Abstract
The number of variables at play in the expression and purification of a single protein dwarf those involved in sequencing a genome. Although certain trends are apparent, there is no one-size-fits-all approach to the process of purifying proteins. Thus, whereas numerous genome sequencing projects are providing an overwhelming number of interesting open reading frames for structural biologists to study, fully realizing the potential of this resource is still only a distant hope. We will discuss several current approaches to high throughput expression and purification as well as strategies that have served us well to quickly identify lead protein expression constructs in the context of a core service protein expression and purification laboratory. The use of the baculovirus expression vector system and implementation of a purification screening method will be emphasized.
Keywords: Protein purification, baculovirus, high throughput
Introduction
Expressing and purifying human proteins, especially in Escherichia coli, the traditional host organism for high throughput (HTP) protein expression and purification, continues to be problematic for researchers. The low success rate (2–20% when expressing eukaryotic proteins in E. coli (Service) stems from several well known problems including low yields due to toxicity, recombinant protein insolubility, and protein aggregation. It is this low success rate that drives the use of alternative expression systems, a wide variety of expression constructs, and numerous HTP approaches.
The baculovirus expression vector system (BEVS) has become an indispensable expression system for the production of proteins (Hunt, Aricescu). There are several advantages of insect cells over E. coli including improved solubility, incorporation of some post-translational modification and higher yields for secreted proteins (Jarvis). The use of viral promoters leads to soluble expression levels often equal to or greater than those reached in E. coli.
For the past seven years we in the Protein Expression Laboratory (PEL) have striven to deliver a wide variety of eukaryotic proteins to researchers at the NIH. While there are no "magic bullets", we have evolved a pipeline using baculovirus expression in insect cells and micro scale protein purification testing that allows us to accurately predict success or failure quickly and cheaply. Combined with multiple expression formats that can be compared in parallel, our recent success at scale-up has risen to 90%. This review will focus on our experiences and is by no means a comprehensive review of all available techniques.
The PEL approach
It should be noted at the outset that this review is intended to convey useful information to laboratories that might not have the resources for 'high-end' automation. Our lab falls into this category as do most of the NIH scientists to whom we provide services. Thus, many of the results we discuss were developed with minimal or no automation. The important exception to this will be discussed in detail, however, even in this case the cost was a fraction (~10%) of many fully automated platforms.
In part due to this financial limitation, but more importantly because we observed very quickly after our inception that success in protein production can be improved by embracing diversity in terms of constructs, expression systems, expression conditions, and protein homologs, the PEL has approached the protein production problem in an incremental fashion on many fronts. Accordingly, our processes have changed over time and it is the goal of the review to discuss what has, and equally important, what has not worked in our hands.
The most broad reaching and influential of these improvements has been the shift away from screening by expression profiling to screening by purification, a platform we refer to as purify first, or PF. It is this partially automated approach that has had a dramatic impact on our success by reducing the effort we spend on 'dead-end' constructs and in a synergistic fashion, has allowed us to 'widen the bottle neck' of our process. Specifically, we learn very quickly and for little cost what constructs/expression systems/expression conditions lead to purified protein. The higher throughput and efficiency of PF allows us to test a wider diversity of constructs/expression systems/expression conditions which translates into higher chance of success for any given protein.
BEVS cloning
Cloning genes for expression in the BEVS is a two step process: 1) creating a construct with the proper context (e.g. promoters, affinity and/or solubility tags) using standard cloning techniques for propagation in E. coli and 2) transferring the expression construct to insect cells for expression. Recombinational cloning has largely replaced restriction enzyme/ligase based cloning for the first step of this process (Walhout, Hartley). There are several strategies available for transfer of the expression construct to the insect cells either via a bacmid DNA (e.g. Bac-to-Bac, Invitrogen) or directly into the viral genome (e.g. BacMagic, EMD Biosciences) (Koehn, Hunt). Although the PEL creates many constructs for BEVS expression (~ 150 DNAs/year), the numbers are too low to justify the expense of automation. Rather, we have modified several aspects of the BEVS system to speed the process and increase the likelihood of finding optimal expression conditions.
BEVS expression
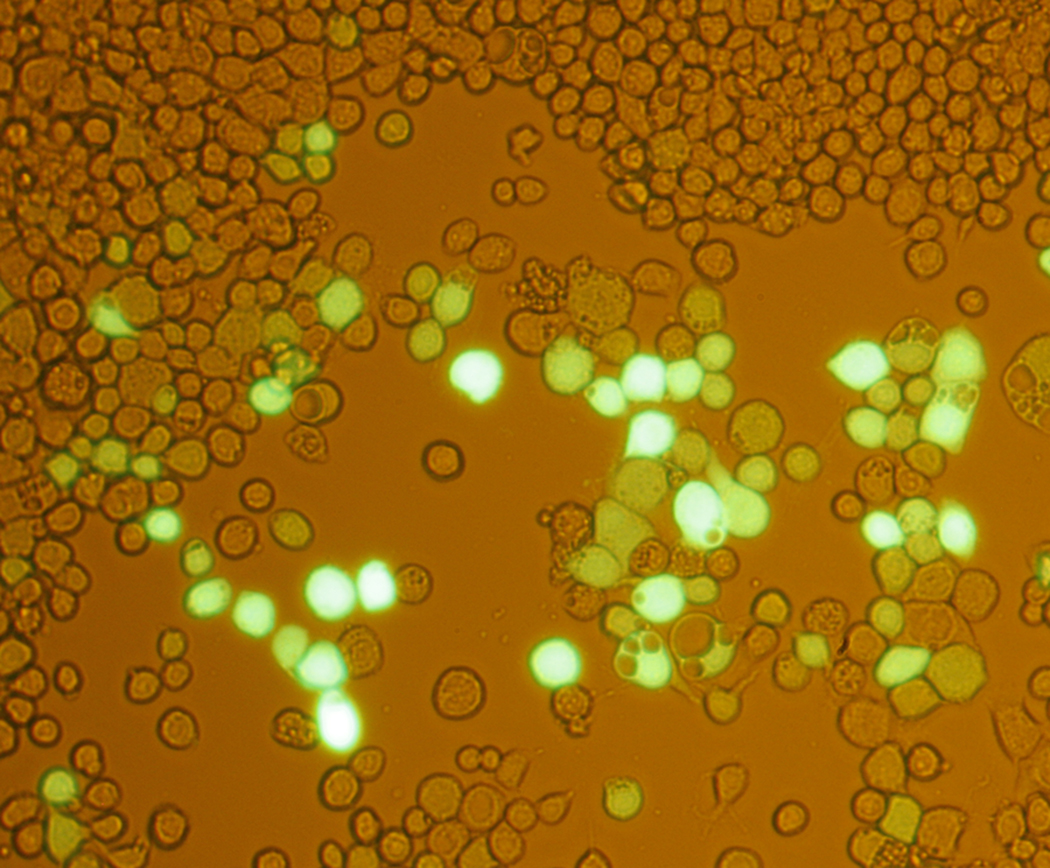
We use the Bac-to-Bac system to transfer expression constructs from E. coli to insect cells via an intermediate bacmid DNA. Once transfected, the bacmid DNA leads to the production of active virus in a lytic cycle. Using a modification to the transfection protocol, we bypass the traditional amplification step that may require 1–2 virus passages to achieve a high titer stock. In what we term 'large-scale direct' (LSD) transformation (for an outline of the protocol, see Figure 1), a 100 ml high titer BV stock is created in four days (manuscript in preparation). Determining the titer of the viral stock allows for consistency in scale up expression experiments and we have greatly simplified this step by using a modified cell line which contains a fluorescent protein under the control of the viral polyhedrin promoter (Hopkins). Virus titers can be determined in 3 days compared with the 6–8 days required for the cytopathic effect method. Also the detection of GFP-positive wells in an end-point dilution assay is much easier than detection of cell death, especially for the novice (Figure 2).
Figure1.
Figure2.
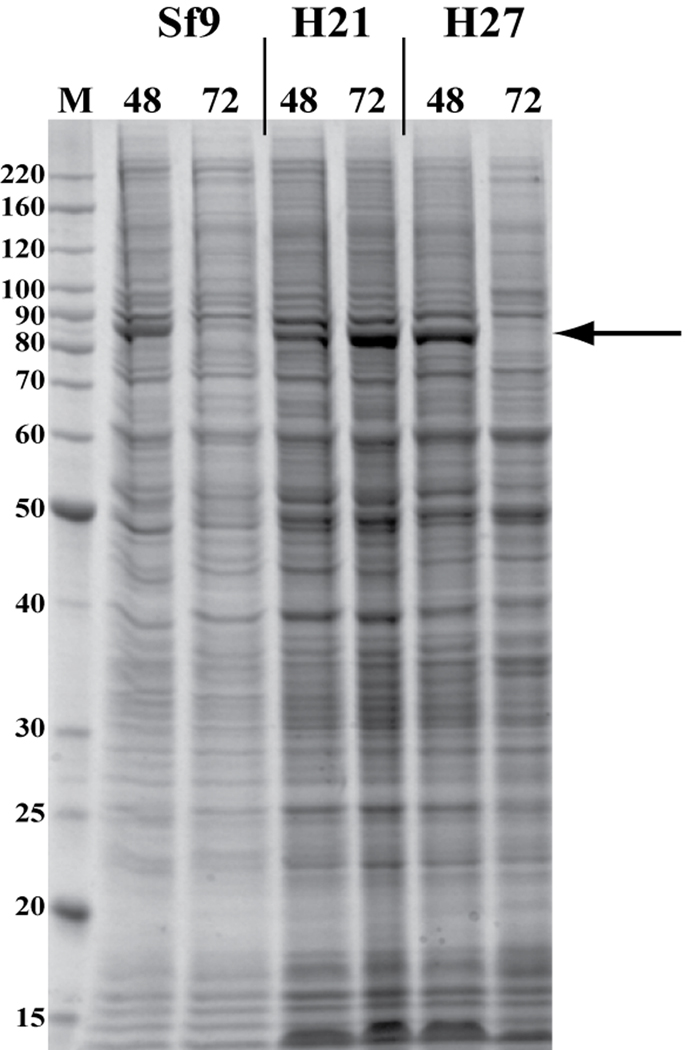
Several years ago we observed distinct qualitative and quantitative differences between insect cell lines in terms of protein expression levels and the extent of proteolysis. Differences were also observed under certain culture conditions. Our lab routinely uses Sf9 cells and High Five™ (Invitrogen) cells for expression testing. In general, we find proteins expressed in High Five™ cells suffer more proteolysis, but this can sometimes be mitigated by lowering the incubation temperature from 27°C to 21°C. Protein expression levels are also frequently 2–5 fold higher in High Five™ cells than in Sf9 cells. However, this observation is not universal among researchers and several labs report good results with Sf9 and Sf21 cell lines. From these observations we developed a three culture (Sf9 cells incubated at 27°C, High Five™ cells incubated at 21°C, and High Five™ cells incubated at 27°C), six sample time course (two harvests/per culture, 48 and 72 hr post-infection) optimization method to determine the best conditions for protein expression (see Figure 3 for a typical example). After using this platform to analyze the expression pattern of hundreds of proteins it became apparent that at least 90% of the time a single set of conditions was optimal: High Five™ cells, incubated at 21°C and harvested 72 hr post-infection. It should be noted that although incubation of High Five™ cells at the standard temperature of 27°C does frequently give high expression levels, the window of time before the target protein becomes degraded is shorter than at 27°C and this poses problems especially in scale up. This point is easy to observe in Figure 3 as the target protein is essentially absent at 27°C at 72 hr. Given that the titers of the high titer stock were commonly within a fairly limited range (2–4 ×108 PFU/ml), we reasoned that we could have high confidence in a single condition expression test using non-titered virus: a process we refer to as 'early detection' (ED). For an outline of the protocol, see Figure 1. Thus, combining LSD and ED methods we reduce the time it takes to go from bacmid to samples for analysis from 3 weeks to 1 week.
Figure3.
Insect cells can also be used to express a secreted protein although the yields are not as high as obtained from Pichia pastoris or Kluyveromyces lactis. Nevertheless, we have observed yields up to 30 mg/liter from insect cells and routinely use this as an option when working with secreted proteins. The major drawback to the approach is the incompatibility of insect cell culture media with immobilized metal ion affinity chromatography (IMAC). The use of a multi-histidine tag is extremely common as it affords a simple and relatively cheap affinity chromatography step. Purification of his-tagged secreted proteins from insect cell culture requires a buffer exchange step prior to chromatography (Hunt). Since signal peptides and cell lines have been reported to affect expression levels (Futatsumori-Sugai, Hunt), we routinely evaluate GP67, honey bee melittin, and native signal peptides for secretion constructs. As of yet, we have detected no predictable pattern and thus we screen these signal peptides as expression levels can be enhanced up to 3–4 fold.
Mammalian expression
Transient transfection of DNA constructs for cytoplasmic expression of proteins in mammalian cells has long been overlooked as a source of protein production. In a recent study (manuscript in preparation) we examined the effects of promoters, enhancers, UTR elements, other vector backbone components and fusion tags on the expression of different genes in mammalian hosts. The results validate our approach as certain combinations of elements led to a dramatic improvement in protein expression levels. Thus screening these conditions may lead to significant increases in protein production. Also, we observed that fusion to the green fluorescent protein (eGFP) can serve as a readout for protein expression which is highly scaleable for many proteins to large-scale production. Finally, we hope that given enough data, we may start to see trends in the “best” promoter or tag, which might limit the cloning options to a smaller initial set of test clones for a given protein.
Widening the bottleneck
Background
It is well established that protein production is the limiting factor in many biological experiments and especially so for large scale structure based initiatives. A common strategy has been to choose a limited number of expression clones (and many times single) to simplify screening and purification. The success rate of this approach is typically less than 10% when applied to eukaryotic proteins expressed in E. coli (Chandonia). This strategy allows the analysis of thousands of clones relatively quickly, and is useful in identifying the so-called 'low hanging fruit' when screening an ORFeome. The approach has also been used successfully to analyze truncation variants (Lesley). However, the low success rate inherent in this approach requires creating, expressing and analyzing large numbers of constructs that is prohibitive for all but extremely well-funded laboratories. These approaches leave little, if any, room for optimization aside from manipulating expression conditions or processing buffers. Varying these parameters will rescue a small percentage of targets from insolubility but the majority of targets remain either poorly expressed, insoluble or fail to adopt a native structure.
The Protein Expression Laboratory (PEL) is a core service lab that serves the protein expression and purification needs of the National Cancer Institute. The proteins we are asked to produce are typically mammalian cytosolic or secreted proteins, however our general approach is applicable to the production of membrane proteins as well. Faced with the limited success of the typical HTP approach outlined above, our strategy since the PEL's inception in 2001 has been to investigate multiple variables in order to improve our protein delivery success rate without resorting to costly automation. Initially, this effort was focused on improving protein solubility both through construct design using solubility fusion tags (Esposito) and optimizing expression conditions. By so doing, we greatly increased our success (calculated as the frequency that a given protein could be expressed in soluble form) in both E. coli and BEVS.
However, this improvement came with a price: increased sample numbers. Although the implementation of the ED protocol has recently reduced the number of BEVS samples we process from ~900 to 150 per year, we receive an ever increasing number of samples from E. coli, mammalian cell culture, hybridoma cell lines, transient insect cell transfection, and K. lactis expression systems. It became clear that our downstream purification abilities were restricting the number of variables we could assess due to failures during the scale up purification process. These failures can occur at many steps in the pathway including expression, protein aggregation (which could also lead to poor purity), poor protease cleavage of cleavable affinity/solubility tags, protein instability after removal of affinity/solubility tags, or difficulties in separating target proteins from cleaved affinity/solubility tags. These failures (at a scale of 1–60 liters for the expression culture) were expensive and time consuming and thus a solution was needed that did not involve an increase in personnel. Because there is no reliable way to predict if or when a protein will fail to purify, the solution to the problem was to create a higher throughput process that replicated the behavior of the scale up system, in our case, FPLC-based protein purification. As with any system that attempts to apply a single approach to a diverse set of starting materials, the challenge is to limit the number of false negatives while maintaining a stringent screen that clearly separates worthwhile targets from poor ones.
A note on the PEL throughput and capacity
To provide context for the reader, it is useful to elaborate in some detail on the PEL workload mentioned above. When we were considering higher throughput alternatives, ~80 projects were completed in the previous year for the protein purification portion of the lab (four FTEs). Although the range was wide (1–30), the average number of constructs per project was four. The average number of samples developed per construct was four, thus over the course of the year, ~1280 samples were received, of which ~1200 were small scale expression testing samples.
Additional factors also influenced our investigation of HTP platform. The PEL uses Gateway recombinational cloning (Hartley et al.) and can easily, and with little added expense, increase the number of constructs by several fold. We expected that this would increase (and indeed it did) because in previous work the processing/analysis bottleneck had limited the construct number decision during project set up. Also, the amount of work requested of the PEL has increased an average of 20% each year since our inception (9 years ago) and there are no signs that this trend will abate. In fact, we also anticipated that during lean budget climates, such as we have experienced recently, NIH principal investigators will be exploring ways to stretch their laboratory budgets. This assumption has also proven valid. Thus, not only would a system need to immediately improve our throughput but it would need to be capable of expanding for future workloads. While these specifications set the bar relatively high, we expected both direct and indirect benefits from such a change. The direct benefits would be a much improved screening method based on purification rather than just solubility as well as a far less costly and risky approach for our NIH investigators. Indirect benefits derived from the anticipated ability to screen more samples (thus allowing more constructs and conditions to be screened which have nearly always translated into increased success for the investigators) while enabling the protein purification portion of the lab to return to their mandate of technology development that had been severely compromised with the increased workload.
Initial attempts
We began our search for an HTP method by focusing on the smallest scale columns available (one ml) for our existing FPLCs (GE Healthcare). While this approach did provide reproducible and scalable data, the increased throughput was insufficient to shift or enlarge the bottleneck significantly. Thus, we began to evaluate parallel systems of purification. Our lab had observed poor correlation between results obtained with vacuum or spin-column based approaches and results from scale up purification on our FPLC instruments, largely due to limited contact time between the protein and the purification resin, and thus we did not consider these further. Magnetic bead based approaches are currently used in several large scale HTP projects (Chambers). However, this approach is not scalable and while it is successful as a HTP screen for purification, we were hoping to find a system that would allow the use of the same resin as in the scale up columns as well as providing information about the chromatography, specifically, information about resin affinity and contaminants. We also evaluated an instrument (BioOptix10, Teledyne ISCO) capable of 10 parallel purifications. In our hands, this machine turned out to be more suitable for larger columns.
Current platform
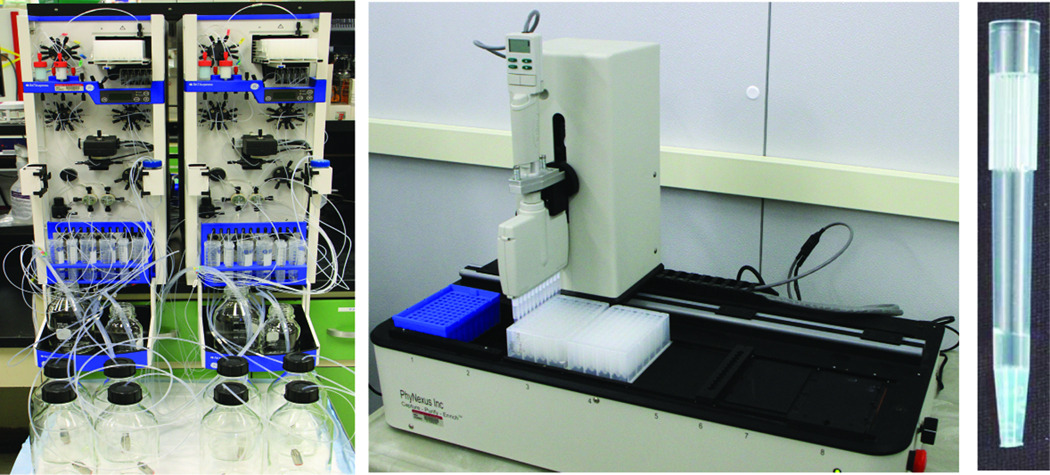
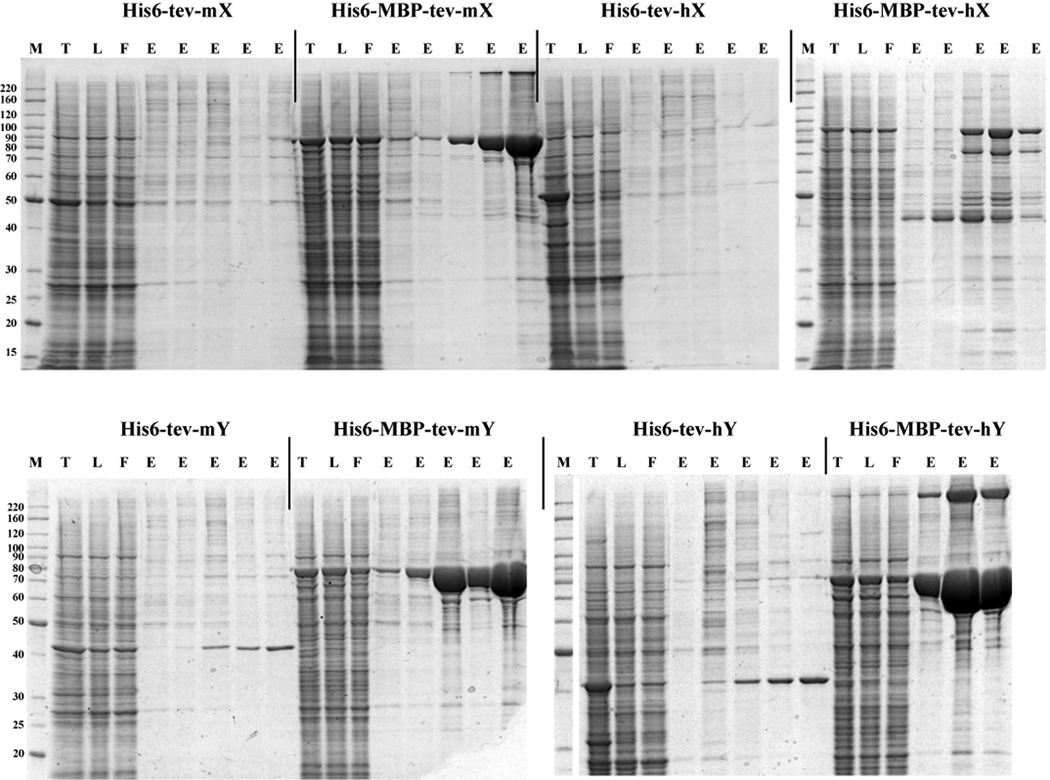
We evaluated and eventually adopted a parallel purification platform (developed by PhyNexus, Inc.) based on the packing of small volumes (5 – 320 microliters) of chromatography resins into a modified disposable plastic pipette tips with frits to contain the resin. The tips are mounted on a 12-channel pipette (Rainin) that is controlled by a software program. The pipette is mounted on a robotic workstation (MEA, PhyNexus, Inc.) also controlled by the software program (Figure 4). The PhyTips are lowered into wells of a 96-well plate into which the user has added the appropriate buffers for the chromatography. The user controls the volumes and flow rates of the liquid handler via the software. An example of typical results are shown in Figure 5, in which several samples from the baculovirus expression system were screened using this parallel purification approach.
Figure4.
Figure5.
Benefits
Predictive and useful chromatographic data. The user has total control of flow rate and thus residence time, both critical parameters that must be considered in any scale up work. This leads to results that are predictive both quantitatively and qualitatively for scale up work on an FPLC. Because most standard chromatographic principles are applicable in this micro scale parallel platform, the judicious choice of buffers and sample volumes can lead to useful information that can be applied during scale up. This information includes the actual protein binding capacity of the purification resin, affinity of the target (and contaminants) for the resin, and the ability to calculate a purification yield. Because the results are predictive, we frequently customize our scale up chromatography (e.g. shape of the elution profile, concentration of elution buffers) based on the micro scale results.
Reduction in scale up failures. The adoption of this 'purify first' method for screening has reduced our scale up purification failure rate; we know at small scale what will and won't work (Figure 5). This is achieved at a fraction of the cost and time that we previously incurred. We do not proceed to scale up unless we have a method developed in the micro scale platform. Frequently the method developed involves 2–3 chromatographic steps. The time previously spent on troubleshooting at scale up can now be much more efficiently used to pursue other options, and, as noted above, redirected toward technology development. It should be noted that this platform does not improve chromatography per se, but provides the opportunity to optimize the chromatography and to avoid ineffective steps.
HTP. The small amount of resin packed into the tips (5 – 320 microliter), not only allows for faster chromatography, but has ramifications throughout a protein expression lab. Smaller culture volumes are necessary which feeds back to more constructs/conditions that can be tested. Thus close coordination with the cloning and protein expression units of a lab is critical when evaluating and implementing an HTP platform for purification.
Assessment of proteolysis. One valuable insight gained is the extent of proteolysis of the target protein. This is much more difficult to obtain by a solubility assessment, usually requiring Western analysis, a decidedly low throughput process. This knowledge can be critical to a scale up purification and may, if not precluding the choice of conditions/constructs which exacerbate proteolysis, would substantially affect the purification method.
Flexibility. PhyTips are flexible with regards to resins, both type and volumes, the sole resin limitation being a minimum bead size of 30 microns. We routinely perform IMAC, IEX and HIC. Additional chromatography modes that we have performed include GST, MBP, Strep2, Protein A, and Protein G affinity purification. Individual methods must be created for each style of chromatography to accommodate the unique aspects of the individual chromatography. We often customize the purification to take advantage of smaller (10 microliter) or larger (360 microliter) PhyTips, depending on the application. Since the MEA is a liquid handler, samples can be moved around the deck and with a modified protocol and special tip, samples can be desalted as well.
Simplicity. Compared to an FPLC instrument capable of parallel purification, the MEA is a model of simplicity, with no valves, pumps, tubing, or fraction collector (Figure 3). Accordingly, we experience a much more trouble free operation (e.g. no leaks or detector failures). Our instrument has not required a repair over a four year period, whereas our FPLCs require yearly preventative maintenance and additional costly repairs (or service contract) of at least one per instrument per year.
Screening. Screening of many types, in addition to the straightforward plus/minus purification result, is straightforward. One can screen resins simply by the choice of tips and chromatography buffers by 96-well plate set-up. This is extremely useful when scouting IEX and HIC resins and buffers.
Drawbacks/Issues
Data collection/transfer. Increasing throughput is a double edged sword in that it increases the need for error-less data management not only during purification, but also in the other units of the lab that now are producing more materials for each project. An electronic data storage and retrieval system becomes absolutely necessary in this transition. We use a relatively low-end approach of a FileMaker Pro database developed in house.
Data analysis. The gold standard of protein analysis during chromatography has traditionally been a Coomassie-stained SDS-PAGE gel. This has now become the rate limiting step (with its attendant photography and subsequent electronic editing) and should be considered when contemplating an increased throughput platform. Several labs have converted to an automated microfluidic separation instrument (GXII, Caliper Lifesciences). The cost of consumables for this approach is considerably more than a gel based platform, however the labor savings and the benefits of searchable electronic data may offset this.
Equipment requirements/limitations. Although the liquid handler used in our lab is relatively inexpensive relative to other units available, it will still be beyond the reach of many labs. The instrument is also limited to 96-well plates. Although PhyTips are produced for several common liquid handlers, compatibility with existing liquid handlers should be checked. PhyTips are compatible with Rainin LTS manual pipettes and thus manual operation is possible. This can be quite useful when a quick answer to the question, “will the protein bind?” is required, but this obviously subtracts from the benefits of high throughput. Because the buffers are introduced to the column from the bottom of the tip and then expelled in the reverse direction, gradient elutions are not possible and the target protein will be diluted based on the volume of the buffer in the well. For our applications, these are not serious drawbacks, and can be overcome with judicious choice of elution buffers and volumes. Problems such as poor yield arise mainly when working with limited amounts of target protein, or proteins that have low affinity for the resin. However, these are protein-specific problems encountered in any affinity-based screening strategy.
Summary
The need for higher throughput, predictive approaches to protein purification is becoming greater as the number of potential protein targets, expression samples, and constructs increase. The Protein Expression Laboratory has adopted a higher throughput micro scale parallel platform that screens by purification. The result has been a sea-change in how a protein is purified in our lab. Proteins are now candidates for scale up only when a method has been determined in the micro scale platform. The efficiencies gained by the approach lead to the analysis of many more samples which in turn increases the likelihood that the combination of construct and expression conditions that lead to successful protein purification for a given protein will be discovered.
Acknowledgements
The authors wish to thank Jim Hartley for critical comments on the manuscript.
Abbreviations
- HTP
high throughput
- BEVS
baculovirus expression vector system
- IMAC
immobilized metal ion affinity chromatography
- LSD
large scale direct
- ED
early detection
- PEL
Protein Expression Laboratory
- GFP
green fluorescent protein
- IEX
ion-exchange chromatography
- HIC
hydrophobic interaction chromatography
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
References
- Chambers SP, Fulghum JR, Austen DA, Lu F, Swalley SE. E. coli and insect cell expression, automated purification and quantitative analysis. Methods Mol Biol. 2009;498:143–156. doi: 10.1007/978-1-59745-196-3_10. [DOI] [PubMed] [Google Scholar]
- Chandonia JM, Brenner SE. The Impact of Structural Genomics: Expectations and Outcomes. Science. 2006;311:347–351. doi: 10.1126/science.1121018. [DOI] [PubMed] [Google Scholar]
- Futatsumori-Sugai M, Tsumoto K. Signal peptide design for improving recombinant protein secretion in the baculovirus expression vector system. Biochem Biophys Res Commun. 2010;391(1):931–935. doi: 10.1016/j.bbrc.2009.11.167. [DOI] [PubMed] [Google Scholar]
- Hartley JL. Use of the gateway system for protein expression in multiple hosts. Curr Protoc Protein Sci. 2003:5.17. doi: 10.1002/0471140864.ps0517s30. [DOI] [PubMed] [Google Scholar]
- Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10(11):1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins R, Esposito D. A rapid method for titrating baculovirus stocks using the Sf-9 Easy Titer cell line. Biotechniques. 2009 Sep;47(3):785–788. doi: 10.2144/000113238. [DOI] [PubMed] [Google Scholar]
- Hunt I. From gene to protein: a review of new and enabling technologies for multi-parallel protein expression. Protein Expr Purif. 2005 Mar;40(1):1–22. doi: 10.1016/j.pep.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Jarvis DL. Baculovirus-insect cell expression systems. Methods Enzymol. 2009;463:191–222. doi: 10.1016/S0076-6879(09)63014-7. [DOI] [PubMed] [Google Scholar]
- Koehn J, Hunt I. High-Throughput Protein Production (HTPP): a review of enabling technologies to expedite protein production. Methods Mol Biol. 2009;498:1–18. doi: 10.1007/978-1-59745-196-3_1. [DOI] [PubMed] [Google Scholar]
- Lesley SA. Parallel methods for expression and purification. Methods Enzymol. 2009;463:767–785. doi: 10.1016/S0076-6879(09)63041-X. [DOI] [PubMed] [Google Scholar]
- Service RF. Structural genomics. Tapping DNA for structures produces a trickle. Science. 2002;298:948–950. doi: 10.1126/science.298.5595.948. [DOI] [PubMed] [Google Scholar]
- Walhout AJ, Temple GF, Brasch MA, Hartley JL, Lorson MA, van den Heuvel S, Vidal M. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]