999 [PubMed] [Google Scholar]
- 36.Weiler-Normann C, Herkel J, Lohse AW. Mouse models of liver fibrosis. Z Gastroenterol 45: 43– 50, 2007 [DOI] [PubMed] [Google Scholar]
- 37.Xu D, Thiele GM, Beckenhauer JL, Klassen LW, Sorrell MF, Tuma DJ. Detection of circulating antibodies to malondialdehyde-acetaldehyde adducts in ethanol-fed rats. Gastroenterology 115: 686– 692, 1998 [DOI] [PubMed] [Google Scholar]
- 38.Xu D, Thiele GM, Kearley ML, Haugen MD, Klassen LW, Sorrell MF, Tuma DJ. Epitope characterization of malondialdehyde-acetaldehyde adducts using an enzyme-linked immunosorbent assay. Chem Res Toxicol 10: 978– 986, 1997 [DOI] [PubMed] [Google Scholar]
- 39.Zima T, Kalousova M. Oxidative stress and signal transduction pathways in alcoholic liver disease. Alcohol Clin Exp Res 29: 110S– 115S, 2005 [DOI] [PubMed] [Google Scholar]