Abstract
Progress in the management of patients with myelodysplastic syndromes (MDS) has been hampered by the inability to detect cytogenetic abnormalities in 40-60% of cases. We prospectively analyzed matched pairs of bone marrow and buccal cell (normal) DNA samples from 51 MDS patients by single nucleotide polymorphism (SNP) arrays, and identified somatically acquired clonal genomic abnormalities in 21 patients (41%). Among the 33 patients with normal bone marrow cell karyotypes, five (15%) had clonal, somatically acquired aberrations by SNP array analysis, including four with segmental uniparental disomies (UPD) and one with three separate microdeletions. Each abnormality was detected more readily in CD34+ cells then in unselected bone marrow cells. Paired analysis of bone marrow and buccal cell DNA from each patient was necessary to distinguish true clonal genomic abnormalities from inherited copy number variations and regions with apparent LOH. UPDs affecting chromosome 7q were identified in two patients who had a rapidly deteriorating clinical course despite a low-risk International Prognostic Scoring System score (IPSS). Further studies of larger numbers of patients will be needed to determine whether 7q UPD detected by SNP array analysis will identify higher-risk MDS patients at diagnosis, analogous to those with 7q cytogenetic abnormalities.
Keywords: Myelodysplastic Syndrome, SNP array, Uniparental Disomy
Introduction
The myelodysplastic syndromes (MDS) are predominantly diseases of the elderly (median age at diagnosis, 70 years) and are among the most common hematologic malignancies, affecting 1 in 500 persons over 65 years of age (1-3). Patients present with cytopenias involving one or more blood cell lineages, bone marrow morphologic findings indicative of mono-, di- or tri-lineage dysplasia, and a variable risk of transformation to acute leukemia. Currently, allogeneic stem cell transplantation offers the only curative therapy for MDS (4), but is not a realistic option for most elderly patients.
Cases of MDS are thought to arise from the clonal acquisition of mutations in multipotent hematopoietic stem cells, but the precise molecular abnormalities remain poorly defined. Bone marrow cytogenetic studies are helpful in this regard, revealing clonal abnormalities in 30 to 50% of cases, most frequently deletions of chromosomes 5, 7, and 20 and/or trisomy 8 (5). Cases with del(5q) as the sole abnormality, the so-called 5q syndrome, have shown remarkable responses to lenalidomide (6), and haploinsufficiency for the ribosomal protein 14 (RPS14) gene appears to account for many of the clinical features of this syndrome (7). This suggests that improved detection of clonal genomic aberrations in MDS could identify additional patients likely to respond well to lenalidomide or perhaps to DNA methyltransferase inhibitors, such as 5-azacitidine and decitabine (8, 9). However, this leaves an estimated 50 to 70% of cases with normal bone marrow cell karyotypes for whom there are no molecular tests that could be used to distinguish between MDS and benign bone marrow cell diseases.
Recent studies of human cancers have indicated that small regions of specific clonal genomic abnormalities can be detected with high-density single nucleotide polymorphism (SNP) arrays, which provide a detailed structural examination of the cancer genome, including high-resolution analysis for loss of heterozygosity (LOH) (10). Two groups have applied this technology to the study of archival bone marrow samples from MDS patients, reporting a high frequency of detection of uniparental disomy (UPD) and copy number alterations, even in cases with a normal karyotype (11-15). We were troubled by these reports, because the MDS bone marrow samples were analyzed without a paired buccal or other normal sample from the same patient. Instead, results were compared with those obtained from the blood cells of unrelated normal individuals. Without a paired analysis of bone marrow DNA with normal control DNA for each patient, we suspected that inherited copy number variations (CNVs) and genomic regions with apparent homozygosity due to linkage disequilibrium or consanguinity might confound the detection of somatically acquired abnormalities linked to the pathogenesis of the malignant clone. Thus, we undertook a prospective study of adults with MDS in which SNP array findings from bone marrow cell DNA were compared with those from matched control normal DNA extracted from autologous buccal swabs. We identified unambiguous, somatically acquired abnormalities with this approach in patients whose bone marrow cells had a normal karyotype, although at a lower frequency than has been previously reported (11-15). Importantly, we found that paired analysis of bone marrow and normal (buccal) DNA from each patient was necessary to distinguish acquired clonal abnormalities from inherited genomic variations.
Materials and Methods
Patients and clinical samples
Fifty-one patients were consecutively enrolled in this study seen at M.D. Anderson Cancer Center (MDACC) for a diagnosis and classification of MDS according to the WHO, FAB and IPSS systems (16-18) from June 21, 2006 to August 8, 2007. Patient characteristics are reported in Table 1 and Supplementary Tables 1 and 2. Buccal swabs and bone marrow aspirates were obtained with informed consent. Mononuclear cells were isolated from bone marrow aspirates by Ficoll centrifugation, and when sufficient cells were available, two thirds of these cells were further purified by magnetic beads (Milentyi) to obtain the CD34+ fraction. Cells were lysed in Gentra Puregene buffer (Qiagen). DNA from buccal swabs and the bone marrow cell lysates were prepared with the Gentra Puregene kit (Qiagen) and analyzed by spectrometry (Nanodrop Technologies).
Table 1. Characteristics of the patient cohort (n=51)*.
| Median Age (range) | 66 (43-83) | |||
|---|---|---|---|---|
| number | percentage | |||
| Cytogenetics | normal karyotype | 33 | 64.7 | |
| aberrant karyotype | 18 | 35.3 | ||
| Sex | female | 20 | 39.2 | |
| male | 31 | 60.8 | ||
| WHO classification | RA | 4 | 7.8 | |
| RCMD | 7 | 13.7 | ||
| RARS | 1 | 2.0 | ||
| RCDM-RS | 4 | 7.8 | ||
| RAEB-1 | 11 | 21.6 | ||
| RAEB-2 | 13 | 25.5 | ||
| unclassified | 11 | 21.6 | ||
| FAB classification | RA | 15 | 29.4 | |
| RARS | 5 | 9.8 | ||
| RAEB | 21 | 41.2 | ||
| RAEB-t | 5 | 9.8 | ||
| CMML | 3 | 5.9 | ||
| AML evolved from MDS | 2 | 3.9 | ||
| IPSS | 0 | Low risk | 12 | 23.5 |
| 0.5-1.0 | Intermediate 1 | 20 | 39.2 | |
| 1.5-2.0 | Intermediate 2 | 13 | 25.5 | |
| >2.0 | High | 5 | 9.8 | |
| AML evolved from MDS | 1 | 2.0 | ||
see Supplementary Tables 1 and 2 for patient-by-patient characteristics and results
SNP array analysis
Mononuclear bone marrow cell DNA and matched normal DNA from all 51 patients, together with the DNA from CD34+ bone marrow cells from 24 of these patients, was analyzed with Genechip Mapping 250K StyI arrays according to the manufacturer's protocol (Affymetrix). The data were analyzed with dChip (version August 5, 2008) (19). Raw data were normalized and modeled (PM/MM difference). Copy number values were computed by the “batch normalization” and “scaling copy number mode to 2 copies” algorithms. The raw data were inferred by the “median smoothing” algorithm and a window of 10 SNPs. To obtain standardized intensities (copy number values) across all probe sets, the log2 ratio was computed as log2 (signal intensity/mean signal intensity of all normal samples for a given SNP probe set). As a quality control, the standard deviation of the raw log2 ratio for each SNP probe set minus the raw log2 ratio for the adjacent SNP probe set was calculated along chr 1. Only samples with a standard deviation of 0.6 or smaller were included in the analysis. Reported segements of deletion or amplification (Supplementary Tables 1-3) were obtained by the Hidden Markov Model algorithm using a step of 0.2 and a standard deviation of 0.4. Only regions with a coverage of at least 5 consecutive probe sets, an absolute average value of 0.2 and visual confirmation are reported. To identify CNVs, each matched normal sample was analyzed in the same way. LOH was inferred by the Hidden Markov Model. The LOH analysis for each patient was based on the comparison with the matched normal DNA. Only segments with an LOH probability greater then 0.8 were considered as LOH. UPDs were identified as copy-neutral LOH by comparison with the results of the copy number analysis. In addition to the LOH probability criterion, a UPD regions had to have at least 6 LOH calls (all LOH calls minus all conflict calls within that region) to be reported. As a third criterion, regions containing less than 5% informative calls were disregarded. Intervening segments of apparent retention were considered as LOH by comparison to their copy number status or if the number of calls was less then 6 (all retention calls minus all conflict calls within the segment). For the unpaired analysis all normal samples were disregarded. Instead, a reference genotype file containing 60 normal controls from the CEPH subset of the HapMap project (20) was used for the LOH analysis. All other parameters remained unchanged. Since no individual comparative calls are generated, the threshold for a region to be considered was set to at least 50 probe sets (SNPs) and at least 2 Mb in size.
Results
High Frequency of Deletions in MDS patients
We analyzed bone marrow samples from 51 patients with newly diagnosed MDS using high-resolution SNP arrays. Their median age was 66 years (range, 43 to 83 years) and 32 of 51 (63%) had lower-risk disease (low or intermediate-1 IPSS scores). Thirty-three (65%) had normal karyotypes by conventional cytogenetic analysis. The distributions of patients by WHO, FAB and IPSS classifications are given in Table 1. For each patient, we compared the SNP array profiles of bone marrow mononuclear cell DNA with those of matched buccal swab DNA to detect somatically acquired clonal abnormalities of copy number as well as regions of LOH. Because MDS originates in hematopoietic stem cells, we also selected CD34+ cells from bone marrow aspirates of 24 patients and compared the SNP profiles with those of matched bone marrow mononuclear and buccal cells.
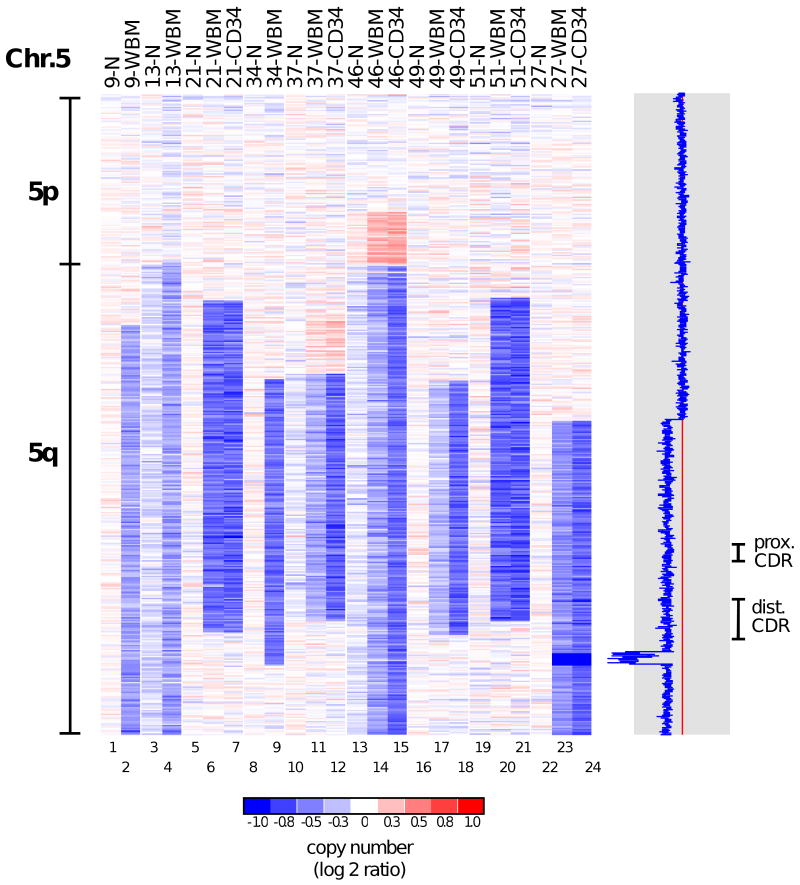
DNA copy number analysis identified chromosomal defects, mainly deletions, in 21 (41%) of the 51 patients (Supplementary Fig. 1, also Supplementary Tables 1 and 2). Chromosomal gains were less frequent than deletions and were found only in MDS cases with complex karyotypes (Supplementary Table 2). The most frequently deleted region (18% of all cases) was located on chromosome 5q, as is typical of cytogenetic findings in MDS(5). Heterozygous deletions were identified in eight patients (16%), together with a focal homozygous deletion of about 5 Mb within the 5q deleted region of patient 27 (Fig. 1, lanes 22-24). By conventional karyotyping, this case had both a del(5q) and a t(5;12)(q33;q12) translocation, accounting for the deletion. Although the homozygously deleted region is located outside the two previously defined critical deleted regions (CDRs) on chromosome 5q(21-23), half of our cases had a del(5q) that extended to the telomere, thus including the 5 Mb homozygously deleted region.
Figure 1. Copy number alterations on chromosome 5.
The copy number values for probe sets on chromosome 5 are shown as log2 ratios. Patients with abnormalities affecting this chromosome are identified at the top of each column together with the DNA source (N, normal cells; WBM, whole bone marrow mononuclear cells; CD34, CD34+ bone marrow cells). White areas indicate no copy number change (log2 ratio = zero), while shades of blue and red designate losses and gains, respectively (see scale at bottom). All blue areas on the q-arm correspond to heterozygous deletions, except in patient 27, where a homozygously deleted area (dark blue) was found within the heterozygous region of loss. A profile view of the copy number for this patient is shown on the right. The red line indicates the expected normal copy number, while the blue line indicates the log2 ratio of the measured copy number along the chromosome. The positions of the two previously described common deleted regions (CDRs) of chromosome 5q are indicated.
Examples of recurrent chromosomal gain were also found in this cohort. Two patients (27 and 49) had small regions of gain on chromosome 8 (Supplementary Fig. 3) that commonly encompassed the 8q24.13-21 locus, which harbors the MYC gene. Four others (patients 34, 37, 46 and 51) had 11q23 gains in association with multiple complex cytogenetic abnormalities, similar to the findings of others (24, 25).
Among the 24 patients with available CD34+ cell DNA, 11 (42%) had detectable chromosomal abnormalities by SNP array analysis. In most of these cases, the same aberrations were more readily detected in the purified CD34+ cells than in mononuclear bone marrow cells (see, for example, Fig.1, lanes 11 vs. 12, 14 vs. 15, 17 vs. 18 and 23 vs. 24), consistent with the origin of MDS in hematopoietic stem cells. Thus, residual normal cells were present in the whole bone marrow mononuclear fractions of MDS patients. A significant fraction of these residual normal cells appeared to be mature T lymphocytes included in the unfractionated bone marrow mononuclear cells, as deletions of the T-cell receptor alpha locus (14q11.2) were detected in the bone marrow mononuclear cells of some cases, but not in either the matched CD34+ cell DNA or the buccal cell DNA (Supplementary Fig. 2). Buccal scrapings from some patients contained a small fraction of clonal blood leukocytes, resulting in a slightly lower-than-expected copy number for the region that was hemizygously deleted in the malignant clone (see, for example, patient 46 in Fig.1, lane 13). Relatively large regions of copy number gain were also observed at the proximal boundaries of some of the deletions (patients 37 and 46; Fig. 1, lanes 11, 12, 14 and 15).
Clonal Abnormalities in Patients with a Normal Bone Marrow Cell Karyotype
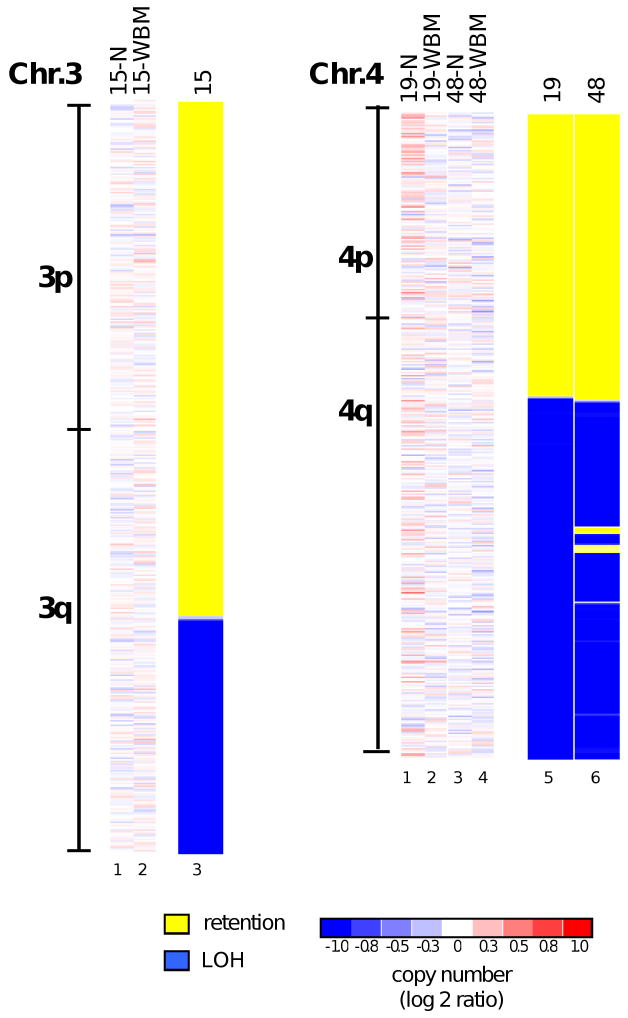
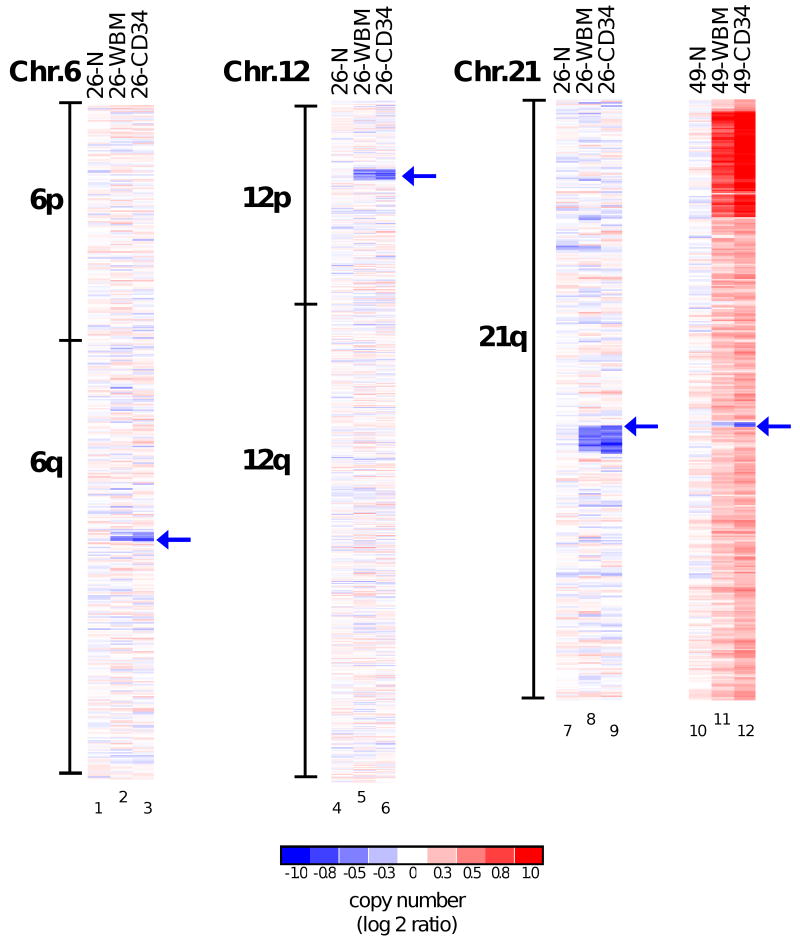
Comparison of DNA genotyping profiles for bone marrow versus buccal cells from each patient identified clonal regions of acquired, copy-neutral LOH (indicative of acquired segmental regions of uniparental disomy, UPD) in MDS bone marrow cells of four of 33 (12%) patients with a normal bone marrow cell karyotype (Figs. 2 and 4). In addition, one (patient 34) with a complex karyotype and one (patient 19) with a t(11,19) translocation had UPDs. The UPDs were found within segments of 3q (patient 15), 4q (patients 19 and 48), 7q (patients 5 and 7) and 17p (patient 34) and entirely extended to the telomere, suggesting that they resulted from a single recombination event per chromosome, possibly involving break-induced replication (BIR) (26). Monosomy 7 and del(7q) are established poor-risk cytogenetic findings in the bone marrow cells of MDS patients (17); hence, our detection of 7q UPDs spanning the same critical region in two of 33 cases with a normal karyotype indicates that approximately 6% of patients (90% confidence interval 1% to 18%) who lack other high-risk chromosomal features may harbor defects affecting this critical region of chromosome 7q. Patient 26 had a normal bone marrow cell karyotype by standard cytogenetic analysis, but SNP profiling revealed three small heterozygous deletions (Fig. 3) at 6q21 (3 Mb), 12p13.2 (1.5 Mb) and 21q22.12 (1.2 Mb). The 21q22.12 region contains the RUNX1 gene, and the deletion breakpoint was located within that locus. Alternatively, a cryptic translocation with an accompanying deletion of a small region could have led to the disruption of the intact RUNX1 locus in this patient. Another patient (49) also showed a disruption of the RUNX1 locus by a very small deletion, which was detected in the context of other abnormalities discernible by standard karyotyping. RUNX1 encodes a critical regulator of hematopoietic differentiation and can be inactivated by mutation or translocation in both MDS and AML (27, 28). Thus, SNP array analysis can pinpoint small regions of deletion that harbor tumor suppressor genes, such as RUNX1, which are often mutated in MDS.
Figure 2. Uniparental disomies in cases with a normal bone marrow cell karyotype.
Copy number analysis and paired genotype (LOH) analysis are shown for patients 15, 19 and 48, as indicated at the top of each column together with the DNA source (N, normal cells; WBM, whole bone marrow mononuclear cells). Only informative chromosomes are shown; all remaining chromosomal regions were normal by copy number and LOH analysis. The finding of LOH with a normal copy number reveals the presence of segmental UPD in these patients.
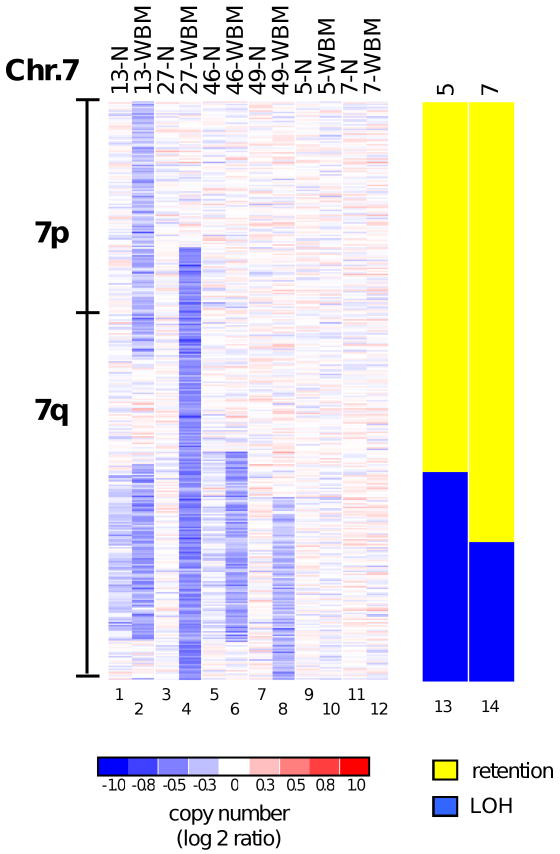
Figure 4. Uniparental disomies and copy number alterations on chromosome 7.
Copy number values along chromosome 7 are shown as log2 ratios for all patients having a copy number change on this chromosome and for patients 6 and 8, who lack a copy number change. Patient identifiers are included at the top of each column together with the DNA source (N, normal cells; WBM, whole bone marrow mononuclear cells). Two additional plots (for patients 5 and 7) show the results of a paired genotype analysis (LOH analysis), which is based on the comparison of mononuclear cell DNA with matched DNA. Yellow areas indicate retention of the genotype, while blue areas indicate LOH. In patients 5 and 7, the finding of LOH extending to the telomere of the long arm of chromosome 7 with a normal copy number is caused by segmental UPD (see text).
Figure 3. Small deletions identified in patients 26 and 49.
Copy number values along chromosomes 6 and 12 (patient 26) and 21 (patient 26 and 49) are shown as log2 ratios. Patient identifiers are designated at the top of each column, together with the DNA source (N, normal cells; WBM, whole bone marrow mononuclear cells). The blue arrows indicate the loci of deletions. On chromosome 21, the arrows indicate the locus of the RUNX1 gene, which is affected by a deletion distal to the centromere in patient 26 and proximal in patient 49. Chromosomes are not drawn to scale.
Association of UPD 7q with a Deteriorating Clinical Course
The short follow-up times for this study group precluded a statistical analysis of the association between SNP array findings and the clinical outcome of therapy. Within the normal karyotype subgroup (n=33), however, we noted that the two patients identified as having UPDs affecting the long arm of chromosome 7 (Fig. 4) experienced a deteriorating clinical course. One patient (no. 5) was a 56-year-old woman with refractory anemia and an IPSS score of zero who had a UPD at 7q22.1-7qtel at presentation. Over the next 10 months, she developed rapidly increasing bone marrow blast counts and worsening cytopenias and died. The other patient (no. 7) was a 67-year-old man with refractory anemia and a hypercellular bone marrow showing multilineage dysplasia and an IPSS score of zero. At presentation he had a UPD at 7q31.33-7qtel. Over the next 12 months, he developed thrombocytopenia, leukocytosis and pleural effusions, and died. The finding of a chromosome 7 cytogenetic abnormality in MDS confers a higher-risk score according to the IPSS criteria (17). In these two patients with normal bone marrow cell karyotypes, our results suggest that UPD 7q detected by SNP array may identify the same subset of higher risk patients as does a cytogenetic abnormality of chromosome 7 (Fig. 4).
Requirement for Matched Normal DNA in SNP Array Analysis
The human genome contains structural variations consisting of thousands of regions in which segments of DNA have been gained or lost. These so-called copy number variations, or CNVs, constitute much of the inherited variation among individuals (29-31). A growing number of the most frequently occurring CNVs have been cataloged in databases (32), and must be considered in the interpretation of SNP array results; however, additional CNVs occur at low frequency in human populations and have not yet been included in existing databases.
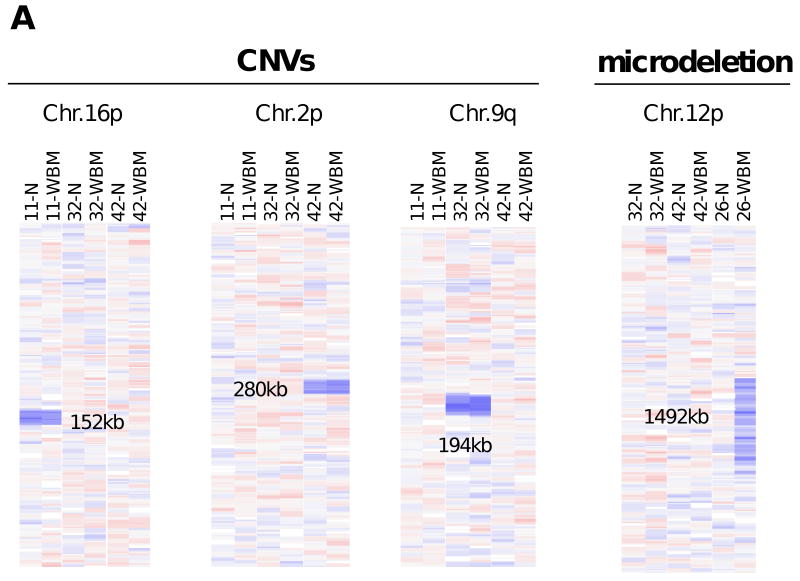
Published SNP array analyses of bone marrow cells from MDS patients (11-15) have relied mainly on CNV databases and DNA samples from unrelated individuals, rather than on paired samples of normal DNA for each patient, to substantiate regions of MDS-related copy number aberrations or UPD. This method can yield spurious results, as low allelic frequencies of inherited CNVs and linkage disequilibrium in the inheritance of linked SNPs can often mimic small regions of deletion or gain, as well as acquired UPDs, in an unpaired analysis. Thus, we reanalyzed the SNP array data from our patient cohort with normal bone marrow cell karyotypes (n=33) to determine the proportion of inherited polymorphisms and CNVs that would be erroneously identified as clonal, somatically acquired genomic abnormalities without the use of individual normal controls. By this approach, we identified 31 non-redundant copy number changes, mainly deletions, in 27 of the patients; their sizes ranged from 0.4 to 1897 kb (minimal coverage 5 consecutive SNPs). Analysis with paired normal DNA samples revealed that only 3 of these lesions were true microdeletions, whereas 28 were CNVs. Even upon reconciliation with a CNV database (32), 6 (21%) of the 28 CNVs we identified would have been falsely identified as microdeletions. Figure 5A shows examples of 3 CNVs that each occurred only once in the analysis of the 33 patients with a normal karyotype. Each of these areas of copy number loss is readily apparent in both the DNA from bone marrow and the paired buccal swab from the same patient, clearly indicating that they are all inherited CNVs. By contrast, the region of copy number loss identified in the bone marrow cells of patient 26 is not present in the matched buccal DNA sample, identifing this region as a somatically acquired microdeletion. Thus, matched normal controls are needed to reliably detect true copy number alterations.
Figure 5.
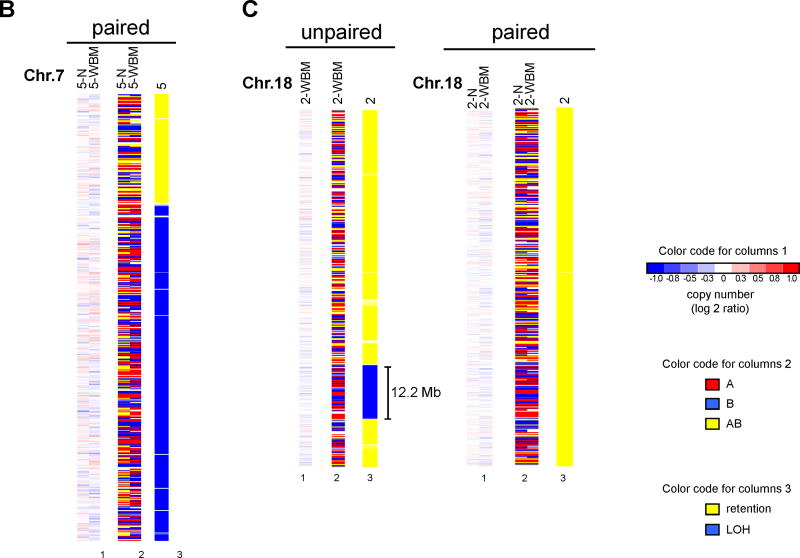
A: Comparison of CNV loci and mircrodeletions. The copy number values are shown as log 2 ratios at high magnification. Different chromosomes are not drawn to the same scale. Patients are identified at the top of each column together with the DNA source (N, normal cells; WBM, whole bone marrow mononuclear cells). The comparative analysis of copy number values with a match normal allows the differentiation of CNVs (left panel) and microdeletions (right panel). CNVs and microdeletions shown here explamplify findings summarized in Supplementary Table 1 and 3. B/C: Detection of true UPDs in paired samples as opposed to findings by unpaired LOH analysis. The results of copy number analysis are shown in the columns labeled “1”and reveal no deletions or gains. The raw genotyping calls are depicted in columns labeled “2” (A in red, B in blue, AB in yellow). The inferred LOH analysis (column 3; yellow signifies retention and blue LOH, respectively) is the result of the comparative genotype analysis. In cases of unpaired analysis, 60 normal controls (not shown) were used (CEPH subset of the HapMap project). Planel B illustrates the detection of a true UPD on chromosome 7p in patient 5. Planel C: Detection of a 12Mb locus with apparent UPD by unpaired analysis in patient 2, that is reveal as strech of inherited homozygosity by paired analysis. The comparison between left and right part of panel C emphasizes the necessity of paired analysis.
Similarly, reanalysis of our LOH results, replacing the matched normal DNA samples with 60 unrelated normal controls (CEPH subset of the HapMap project (20)) identified 110 regions of potential LOH in the 33 patients with a normal karyotype (regions covered by more than 50 SNPs and greater than 2 Mb in size). With use of matched buccal DNA samples, however, we were able to show that only 4 (3.6%) of these 110 regions represented true UPDs. An example of the detection of a true UPD is shown in Figure 5B, in which a large region of chromosome 7q exhibits LOH based on the SNP calls, with retention of normal copy number by comparison of bone marrow and matched normal DNA. By contrast, a region of 12.2 Mb in patient 2 shows apparent LOH based on analysis of 60 unrelated normal controls (Fig. 5C, unpaired). However, analysis of the same patient's bone marrow DNA compared with the matched normal DNA (Fig. 5C, paired) reveals that this region is actually a stretch of apparent homozygosity (only A or B calls), which is maintained in both the normal and the bone marrow DNA of this patient and does not represent a somatically acquired loss and reduplication event leading to UPD (see Supplementary Fig. 4 for an additional example). Thus, failure to include paired normal DNA samples in SNP array analysis of bone marrow cells from MDS patients can lead to vastly overestimated frequencies of UPDs and microdeletions, most of which are not linked to somatic abnormalities that have occurred during the genesis of the MDS clone.
Discussion
This prospective study, the first to apply genome-wide SNP array analysis to paired bone marrow and normal DNA specimens from a large cohort of patients with MDS, identified cryptic chromosomal changes with both clinical and molecular genetic implications. Our findings differ in important aspects from those in retrospective studies based on DNA extracted from archived bone marrow cell samples (11-15). Importantly, the overall rate of genomic abnormalities in MDS patients with normal bone marrow karyotypes was much lower in our analysis than in earlier studies. We attribute this discrepancy to the lack of paired normal DNA samples in most previous analyses resulting in the spurious identification of inherited CNVs or regions of apparent homozygosity as somatically acquired genomic alterations within the MDS clone. Indeed, we demonstrate that the majority of apparent abnormalities detected by analysis of bone marrow cell DNA alone reflect inherited genomic diversity rather than clonal somatic abnormalities. Thus, a straightforward comparison with matched normal DNA is required to identify unequivocally the more subtle aberrations arising from acquired clonal genomic changes. Hence, an attempt to correlate UPD or copy number changes with clinical outcome (14, 15) in patients who have been incorrectly classified due to uncontrolled assessment of genomic variation will not yield valid results.
Aside from its role in assessing bone marrow cell clonality in newly diagnosed cases of MDS, SNP array analysis promises to help unravel the molecular pathogenesis of these complex stem cell diseases. Indeed, identification of discrete clonal molecular changes in MDS by high-density SNP array analysis will likely provide the best starting point for the discovery of new genetic mutations and signal transduction pathways involved in MDS, leading to the development of more effective targeted therapies. We would emphasize that the smallest regions of copy number alteration or LOH are often the most informative in implicating individual genes for detailed sequence analysis, but these small regions are also the most difficult to distinguish from nonpathologic human germline variations. Hence, consistent use of paired bone marrow and normal DNA samples would also be expected to accelerate the pace of disease allele discovery in MDS.
We also addressed the question of whether CD34+ cell selection is required to assess the clonal aberrations in MDS. All of the aberrations found in the CD34+ fraction were also identified in unselected mononuclear cells, indicating that the clonality of the MDS bone marrow is remarkably high. Our results suggest that the cells lacking MDS-associated clonal changes in the bone marrow-derived samples are mainly circulating blood T-lymphocytes (see Supplementary Fig. 2), indicating that depletion of mature T cells might be a useful strategy for removing residual normal cells from the bone marrow aspirate. In any event, SNP array analysis in MDS can be performed with the mononuclear fraction of whole bone marrow cells, and is therefore clinically feasible even when, as is often the case, the MDS bone marrow sample contains low numbers of cells.
We report the identification of UPDs in four of 33 patients with a normal karyotype. Notably, all segmental UPDs discovered in this study were terminal (see Supplementary Fig.5 for summary). This suggests that break-induced replication (BIR (26)) might be a dominant mechanism by which a cell duplicates a somatically acquired event such as a mutation, a microdeletion or an epigenetically suppressed region and consequently become homozygous for this segmental region. BIR appears to be a common repair mechanism at stalled or broken replication forks, however, only the reduplication of a region that contains a genetic or epigenetic alteration conferring a growth advantage to the cell would allow for its clonal dominance and selection, leading to its detection by LOH analysis. In this regard, our findings of nonrandom segmental UPDs help to clarify models purporting to explain the pathogenetic basis of large deletions that are typically observed in MDS cases. Certain large deletions, such as the del(5q), are thought to arise from haploinsufficiency for one or more genes within the targeted regions (21, 23). Indeed, haploinsufficiency for the RPS14 gene has recently been linked to the pathogenesis of MDS associated with the 5q- syndrome(7); homozygous inactivation is not observed in such cases because the RPS14 protein is essential for cellular protein synthesis. Consistent with this model of molecular pathogenesis, we did not observe any UPD affecting 5q in MDS cases. Apparently, deletion of one allele of 5q is sufficient for pathogenesis, and loss of the wild-type alleles with reduplication of mutated or microdeleted target genes on 5q, as would occur with a UPD, is strongly selected against, because it is lethal to the MDS stem cell. By contrast, we did observe UPD affecting the same deleted region that is affected in cases with cytogenetically apparent loss of the long arm of chromosome 7. This implies that at least one mutated or epigenetically suppressed gene in this region is likely reduplicated together with loss of the normal allele in cases with 7q UPD, fulfilling Knudsen's hypothesis for the homozygous inactivation of classical tumor suppressor genes (33).
We hypothesize that genes within the region of UPD on chromosome 7 are likely to harbor inactivating point mutations that will eventually be identified by judicious sequencing of the involved genes in specific cases with UPD. By contrast, other regions of UPD that do not correspond to regions of cytogenetic deletion may harbor activating mutations that are duplicated by UPD and thus provide a growth advantage (34). FLT3 mutations in acute myeloid leukemia (AML) fit the latter category, in that they are frequently identified on both alleles in AML cases with a normal karyotype. Thus, SNP array analysis can provide critical information needed to pinpoint and identify mutated genes and altered signal transduction pathways in MDS.
A major goal of our study was to detect clonal genomic abnormalities in MDS patients that could be used to improve the clinical management of these disorders. Although longer follow-up times are needed to determine the association of our SNP array findings with treatment outcome, several results appear to have immediate relevance to patient management. The ability to detect clonal genomic aberrations in cases with a normal bone marrow cell karyotype using SNP array analysis can distinguish MDS from other causes of bone marrow cell dysplasia and pancytopenia due to the effects of drugs, environmental toxins or aberrant immune responses, thus aiding in the initial diagnosis. Moreover, our identification of UPDs affecting chromosome 7 in two patients with low-risk IPSS scores who later showed rapidly deteriorating clinical courses is intriguing. UPD selects for homozygosity of a specific genomic region, and the same region of chromosome 7q affected by UPD is also very often deleted in MDS cases. When identified by cytogenetic analysis, abnormalities of chromosome 7q by themselves signify an increased IPSS score. Thus, it is possible that the 7q UPDs we identified are equivalent to a deletion in this region and may constitute a high-risk feature in MDS (17). Whether or not UPDs affecting this and other genomic regions convey important prognostic information is an important question to address in additional prospective studies using SNP array technology to evaluate patients with MDS entering clinical therapeutic trials.
Supplementary Material
Acknowledgments
We thank Martin H. Nguyen and Michael Fernandez (MDACC) as well as Nikki Flores and Elena Wang (USCF) for excellent technical assistance and we are grateful to John Gilbert for editorial review and helpful discussions. We also thank Lorna Mangus for administrative support. The work was supported in part by NIH P01 grant CA-108631 from the National Institutes of Health. No potential conflict of interest relevant to this article was reported.
References
- 1.Corey SJ, Minden MD, Barber DL, Kantarjian H, Wang JC, Schimmer AD. Myelodysplastic syndromes: the complexity of stem-cell diseases. Nat Rev Cancer. 2007 Feb;7(2):118–129. doi: 10.1038/nrc2047. [DOI] [PubMed] [Google Scholar]
- 2.Estey E. Acute myeloid leukemia and myelodysplastic syndromes in older patients. J Clin Oncol. 2007 May 10;25(14):1908–1915. doi: 10.1200/JCO.2006.10.2731. [DOI] [PubMed] [Google Scholar]
- 3.Nimer SD. Myelodysplastic syndromes. Blood. 2008 May 15;111(10):4841–4851. doi: 10.1182/blood-2007-08-078139. [DOI] [PubMed] [Google Scholar]
- 4.de Witte T, Oosterveld M, Muus P. Autologous and allogeneic stem cell transplantation for myelodysplastic syndrome. Blood Rev. 2007 Jan;21(1):49–59. doi: 10.1016/j.blre.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Haase D, Germing U, Schanz J, Pfeilstocker M, Nosslinger T, Hildebrandt B, et al. New insights into the prognostic impact of the karyotype in MDS and correlation with subtypes: evidence from a core dataset of 2124 patients. Blood. 2007 Dec 15;110(13):4385–4395. doi: 10.1182/blood-2007-03-082404. [DOI] [PubMed] [Google Scholar]
- 6.List A, Kurtin S, Roe DJ, Buresh A, Mahadevan D, Fuchs D, et al. Efficacy of lenalidomide in myelodysplastic syndromes. N Engl J Med. 2005 Feb 10;352(6):549–557. doi: 10.1056/NEJMoa041668. [DOI] [PubMed] [Google Scholar]
- 7.Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, et al. Identification of RPS14 as a 5q-syndrome gene by RNA interference screen. Nature. 2008 Jan 17;451(7176):335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kantarjian H, Oki Y, Garcia-Manero G, Huang X, O'Brien S, Cortes J, et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood. 2007 Jan 1;109(1):52–57. doi: 10.1182/blood-2006-05-021162. [DOI] [PubMed] [Google Scholar]
- 9.Silverman LR, Demakos EP, Peterson BL, Kornblith AB, Holland JC, Odchimar-Reissig R, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J Clin Oncol. 2002 May 15;20(10):2429–2440. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- 10.Heinrichs S, Look AT. Identification of structural aberrations in cancer by SNP array analysis. Genome Biol. 2007 Jul 31;8(7):219. doi: 10.1186/gb-2007-8-7-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gondek LP, Dunbar AJ, Szpurka H, McDevitt MA, Maciejewski JP. SNP Array Karyotyping Allows for the Detection of Uniparental Disomy and Cryptic Chromosomal Abnormalities in MDS/MPD-U and MPD. PLoS ONE. 2007;2(11):e1225. doi: 10.1371/journal.pone.0001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gondek LP, Haddad AS, O'Keefe CL, Tiu R, Wlodarski MW, Sekeres MA, et al. Detection of cryptic chromosomal lesions including acquired segmental uniparental disomy in advanced and low-risk myelodysplastic syndromes. Exp Hematol. 2007 Nov;35(11):1728–1738. doi: 10.1016/j.exphem.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Gondek LP, Tiu R, Haddad AS, O'Keefe CL, Sekeres MA, Theil KS, et al. Single nucleotide polymorphism arrays complement metaphase cytogenetics in detection of new chromosomal lesions in MDS. Leukemia. 2007 Sep;21(9):2058–2061. doi: 10.1038/sj.leu.2404745. [DOI] [PubMed] [Google Scholar]
- 14.Gondek LP, Tiu R, O'Keefe CL, Sekeres MA, Theil KS, Maciejewski JP. Chromosomal lesions and uniparental disomy detected by SNP arrays in MDS, MDS/MPD, and MDS-derived AML. Blood. 2008 Feb 1;111(3):1534–1542. doi: 10.1182/blood-2007-05-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohamedali A, Gaken J, Twine NA, Ingram W, Westwood N, Lea NC, et al. Prevalence and prognostic significance of allelic imbalance by single-nucleotide polymorphism analysis in low-risk myelodysplastic syndromes. Blood. 2007 Nov 1;110(9):3365–3373. doi: 10.1182/blood-2007-03-079673. [DOI] [PubMed] [Google Scholar]
- 16.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, et al. Proposals for the classification of the myelodysplastic syndromes. Br J Haematol. 1982 Jun;51(2):189–199. [PubMed] [Google Scholar]
- 17.Greenberg P, Cox C, LeBeau MM, Fenaux P, Morel P, Sanz G, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997 Mar 15;89(6):2079–2088. [PubMed] [Google Scholar]
- 18.Harris NL, Jaffe ES, Diebold J, Flandrin G, Muller-Hermelink HK, Vardiman J, et al. World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues: report of the Clinical Advisory Committee meeting-Airlie House, Virginia, November 1997. J Clin Oncol. 1999 Dec;17(12):3835–3849. doi: 10.1200/JCO.1999.17.12.3835. [DOI] [PubMed] [Google Scholar]
- 19.Lin M, Wei LJ, Sellers WR, Lieberfarb M, Wong WH, Li C. dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics. 2004 May 22;20(8):1233–1240. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- 20.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007 Oct 18;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu TX, Becker MW, Jelinek J, Wu WS, Deng M, Mikhalkevich N, et al. Chromosome 5q deletion and epigenetic suppression of the gene encoding alpha-catenin (CTNNA1) in myeloid cell transformation. Nat Med. 2007 Jan;13(1):78–83. doi: 10.1038/nm1512. [DOI] [PubMed] [Google Scholar]
- 22.Boultwood J, Fidler C, Strickson AJ, Watkins F, Gama S, Kearney L, et al. Narrowing and genomic annotation of the commonly deleted region of the 5q- syndrome. Blood. 2002 Jun 15;99(12):4638–4641. doi: 10.1182/blood.v99.12.4638. [DOI] [PubMed] [Google Scholar]
- 23.Le Beau MM, Espinosa R, 3rd, Neuman WL, Stock W, Roulston D, Larson RA, et al. Cytogenetic and molecular delineation of the smallest commonly deleted region of chromosome 5 in malignant myeloid diseases. Proc Natl Acad Sci U S A. 1993 Jun 15;90(12):5484–5488. doi: 10.1073/pnas.90.12.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poppe B, Vandesompele J, Schoch C, Lindvall C, Mrozek K, Bloomfield CD, et al. Expression analyses identify MLL as a prominent target of 11q23 amplification and support an etiologic role for MLL gain of function in myeloid malignancies. Blood. 2004 Jan 1;103(1):229–235. doi: 10.1182/blood-2003-06-2163. [DOI] [PubMed] [Google Scholar]
- 25.Zatkova A, Schoch C, Speleman F, Poppe B, Mannhalter C, Fonatsch C, et al. GAB2 is a novel target of 11q amplification in AML/MDS. Genes Chromosomes Cancer. 2006 Sep;45(9):798–807. doi: 10.1002/gcc.20344. [DOI] [PubMed] [Google Scholar]
- 26.McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 27.Harada H, Harada Y, Niimi H, Kyo T, Kimura A, Inaba T. High incidence of somatic mutations in the AML1/RUNX1 gene in myelodysplastic syndrome and low blast percentage myeloid leukemia with myelodysplasia. Blood. 2004 Mar 15;103(6):2316–2324. doi: 10.1182/blood-2003-09-3074. [DOI] [PubMed] [Google Scholar]
- 28.Imai Y, Kurokawa M, Izutsu K, Hangaishi A, Takeuchi K, Maki K, et al. Mutations of the AML1 gene in myelodysplastic syndrome and their functional implications in leukemogenesis. Blood. 2000 Nov 1;96(9):3154–3160. [PubMed] [Google Scholar]
- 29.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006 Jan;38(1):75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 30.Khaja R, Zhang J, MacDonald JR, He Y, Joseph-George AM, Wei J, et al. Genome assembly comparison identifies structural variants in the human genome. Nat Genet. 2006 Dec;38(12):1413–1418. doi: 10.1038/ng1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature. 2006 Nov 23;444(7118):444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Feuk L, Duggan GE, Khaja R, Scherer SW. Development of bioinformatics resources for display and analysis of copy number and other structural variants in the human genome. Cytogenet Genome Res. 2006;115(3-4):205–214. doi: 10.1159/000095916. [DOI] [PubMed] [Google Scholar]
- 33.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971 Apr;68(4):820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fitzgibbon J, Smith LL, Raghavan M, Smith ML, Debernardi S, Skoulakis S, et al. Association between acquired uniparental disomy and homozygous gene mutation in acute myeloid leukemias. Cancer research. 2005 Oct 15;65(20):9152–9154. doi: 10.1158/0008-5472.CAN-05-2017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.