Abstract
Converting normal prion protein (PrPC) to the pathogenic PrPSc isoform is central to prion disease. We previously showed that, in the presence of lipids, recombinant mouse PrP (rPrP) can be converted into the highly infectious conformation, suggesting a crucial role of lipid-rPrP interaction in PrP conversion. To understand the mechanism of lipid-rPrP interaction, we analyzed the capability of various rPrP mutants to bind anionic lipids and to gain the lipid-induced proteinase K (PK)-resistance. We found that the N-terminal positively charged region contributes to the electrostatic rPrP-lipid binding, but does not affect lipid-induced PK-resistance. In contrast, the highly conserved middle region of PrP, consisting of a positively charged region and a hydrophobic domain, is essential for lipid-induced rPrP conversion. The hydrophobic domain deletion mutant significantly weakened the hydrophobic rPrP-lipid interaction and abolished the lipid-induced C-terminal PK-resistance. The rPrP mutant without positive charges in the middle region reduced the amount of lipid-induced PK-resistant rPrP form. Consistent with a critical role of the middle region in lipid-induced rPrP conversion, both disease-associated P105L and P102L mutations, localized between lysine residues in the positively charged region, significantly affected lipid-induced rPrP conversion. The hydrophobic domain localized 129 polymorphism altered the strength of hydrophobic rPrP-lipid interaction. Collectively, our results suggest that the interaction between the middle region of PrP and lipid is essential for the formation of PK-resistant conformation. Moreover, the influence of disease-associated PrP mutations and 129 polymorphism on PrP-lipid interaction supports the relevance of PrP-lipid interaction to the pathogenesis of prion disease.
Prion diseases are a group of transmissible neurodegenerative disorders including Creutzfeldt-Jakob Disease (CJD) and Gerstmann-Straussler-Scheinker syndrome (GSS) in humans, scrapie in sheep, and bovine spongiform encephalopathy in cows (1–3). The transmission of prion disease is mediated by an unusual infectious agent, which is composed of a pathogenic conformer of the normal prion protein (PrP) (1, 4–6). The conformational change of PrP, converting the normal protease sensitive PrPC to the pathogenic protease resistant PrPSc form, is a critical pathogenic event in prion disease (7–11). We recently reported that in the presence of anionic phospholipid POPG (1-palmitoyl-2-oleoylphosphatidylglycerol) and total mouse liver RNA, bacterially expressed recombinant PrP (rPrP) can be converted in vitro into the infectious conformer, causing prion disease in wild type animals (12). The high prion infectivity associated with the recombinant prion highlights a critical role of POPG and RNA in converting rPrP into the infectious conformation (13).
The facilitating role of factors such as phospholipid and RNA in PrP conversion is consistent with previous reports. It has been shown that polyanions, particularly RNA molecules, enhance the in vitro propagation of naturally occurring prions (14, 15). Using purified hamster PrPC, Deleault et al reported that the infectious prion can be formed de novo in the presence of synthetic polyA RNA (16). Notably, the purified PrPC used in that study contains a stoichiometric amount of co-purified lipid molecules, indicating a role of lipid in PrP conversion (16). The role of lipid in PrP conversion has also been implicated by several previous studies. The highly purified “prion rod” contains lipid molecules (17) and re-incorporation of “prion rod” into lipid vesicles results in a higher prion infectivity (18). Compared to detergent purified PrPSc, the lipid membrane-associated PrPSc infects cultured cells with a higher efficiency (19). Altering the lipid composition of prion-infected cells affects the amount of PrPSc produced in these cells (20, 21). Moreover, a glycosylphosphatidylinositol (GPI)-anchor independent PrP-lipid interaction has been found to be essential for PrP conversion in vitro (22). These observations are consistent with the notion that PrP-lipid interaction plays a critical role in PrP conversion.
The PrP-lipid interaction has been studied mainly using biophysical approaches. It has been shown that rPrP binds to lipids and the binding alters secondary structures (23–27) and destabilizes the C-terminal part of PrP (23). Upon additional treatments, lipid bound rPrP was found to form amyloid fibers (25, 27). It has also been shown that concentrated GPI-anchored PrP on a raft-like membrane domain induces PrP to form intermolecular β-sheets (28). We previously reported that anionic lipid-rPrP interaction converts rPrP to a conformation similar to the pathogenic PrPSc conformation, with increased β-sheet content and C-terminal PK-resistance (29). These findings revealed that PrP binds to certain types of lipids independent of its GPI-anchor, and the lipid-interaction leads to conformational changes of PrP. However, it remains unclear which regions of PrP are involved in the lipid-interaction, which residues of PrP are critical in converting rPrP into the PrPSc-like PK-resistant conformation, and whether disease-associated mutations affect PrP-lipid interaction. Given the role of anionic POPG in converting rPrP into the highly infectious form (12), answers to these questions will help us to elucidate the molecular mechanism of PrP conversion.
In this study, we dissected the lipid-PrP interaction by using various rPrP mutants to compare their lipid binding capability and the lipid-induced C-terminal PK-resistance. We found that the highly conserved middle region of PrP, consisting of a positively charged region with four lysine residues (amino acid 100 – 110) followed by a hydrophobic domain (amino acid 111 – 134), is crucial for lipid induced rPrP conformational change. The middle region localized disease-associated P102L and P105L mutations and the 129 polymorphism have distinct effects on rPrP-lipid interaction. Collectively, these results provide additional support for the relevance of PrP-lipid interaction to the pathogenesis of prion disease.
EXPERIMENTAL PROCEDURES
Primers
23BamHI: 5′-AAAGGATCCAAAAAGCGGCCAAAGCCTGGA-3′
120HindIII: 5′-CCCAAGCTTATACTGCCCCAGCTGCCGCAGC-3′
121BamHI: 5′-TTTGGATCCGTGGGGGGCCTTGGTGGCTAC-3′
230RHindIII: 5′-CCCAAGCTTAGGATCTTCTCCCGTCGTAATA-3′
DELN: 5′-ACGTGCATGCTTGAGGTTGGTTTTTGG-3′
DELC: 5′-ACATGCATGCCATGAGCAGGCCCATGATCC-3′
1MF: 5′-AATCAGTGGAACATACCAGCATACCAAAAACCAACCTC-3′
1MR: 5′-GAGGTTGGTTTTTGGTATGCTGGGTATGTTCCACTGATT-3′
2MF: 5′-CCCAGCATACCAATAACCAACCTCATACATGTGGCAGGG-3′
2MR: 5′-CCCTGCCACATGTATGAGGTTGGTTATTGGTATGCTGGG-3′
Generation of PrP mutants
DNA fragments of mouse PrP23-120 and PrP121-230 were amplified using primers 23BamHI and 120HindIII, and 121BamHI and 230HindIII, respectively. PCR products were then cloned into the expression vector pPROEX-HTb (Invitrogen) using restriction endonucleases BamHI and HindIII. To generate mutant ΔH, mouse PrP23-110 DNA sequence was amplified using primers 23BamHI and DELN and then linked to the EcoRV site of pBluescript (SK) vector. The mouse PrP132-230 DNA sequence was amplified using primers DELC and 230HindIII and then linked to pBluescript (SK)-PrP23-110 via SphI and HindIII sites to generate pBluescript (SK)-ΔH plasmid. ΔH DNA sequence was cut off from the pBluescript vector by BamHI and HindIII and linked to pPROEX-HTb. All mutant DNA sequences in pPROEX-HTb are behind a linker sequence encoding six histidines. The histidine tag is followed by a tobacco etch virus (TEV) protease cleavage site; after TEV protease cleavage of the histidine tag, only one extra amino acid, glycine, remained at the N-terminus of PrP. For K/I mutant, primers 1MF and 1MR were used first to mutate lysine residues at 100, and 103 to isoleucine residues with pPROEX-HTb-mosue PrP23-230 as template; then primers 2MF and 2MR were used to mutate the 105 and 109 lysine residues to isoleucine residues using the previously mutated plasmid as template. The generation of human PrP mutants ΔCC1, P102L, P105L and 129V were described previously (30).
Recombinant PrP expression and purification
Recombinant mouse PrP23-230, PrP23-120, PrP121-230, ΔH, K/I, and recombinant human PrPs were purified as previously described (31, 32). Aliquots of purified PrPs in deionized water were stored at −80°C. The protein concentration used in our experiment was OD280 = 0.25. If necessary, the molar concentration was calculated using the molar extinction coefficient ε280 according to ExPASy Proteomics Server of the Swiss Institute of Bioinformatics.
Preparation of lipid vesicles
The isolation of lipids from N2A cells or mouse brains was described previously (33). Other lipids were purchased from Avanti Polar Lipids Inc. For vesicle preparation, lipids in chloroform were dried under a stream of nitrogen at 42°C and then hydrated in 20 mM Tris-HCl buffer (pH 7.4) to reach a final concentration of 2.5 mg/ml. The hydrated lipids were vortexed and then sonicated in a cup-hold sonicator (Misonix Inc. Model XL2020) until clear. Prepared lipid vesicles were flushed with argon and stored at 4°C.
Gradient analysis
For the discontinuous iodixanol density gradient floatation assay, rPrP and lipid vesicles were mixed together for 10 minutes at room temperature and applied to the high density phase of the iodixanol gradient as previously described (29, 33). For high salt and pH extraction, 0.5 M NaHCO3 (pH 11) or a solution of 1.5 M KCl+10 mM NaOH was added to rPrP-lipid mixtures prior to the gradient analysis. For salt competition assay, rPrP was pre-equilibrated with various concentrations of KCl for 5 minutes at room temperature. Immediately before mixing rPrP with lipids, same concentrations of KCl were added to the lipid vesicles. The mixed rPrP and lipid solution was incubated at room temperature for 10 minutes prior to the iodixanol density gradient analysis. Twelve fractions (200 μl/fraction) were collected from top to bottom of the gradient or as indicated.
PrP lipid incubation and PK digestion
For all analyses, rPrP was first subject to a 1-hour 100,000g centrifugation and only soluble rPrP was used. For rPrP and lipid incubation, 300 μl of rPrP (OD280 = 0.25) was mixed with 100 μl of lipid vesicles (2.5 mg/ml). The incubation was carried out at 37°C for indicated time periods. For all PK digestions, 10 μl of incubated samples were subjected to PK digestion at 37°C for 30 minutes with a PK:rPrP molar ratio of 1:16. The reaction was stopped by adding 5 mM phenylmethyl-sulfonylfluoride (PMSF) and kept on ice for 10 minutes. One tenth of PK digested samples were separated by SDS-PAGE and the PrP was detected by immunoblot analyses with the POM1 anti-PrP antibody (34).
Statistical analysis
Student T test was performed for statistical analysis, and the level of significance was set at p < 0.05.
RESULTS
N-terminal part of PrP initiates electrostatic interaction between PrP and anionic lipid
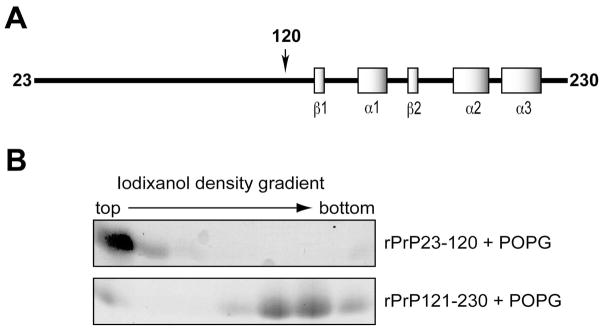
Structural studies revealed that the N-terminal part of PrP is flexible and unstructured, while the C-terminal part is well structured containing 2 β-strands and 3 α-helices (35, 36) (Fig. 1A). In order to determine which part of rPrP is important for its interaction with lipids, we generated two rPrP mutants, the N-terminal rPrP23-120 and the C-terminal rPrP121-230. The mutants were incubated with POPG and then analyzed by the iodixanol gradient. In this discontinuous density gradient, samples were loaded at the bottom, and the lipid-bound protein would migrate to the top of the gradient after ultracentrifugation (29, 33). We found that the N-terminal rPrP23-120 bound strongly to POPG, whereas the majority of the C-terminal rPrP121-230 fragment failed to interact with POPG (Fig. 1B). The N-terminal rPrP23-120 contains two clusters of positively charged amino acid residues: one at the very N-terminus (positively charged cluster 1, designated as CC1) and the other in the middle region (positively charged cluster 2, designated as CC2) (Fig. 2A). Therefore, this result is consistent with our previous finding that the PrP-lipid interaction is initiated by electrostatic contact (29), suggesting that these positively charged regions are important for the initial electrostatic contact between rPrP and anionic POPG.
Figure 1. The N-terminal fragment of rPrP interacts with anionic lipid.
(A) Illustration of mouse rPrP. (B) Iodixanol density gradient analysis of rPrP23-120 + POPG or rPrP121-230 + POPG. Eight fractions (300 μl/fraction) were collected from top to bottom, and the rPrPs were detected by Coomassie Brilliant Blue staining.
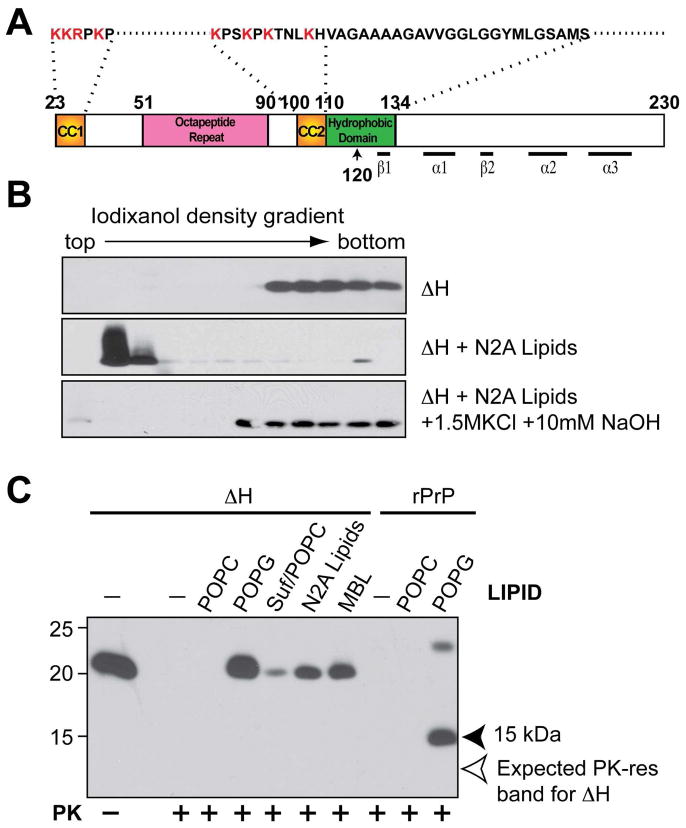
Figure 2. The highly conserved hydrophobic domain of PrP is required for hydrophobic rPrP-lipid interaction.
(A) Illustration of mouse PrP23-230. The amino acids were numbered according to mouse PrP sequence. (B) Iodixanol density gradient analyses of mouse rPrPΔH alone, rPrPΔH + total lipids isolated from N2A neuroblastoma cells (N2A lipids), rPrPΔH + N2A lipids extracted with the 1.5 M KCl plus 10 mM NaOH solution. (C) Mouse rPrP or rPrPΔH were incubated with indicated lipids for 1 hour and subjected to PK digestion. Solid arrowhead points at the 15 kDa PK-resistant band generated from wild-type rPrP. Empty arrow points at the expected position of PK-resistant band of rPrPΔH. Suf, Sulfatide; Suf/POPC, sulfatide and POPC at a mass ratio of 1:1; MBL, mouse brain lipids. PrP was detected by immunoblot analysis with POM1 antibody.
The conserved hydrophobic domain of PrP is required for hydrophobic rPrP-lipid interaction
In addition to the electrostatic interaction, rPrP also interacts with anionic lipids hydrophobically (29). To determine whether the hydrophobic domain (amino acid 111-134) of PrP (Fig. 2A) is involved in the hydrophobic lipid-PrP interaction, we generated a mouse rPrP mutant in which the hydrophobic domain was deleted (amino acid 111-131 deletion, designated as: ΔH). To rule out the possibility that the deletion mutant alters the density and interferes with the gradient analysis, we subjected the ΔH mutant to the iodixanol gradient analysis (29, 33). After ultracentrifugation, ΔH remained at the bottom of the gradient, revealing that the hydrophobic domain deletion did not drastically alter the rPrP density (Fig. 2B, top panel). When ΔH was mixed with lipids, it bound to the lipids and migrated to the top of the gradient (Fig. 2B, middle panel). In contrast to wild-type rPrP-lipid interaction (29), the ΔH-lipid binding was almost completely disrupted by the extraction with a high salt and high pH solution (Fig. 2B, bottom panel), indicating a significantly weakened hydrophobic interaction.
Next, we determined whether the ΔH mutant was able to gain the lipid-induced C-terminal PK-resistance. We previously showed that the anionic lipid interaction causes rPrP to gain different PK-resistant forms (29). Because the C-terminal 15 kDa PK-resistant fragment is similar to the PK-resistant core of recombinant prion in size and pattern (12), we used it as a marker to monitor the rPrP-lipid interaction and the subsequent conformational change. After incubation with various anionic lipids that are capable of converting rPrP ((29) and Fig. 2C, last lane), we found that no C-terminal PK-resistant fragment of mutant ΔH was detected in any of the samples (Fig. 2C, pointed by an empty arrow).
The POM1 antibody used in this experiment recognizes a discontinuous epitope in the globular domain of PrP (amino acid 121-230) (34). The ΔH mutant deleted a fragment within this region, raising the possibility that the lack of the C-terminal PK-resistant band were due to the loss of POM1 epitope. To rule out this possibility, we performed immunoblot analysis with 8H4 antibody, which has a more defined C-terminal epitope (amino acid 175-195) outside of the hydrophobic domain. Our result verified that the lack of C-terminal PK-resistant rPrP fragment was not due to the loss of POM1 epitope (data not shown). Together, our results support that the hydrophobic domain of PrP is required for the hydrophobic rPrP-lipid interaction, which is important for rPrP to gain the C-terminal PK-resistance.
Although the C-terminal PK-resistant form was not detected, various levels of full-length PK-resistant ΔH was detected in samples incubated with anionic lipids, but not in control samples in which ΔH was incubated with or without zwitterionic POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) (Fig. 2C). Based on the fact that ΔH was completely digested in control samples and the wild-type rPrP showed a correct PK-resistant pattern after incubation with anionic POPG (Fig. 2C, last lane) (29), we concluded that the appearance of full-length PK-resistant ΔH was not due to a insufficient PK-digestion. Actually, the separation of full-length and C-terminal PK-resistant forms in these samples and in experiments described below supports that, as we previously suggested (29), these are different rPrP forms that resulted from the rPrP-lipid interaction.
Middle region localized lysine residues affect rPrP-lipid interaction
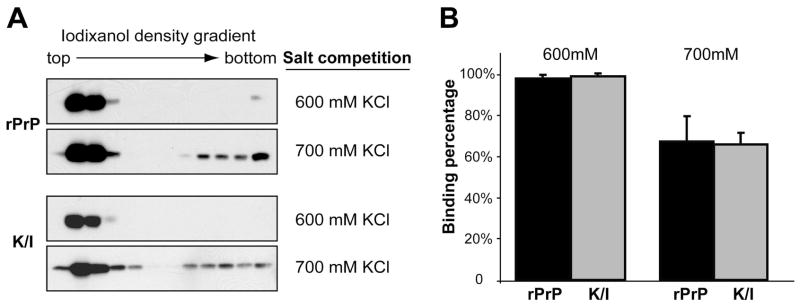
The finding that the hydrophobic domain plays an important role in rPrP-lipid interaction led us to predict that CC2 (Fig. 2A), a positively charged region immediately preceding the hydrophobic domain, will likely modulate rPrP-lipid interaction. To test this hypothesis, we generated a mouse rPrP mutant in which all four lysine residues in CC2 (residue 100, 103, 105, and 109 of mouse PrP) were replaced by isoleucine (designated as K/I). Taking advantage of the fact that high concentrations of salt inhibit the initial electrostatic rPrP-lipid contact (29), we performed a salt competition analysis to compare the strength of electrostatic rPrP-lipid interaction between wild-type and mutant rPrPs. In the presence of 600 mM KCl, the majority of wild-type rPrP bound to POPG. When the salt concentration increased to 700 mM KCl, only about 70% of rPrP remained lipid bound (Fig. 3A, top panels, and Fig. 3B). Interestingly, although the K/I mutant lost quite a few positive charges, the pattern of K/I mutant binding to POPG was similar to that of wild-type rPrP in the salt competition assay (Fig. 3). This result indicates that the loss of positive charges in CC2 does not significantly alter the strength of electrostatic rPrP-lipid interaction.
Figure 3. Positively charged lysine residues in the middle region do not affect electrostatic interaction between rPrP and anionic lipids.
(A) Salt competition assays for wild type rPrP and the K/I mutant. The rPrPs and POPG were pre-equilibrated with KCl at indicated concentrations and then mixed together. After a 10 minute incubation at room temperature, rPrP-lipid mixtures were subjected to iodixanol density gradient separation. PrP was detected by immunoblot analysis with POM1 antibody. (B) Densitometric analyses of results in (A). The density sum of all 12 fractions in each experiment was used as the total amount of rPrP, and the density sum of the first six fractions was used as the amount of lipid bound rPrP. The binding assays were repeated 3 times for each sample under indicated conditions. Error bar represents the standard deviation.
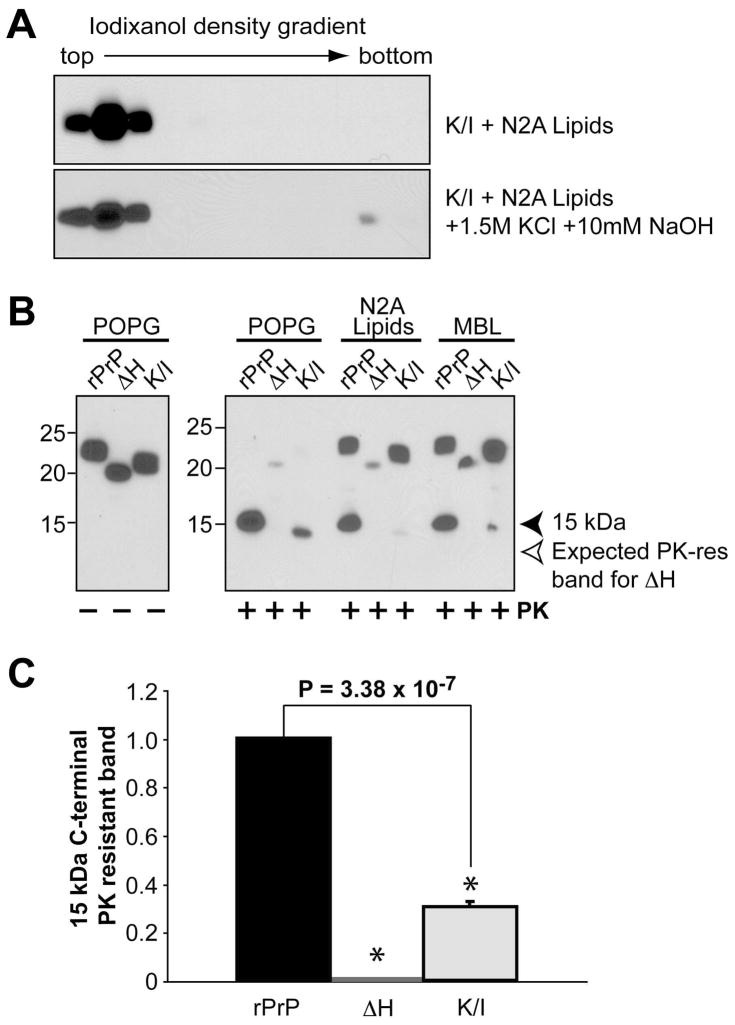
When the K/I mutant-lipid complex was extracted with a high salt and high pH solution, the K/I mutant remained lipid-bound and migrated to the top fractions (Fig. 4A), suggesting a hydrophobic interaction between the K/I mutant and lipids. Intriguingly, while the elimination of these four lysine residues did not seem to alter the strength of electrostatic or hydrophobic lipid-rPrP interaction, it did significantly reduce the lipid-induced C-terminal PK-resistant rPrP (Fig. 4B & C). This finding suggests that the lysine residues in CC2 modulate the rPrP-lipid interaction in such a manner that assists the generation of C-terminal PK-resistant rPrP form.
Figure 4. Middle region localized lysine residues affect lipid-induce rPrP conformational change.
(A) Iodixanol density gradient analysis of mouse rPrP K/I + N2A lipids or K/I + N2A lipids extracted with a 1.5 M KCl plus 10 mM NaOH solution as indicated. (B) Wild type mouse rPrP, ΔH, or K/I were incubated with indicated lipids for 1 hour and subjected to PK digestion. MBL, mouse brain lipids. PrP was detected by immunoblot analysis with POM1 antibody. (C) Densitometric analysis of the 15 kDa PK-resistant band. The density of the 15 kDa band generated from POPG incubated rPrP was set as 1. The density of the 15 kDa band generated from POPG incubated ΔH or K/I mutant was used to calculate the PK resistance. The PK-digestion assay was repeated 3 times for each sample. The error bar represents the standard deviation and the asterisk indicates a significant difference.
Disease-associated P105L and P102L mutations and N-terminal positively charged region affect rPrP-lipid interaction in different manners
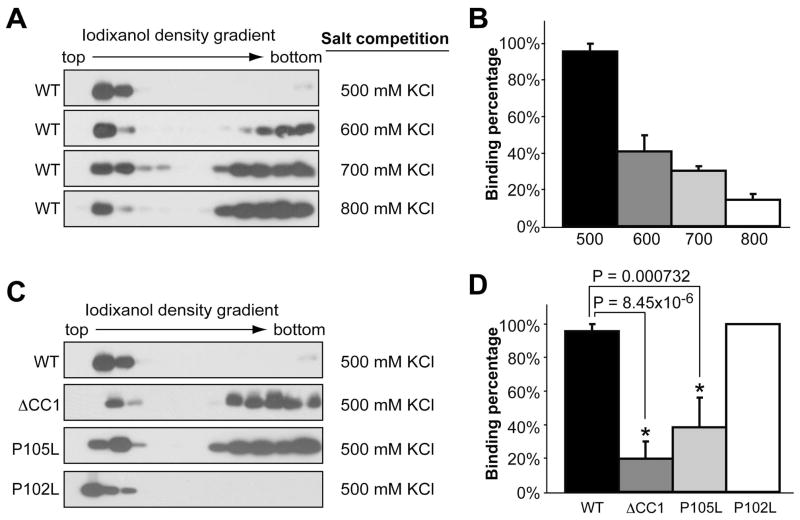
To determine the influence of disease-associated mutation and the N-terminal positively charged region on rPrP-lipid interaction, we analyzed two CC2 localized disease-associated mutations, P102L and P105L, and a PrP mutant without CC1, the very N-terminal positively charged amino acid cluster (amino acid 23-27 deletion, designated as: ΔCC1). Since P102L and P105L are mutations causing human disease, these mutants were made with recombinant human PrP and wild-type human rPrP (designated as: rhPrP) was included as a control to assess the influences of these mutations.
First, we performed the salt competition assay with rhPrP. The majority of rhPrP bound to POPG in the presence of 500 mM KCl, and increasing salt concentrations above 500 mM reduced the amounts of lipid bound rhPrP (Fig. 5A and 5B). Next, we analyzed the lipid-binding capability of various rhPrP mutants in the presence of 500 mM KCl. As expected, a significant portion of ΔCC1 mutant failed to bind POPG in the presence of 500 mM KCl (Fig. 5C, second panel, and Fig. 5D, p = 8.45×10−6), which is likely due to its reduced positive charges at the N-terminus. Surprisingly, the P105L mutant, which retains all the positively charged amino acids, exhibited a significantly reduced POPG binding as well (Fig. 5C, third panel, and Fig. 5D, p = 0.000732). In contrast, the P102L mutant, which is only 3 amino acids upstream and has exactly the same amino acid replacement as the P105L mutant, bound to POPG equally as well as rhPrP (Fig 5C, 4th panel and Fig. 5D).
Figure 5. Middle region localized P105L and P102L mutations and positive charges at N-terminus have different effects on electrostatic rPrP-lipid interaction.
(A) Salt competition assays for wild type human rPrP incubated with POPG in the presence of indicated salt concentrations. After binding, the POPG-rPrP complex was separated by the iodixanol density gradient. (B) Densitometric analyses of results in (A). (C) Salt competition assays for human rPrP mutants ΔCC1, P105L, or P102L incubated with POPG in the presence of 500 mM KCl. PrP was detected by immunoblot analysis with POM1 antibody. (D) Densitometric analyses of results in (C). For densitometric analyses, the density sum of all 12 fractions in each experiment was used as the total amount of rPrP, and the density sum of the first six fractions was used as the amount of lipid bound rPrP. The binding assays were repeated 3 times for each sample. The error bar represents the standard deviation and the asterisk indicates a significant difference.
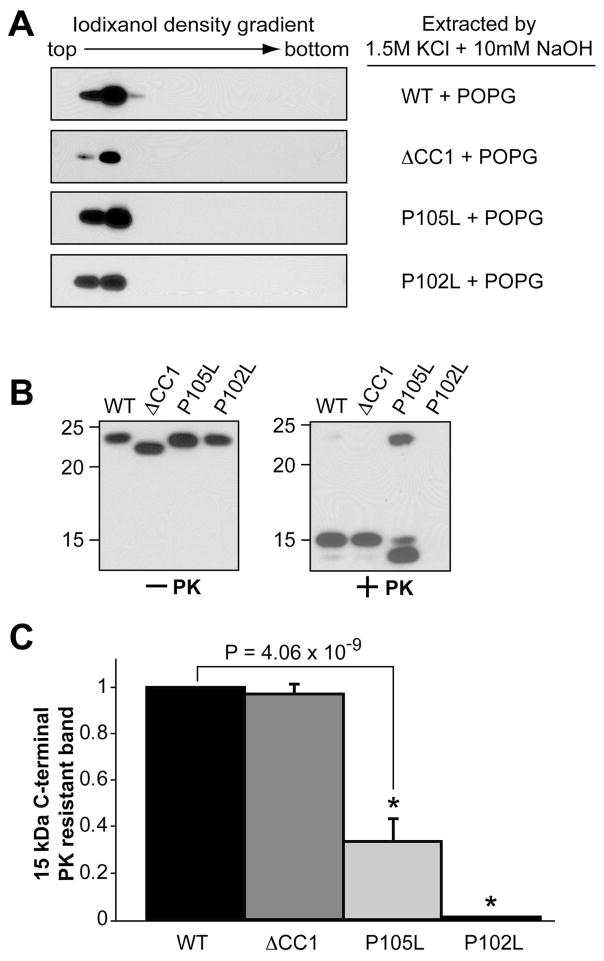
Despite different behaviors in the salt competition assay, all mutants were 100% POPG bound at physiological salt concentration (data not shown). Moreover, they remained lipid bound after extraction with a high salt and high pH solution (Fig. 6A). This result indicates that these mutations do not significantly affect the strength of hydrophobic rPrP-lipid interaction.
Figure 6. Disease-associated P105L and P102L mutations and N-terminal positively charged region affect lipid-induced rPrP conformational change in different manners.
(A) Wild type human rPrP, ΔCC1, P105L, or P102L mutant was incubated with POPG and then extracted by an alkaline solution of 1.5M KCl and 10mM NaOH prior to the iodixanol density gradient analysis. (B) Wild type human rPrP, ΔCC1, P105L, or P102L were incubated with POPG for 1 hour and subjected to PK digestion. PrP was detected by immunoblot analysis with POM1 antibody. (C) Densitometric analyses of PK digestion results in (B). The density of the 15 kDa PK-resistant band of wild-type rPrP was set as 1. The density of the 15 kDa band of ΔCC1, P105L or P102L mutant was used to calculate the PK resistance. The PK-digestion assay was repeated 3 times for each sample. The error bar represents the standard deviation and the asterisk indicates a significant difference.
PK digestion was performed to analyze the influence of these mutations on lipid-induced C-terminal PK-resistance and dramatic differences were observed (Fig. 6B). After a 1-hour incubation with POPG at physiological salt concentration (150 mM NaCl), the ΔCC1 mutant generated a similar amount of the 15 kDa C-terminal PK-resistant fragment as wild-type PrP (Fig. 6B, +PK, the equal amounts of rhPrPs input were verified by immunoblot analysis with samples without PK digestion, Fig. 6B, −PK). However, a much weaker 15 kDa PK-resistant band was detected with the P105L mutant under the same condition (Fig. 6B and 6C, p = 4.06×10−9), while a PK-resistant fragment with slightly smaller molecular weight (~14 kDa) was increased. Surprisingly, the P102L mutant, which interacts with lipids similarly to wild-type PrP, completely failed to generate any PK-resistant band (Fig. 6B and 6C). Together with lipid binding data, these results suggest that both the positively charged CC1 and CC2 regions contribute to rPrP-lipid interaction. Yet the generation of C-terminal PK-resistant rPrP form is mainly influenced by how the CC2 interacts with lipids.
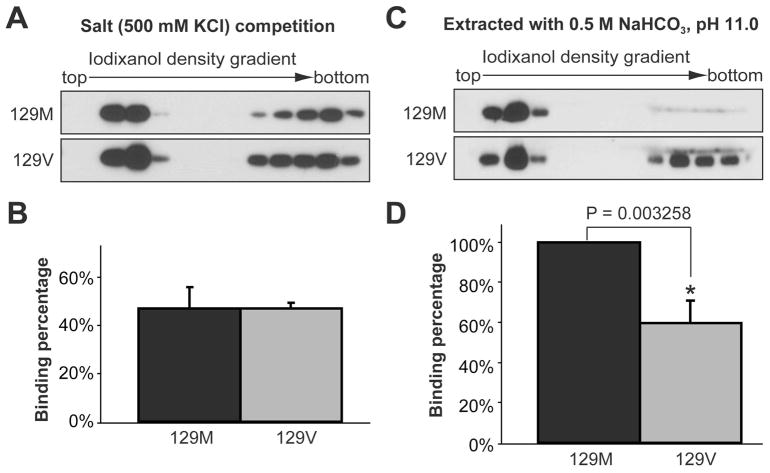
The middle region localized 129 polymorphism affects rPrP-lipid interaction
The 129 ploymorphism, which is localized in the hydrophobic domain, has a significant influence on the pathogenesis of prion disease (37). We tested its influence on rPrP-lipid interaction by generating recombinant human PrP with either methionine or valine at residue 129 (designated as 129M and 129V). The salt competition assay was performed with POPG or mixed lipids isolated from mouse brain (MBL). When POPG was used, almost 100% of 129M and 129V were lipid-bound in the presence of 500 mM KCl (Fig. 5A and data not shown). When MBL was used, the overall capability of rhPrP to bind MBL was reduced (comparing Fig. 7A and Fig. 5A), and some rhPrP remained at the bottom of the gradient. However, no significant difference was observed between 129M and 129V, suggesting that the 129 polymorphism does not significantly alter the strength of electrostatic rPrP-lipid interaction (Fig. 7A and 7B). The high salt and high pH extraction analysis was performed to determine whether the 129 polymorphism alters the strength of hydrophobic rhPrP-lipid interaction. While no difference was observed when POPG was used (data not shown), a significant difference between 129M and 129V was detected when the rhPrP-MBL complexes were extracted by 0.5 M NaHCO3 (pH 11.0). As shown in figure 7C, a significant portion of 129V was extracted from the complex, while the majority of 129M remained lipid bound (Fig. 7C and 7D, p = 0.003258). This result suggested that the 129 polymorphism alters the hydrophobic strength of rPrP binding to certain types of lipids.
Figure 7. The middle region localized 129 polymorphism affects hydrophobic rPrP-lipid interaction.
(A) Salt competition assay for human rPrP 129M or 129V incubated with mouse brain lipids (MBL) in the presence of 500 mM KCl. (B) Densitometric analyses of results in (A). (C) Human rPrP 129M+MBL or 129V+MBL complex were extracted by a 0.5 M NaHCO3, pH 11.0 solution prior to the iodixanol density gradient analysis. PrP was detected by immunoblot analysis with POM1 antibody. (D) Densitometric analyses of results in (C). For densitometric analysis, the density sum of all 12 fractions in each experiment was used as the total amount of rPrP, and the density sum of the first six fractions was used as the amount of lipid bound rPrP. The binding assays were repeated 3 times for each sample. The error bar represents the standard deviation and the asterisk indicates a significant difference.
DISCUSSION
The significance of PrP-lipid interaction in the pathogenesis of prion disease has been implicated by previous studies (17–21). The successful conversion of rPrP into a highly infectious prion in the presence of lipids (12) supports an important role of the PrP-lipid interaction in this process. We previously showed that the anionic lipid-PrP interaction is mediated by both electrostatic and hydrophobic contacts, and that the lipid interaction converts rPrP into a conformation with a 15 kDa C-terminal PK-resistant fragment. The efficiency of lipid-induced rPrP conversion depends on the structure of lipid headgroups and/or the presentation of these headgroups on the surface of lipid vesicles (29). In this study, we analyzed PrP domains involved in the PrP-lipid interaction and revealed different roles of the N-terminal and middle regions of PrP. Moreover, our results of disease-associated P102L and P105L mutations and the 129 polymorphism are consistent with the notion that the PrP-lipid interaction plays a role in the pathogenesis of prion disease.
Our results of the salt competition study of wild-type and ΔCC1 mutant PrPs (Fig. 5C and 5D) clearly showed that the N-terminal positively charged region is involved in the electrostatic interaction. The fact that the ΔCC1 mutant behaves similarly to the wild-type PrP in high salt and high pH extraction and in PK-digestion experiments (Fig. 6A and 6B) indicates that the N-terminal CC1 region does not significantly contribute to the hydrophobic PrP-lipid interaction and the subsequent conformational change of PrP, supporting that the lipid-induced PrP conformational change mainly involves the middle and C-terminal parts of PrP (38).
The highly conserved middle region contains a cluster of four positively charged lysine residues and a hydrophobic domain (39, 40). The ΔH mutant analyses suggest that the hydrophobic domain is the major PrP domain responsible for the hydrophobic PrP-lipid interaction. Moreover, the inability of the ΔH mutant to gain the C-terminal PK-resistance after incubation with a variety of anionic lipids (Fig. 2C and 4B) suggests that the hydrophobic PrP-lipid interaction is essential for its conversion to the PK-resistant form. In addition to the hydrophobic domain, our results also reveal that the four lysine residues in the CC2 region influence the PrP conformational change. The K/I mutant significantly reduced the amount of lipid-induced C-terminal PK-resistance (Fig. 4B). Both middle region localized P102L and P105L mutants reduced the amount of C-terminal PK-resistant form (Fig. 6B). Notably, both 102 and 105 proline residues are flanked by lysine residues in the middle region. Since proline is conformationally restrained, the replacement of proline with leucine would alter the presentation of positively charged lysine residues. Together, our results suggest that the four lysine residues in CC2 region are crucial in orienting the hydrophobic PrP-lipid contact in such a manner that leads rPrP to convert to the C-terminal PK-resistant conformation.
The reduced C-terminal PK-resistant bands associated with the P102L and P105L mutations are consistent with several previous reports of P105L associated GSS cases or transgenic mice expressing P102L mutant, which showed severe neurodegeneration without detectable amount of PK-resistant PrP (41–43). The toxic mechanism of these mutations may involve disruptions in normal PrP folding, in PrP metabolism, or in PrP’s interaction with other ligands for its normal function. Our results, that these two mutants differ drastically from wild-type PrP in lipid binding, may suggest a role of the PrP-lipid interaction in these processes.
Some paradoxical results were obtained comparing the K/I mutant and the P102L or P105L mutant. The K/I mutant, with all 4 middle region localized lysines mutated, did not appear to alter the strength of either the electrostatic or hydrophobic PrP-lipid interaction (Fig. 3A and 4A). However, the P105L mutant did significantly alter the strength of electrostatic PrP-lipid interaction (Fig. 5C). Moreover, although the P102L mutant did not alter the rPrP-lipid binding properties in our assays (Fig. 5C), it completely failed to convert to the PK-resistant form (Fig. 6B). However, with the K/I and P105L mutants, the 15 kDa C-terminal PK-resistant band was reduced, but still detectable (Fig. 4B and 6B). These paradoxical results suggest that, besides their effects on flanking lysine residues, the P105L and P102L mutations likely have an impact on the global structure of rPrP as well (30), which could affect rPrP binding to lipid or the lipid-induced rPrP conformation. Nevertheless, our results clearly reveal the importance of the CC2 region in PrP-lipid interaction and in lipid-induced PrP conversion.
The 129 polymorphism, localized in the hydrophobic domain, significantly affects the susceptibility and pathogenesis of prion disease (37). However, structural studies revealed similar conformation and stability between PrP variants bearing M or V at residue 129 (44), suggesting that it is not their influence on PrP structure, but rather their differences in interacting with other ligands may contribute to the different pathogenic changes. We found that the M and V variants differ in their hydrophobic interactions with lipids and a stronger hydrophobic interaction was detected for 129M (Fig. 7C and D). Although valine is more hydrophobic than methionine, the hydrophobic PrP-lipid interaction does not rely only on residue 129, but involves multiple hydrophobic amino acids. Substituting methionine with valine increases the hydrophobicity and likely causes a tighter interaction between residue 129 and fatty acyl chains. The tight lipid interaction at residue 129 could alter the interaction between lipids and other surrounding hydrophobic amino acids. As a result, the total strength of the hydrophobic lipid interaction is reduced for 129V.
Collectively, our results provide novel insights into the PrP-lipid interaction and indicate a significant role of the highly conserved middle region in lipid-induced PrP conversion. The facts that disease-associated mutations and the 129 polymorphism significantly alter PrP-lipid interaction support the relevance of PrP-lipid interaction in the pathogenesis of prion disease.
Acknowledgments
We thank Dr. Adriano Aguzzi for providing the POM1 antibody and Kate Lorenzetti for proofreading the manuscript.
Abbreviations
- PrP
prion protein
- PK
proteinase K
- rPrP
recombinant mouse PrP
- CJD
Creutzfeldt-Jakob Disease
- GSS
Gerstmann-Straussler-Scheinker
- GPI
glycosylphosphatidylinositol
- CC1
positively charged cluster 1
- CC2
positively charged cluster 2
- ΔH
recombinant mouse PrP with amino acid residues 111-131 deletion
- K/I
recombinant mouse PrP replacing lysine residues at 100, 103, 105 and 109 with isoleucines
- rhPrP
recombinant human PrP
- ΔCC1
recombinant human PrP without the very N-terminal 5 amino acids
- P105L
recombinant human PrP with a leucine at residue 105
- P102L
recombinant human PrP with a leucine at residue 102
- MBL
total mouse brain lipids
- POPG
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt)
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- Suf
Sulfatides (Brain): Cerebroside (Brain, Porcine) (ammonium salt)
Footnotes
This work is supported by grants from the Ellison Medical Foundation and National Institutes of Health (NIH) R01NS060729.
References
- 1.Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caughey B, Chesebro B. Prion protein and the transmissible spongiform encephalopathies. Trends in Cell Biol. 1997;7:56–62. doi: 10.1016/S0962-8924(96)10054-4. [DOI] [PubMed] [Google Scholar]
- 3.Watts JC, Balachandran A, Westaway D. The expanding universe of prion diseases. PLoS Pathog. 2006;2:e26. doi: 10.1371/journal.ppat.0020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collinge J. Prion diseases of humans and animals: their causes and molecular basis. Annu Rev Neurosci. 2001;24:519–550. doi: 10.1146/annurev.neuro.24.1.519. [DOI] [PubMed] [Google Scholar]
- 5.Aguzzi A. Prion diseases of humans and farm animals: epidemiology, genetics, and pathogenesis. J Neurochem. 2006;97:1726–1739. doi: 10.1111/j.1471-4159.2006.03909.x. [DOI] [PubMed] [Google Scholar]
- 6.Caughey B, Baron GS. Prions and their partners in crime. Nature. 2006;443:803–810. doi: 10.1038/nature05294. [DOI] [PubMed] [Google Scholar]
- 7.Cohen FE, Prusiner SB. Pathologic conformations of prion proteins. Annu Rev Biochem. 1998;67:793–819. doi: 10.1146/annurev.biochem.67.1.793. [DOI] [PubMed] [Google Scholar]
- 8.Baskakov IV, Breydo L. Converting the prion protein: what makes the protein infectious. Biochim Biophys Acta. 2007;1772:692–703. doi: 10.1016/j.bbadis.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- 10.Aguzzi A, Baumann F, Bremer J. The prion’s elusive reason for being. Annu Rev Neurosci. 2008;31:439–477. doi: 10.1146/annurev.neuro.31.060407.125620. [DOI] [PubMed] [Google Scholar]
- 11.Caughey B, Baron GS, Chesebro B, Jeffrey M. Getting a grip on prions: oligomers, amyloids, and pathological membrane interactions. Annu Rev Biochem. 2009;78:177–204. doi: 10.1146/annurev.biochem.78.082907.145410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang F, Wang X, Yuan CG, Ma J. Generating a prion with bacterially expressed recombinant prion protein. Science. 2010;327:1132–1135. doi: 10.1126/science.1183748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Supattapone S. Biochemistry. What makes a prion infectious? Science. 2010;327:1091–1092. doi: 10.1126/science.1187790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deleault NR, Lucassen RW, Supattapone S. RNA molecules stimulate prion protein conversion. Nature. 2003;425:717–720. doi: 10.1038/nature01979. [DOI] [PubMed] [Google Scholar]
- 15.Deleault NR, Geoghegan JC, Nishina K, Kascsak R, Williamson RA, Supattapone S. Protease-resistant prion protein amplification reconstituted with partially purified substrates and synthetic polyanions. J Biol Chem. 2005;280:26873–26879. doi: 10.1074/jbc.M503973200. [DOI] [PubMed] [Google Scholar]
- 16.Deleault NR, Harris BT, Rees JR, Supattapone S. From the Cover: Formation of native prions from minimal components in vitro. Proc Natl Acad Sci U S A. 2007;104:9741–9746. doi: 10.1073/pnas.0702662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein TR, Kirsch D, Kaufmann R, Riesner D. Prion rods contain small amounts of two host sphingolipids as revealed by thin-layer chromatography and mass spectrometry. Biol Chem. 1998;379:655–666. doi: 10.1515/bchm.1998.379.6.655. [DOI] [PubMed] [Google Scholar]
- 18.Gabizon R, McKinley MP, Prusiner SB. Purified prion proteins and scrapie infectivity copartition into liposomes. Proc Natl Acad Sci U S A. 1987;84:4017–4021. doi: 10.1073/pnas.84.12.4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baron GS, Magalhaes AC, Prado MA, Caughey B. Mouse-adapted scrapie infection of SN56 cells: greater efficiency with microsome-associated versus purified PrP-res. J Virol. 2006;80:2106–2117. doi: 10.1128/JVI.80.5.2106-2117.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taraboulos A, Scott M, Semenov A, Avrahami D, Laszlo L, Prusiner SB. Cholesterol depletion and modification of COOH-terminal targeting sequence of the prion protein inhibit formation of the scrapie isoform. J Cell Biol. 1995;129:121–132. doi: 10.1083/jcb.129.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naslavsky N, Shmeeda H, Friedlander G, Yanai A, Futerman AH, Barenholz Y, Taraboulos A. Sphingolipid depletion increases formation of the scrapie prion protein in neuroblastoma cells infected with prions. J Biol Chem. 1999;274:20763–20771. doi: 10.1074/jbc.274.30.20763. [DOI] [PubMed] [Google Scholar]
- 22.Baron GS, Caughey B. Effect of glycosylphosphatidylinositol anchor-dependent and -independent prion protein association with model raft membranes on conversion to the protease-resistant isoform. J Biol Chem. 2003;278:14883–14892. doi: 10.1074/jbc.M210840200. [DOI] [PubMed] [Google Scholar]
- 23.Morillas M, Swietnicki W, Gambetti P, Surewicz WK. Membrane environment alters the conformational structure of the recombinant human prion protein. J Biol Chem. 1999;274:36859–36865. doi: 10.1074/jbc.274.52.36859. [DOI] [PubMed] [Google Scholar]
- 24.Sanghera N, Pinheiro TJ. Binding of prion protein to lipid membranes and implications for prion conversion. J Mol Biol. 2002;315:1241–1256. doi: 10.1006/jmbi.2001.5322. [DOI] [PubMed] [Google Scholar]
- 25.Kazlauskaite J, Sanghera N, Sylvester I, Venien-Bryan C, Pinheiro TJ. Structural changes of the prion protein in lipid membranes leading to aggregation and fibrillization. Biochemistry. 2003;42:3295–3304. doi: 10.1021/bi026872q. [DOI] [PubMed] [Google Scholar]
- 26.Critchley P, Kazlauskaite J, Eason R, Pinheiro TJ. Binding of prion proteins to lipid membranes. Biochem Biophys Res Commun. 2004;313:559–567. doi: 10.1016/j.bbrc.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 27.Luhrs T, Zahn R, Wuthrich K. Amyloid formation by recombinant full-length prion proteins in phospholipid bicelle solutions. J Mol Biol. 2006;357:833–841. doi: 10.1016/j.jmb.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 28.Elfrink K, Ollesch J, Stohr J, Willbold D, Riesner D, Gerwert K. Structural changes of membrane-anchored native PrP(C) Proc Natl Acad Sci U S A. 2008;105:10815–10819. doi: 10.1073/pnas.0804721105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang F, Yang F, Hu Y, Wang X, Jin C, Ma J. Lipid interaction converts prion protein to a PrPSc-like proteinase K-resistant conformation under physiological conditions. Biochemistry. 2007;46:7045–7053. doi: 10.1021/bi700299h. [DOI] [PubMed] [Google Scholar]
- 30.Yin S, Pham N, Yu S, Li C, Wong P, Chang B, Kang SC, Biasini E, Tien P, Harris DA, Sy MS. Human prion proteins with pathogenic mutations share common conformational changes resulting in enhanced binding to glycosaminoglycans. Proc Natl Acad Sci U S A. 2007;104:7546–7551. doi: 10.1073/pnas.0610827104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zahn R, von Schroetter C, Wuthrich K. Human prion proteins expressed in Escherichia coli and purified by high-affinity column refolding. FEBS Lett. 1997;417:400–404. doi: 10.1016/s0014-5793(97)01330-6. [DOI] [PubMed] [Google Scholar]
- 32.Yin SM, Zheng Y, Tien P. On-column purification and refolding of recombinant bovine prion protein: using its octarepeat sequences as a natural affinity tag. Protein Expr Purif. 2003;32:104–109. doi: 10.1016/S1046-5928(03)00195-5. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Wang F, Arterburn L, Wollmann R, Ma J. The interaction between cytoplasmic prion protein and the hydrophobic lipid core of membrane correlates with neurotoxicity. J Biol Chem. 2006;281:13559–13565. doi: 10.1074/jbc.M512306200. [DOI] [PubMed] [Google Scholar]
- 34.Polymenidou M, Stoeck K, Glatzel M, Vey M, Bellon A, Aguzzi A. Coexistence of multiple PrPSc types in individuals with Creutzfeldt-Jakob disease. Lancet Neurol. 2005;4:805–814. doi: 10.1016/S1474-4422(05)70225-8. [DOI] [PubMed] [Google Scholar]
- 35.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wuthrich K. NMR structure of the mouse prion protein domain PrP(121-321) Nature. 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 36.Riek R, Hornemann S, Wider G, Glockshuber R, Wuthrich K. NMR characterization of the full-length recombinant murine prion protein, mPrP(23-231) FEBS Lett. 1997;413:282–288. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- 37.Ironside JW, Ritchie DL, Head MW. Phenotypic variability in human prion diseases. Neuropathol Appl Neurobiol. 2005;31:565–579. doi: 10.1111/j.1365-2990.2005.00697.x. [DOI] [PubMed] [Google Scholar]
- 38.Cobb NJ, Sonnichsen FD, McHaourab H, Surewicz WK. Molecular architecture of human prion protein amyloid: a parallel, in-register beta-structure. Proc Natl Acad Sci U S A. 2007;104:18946–18951. doi: 10.1073/pnas.0706522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hegde RS, Mastrianni JA, Scott MR, DeFea KA, Tremblay P, Torchia M, DeArmond SJ, Prusiner SB, Lingappa VR. A transmembrane form of the prion protein in neurodegenerative disease. Science. 1998;279:827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]
- 40.Aguzzi A, Sigurdson C, Heikenwaelder M. Molecular mechanisms of prion pathogenesis. Annu Rev Pathol. 2008;3:11–40. doi: 10.1146/annurev.pathmechdis.3.121806.154326. [DOI] [PubMed] [Google Scholar]
- 41.Telling GC, Haga T, Torchia M, Tremblay P, DeArmond SJ, Prusiner SB. Interactions between wild-type and mutant prion proteins modulate neurodegeneration in transgenic mice. Genes Dev. 1996;10:1736–1750. doi: 10.1101/gad.10.14.1736. [DOI] [PubMed] [Google Scholar]
- 42.Yamada M, Itoh Y, Inaba A, Wada Y, Takashima M, Satoh S, Kamata T, Okeda R, Kayano T, Suematsu N, Kitamoto T, Otomo E, Matsushita M, Mizusawa H. An inherited prion disease with a PrP P105L mutation: clinicopathologic and PrP heterogeneity. Neurology. 1999;53:181–188. doi: 10.1212/wnl.53.1.181. [DOI] [PubMed] [Google Scholar]
- 43.Collins S, McLean CA, Masters CL. Gerstmann-Straussler-Scheinker syndrome, fatal familial insomnia, and kuru: a review of these less common human transmissible spongiform encephalopathies. J Clin Neurosci. 2001;8:387–397. doi: 10.1054/jocn.2001.0919. [DOI] [PubMed] [Google Scholar]
- 44.Hosszu LL, Jackson GS, Trevitt CR, Jones S, Batchelor M, Bhelt D, Prodromidou K, Clarke AR, Waltho JP, Collinge J. The residue 129 polymorphism in human prion protein does not confer susceptibility to Creutzfeldt-Jakob disease by altering the structure or global stability of PrPC. J Biol Chem. 2004;279:28515–28521. doi: 10.1074/jbc.M313762200. [DOI] [PubMed] [Google Scholar]