Abstract
The fourth chromosome of Drosophila melanogaster has a number of unique properties that make it a convenient model for the study of chromatin structure. Only 4.2 Mb overall, the 1.2 Mb distal arm of chromosome four seen in polytene chromosomes combines characteristics of heterochromatin as well as euchromatin. This domain has a repeat density of ca. 35%, comparable to some pericentric chromosome regions, while maintaining a gene density similar to that of the other euchromatic chromosome arms. Studies of position effect variegation have revealed that heterochromatic and euchromatic domains are interspersed on chromosome four, and both cytological and biochemical studies have demonstrated that chromosome four is associated with heterochromatic marks such as HP1 and histone 3 lysine 9 methylation. Chromosome four is also marked by POF, a chromosome four-specific chromosomal protein, and utilizes a dedicated histone methyltransferase, EGG. Studies of chromosome four have helped to shape our understanding of heterochromatin domains, their establishment and maintenance. In this review we provide a synthesis of the work to date and an outlook to the future.
Keywords: chromatin, heterochromatin, dot chromosome, Drosophila, silencing, PEV
Introduction
The various genome sequencing projects carried out during the last 15 years have vastly increased our understanding of the organization and structure of eukaryotic genomes. However, while the DNA is the primary information carrier, the proper utilization of this information is dependent on higher order structure imposed by chromatin and the action of a diverse array of proteins, including transcription factors, polymerases, and chromatin remodelers. To allow for the complex life cycles of eukaryotic organisms, chromatin structure and access to the primary sequence must be adjusted in a dynamic fashion to facilitate the execution of appropriate developmental programs and to keep inappropriate gene expression, including that of transposons, in check. Thus, insights into the establishment, maintenance, and alteration of chromatin structure are of utmost importance to garner a complete understanding of developmental gene regulation and ultimately to develop models for how access to and integrity of genomes are controlled.
Chromatin, the composite of DNA and associated proteins, is historically classified into two forms, heterochromatin and euchromatin. Originally, this classification was based on nucleic acid staining patterns in interphase nuclei, with heterochromatin retaining its densely staining character throughout the cell cycle, while euchromatin stained only weakly during interphase (Heitz 1928). Today, heterochromatin and euchromatin are understood to represent two kinds of molecular structures, each with a set of opposing characteristics (for a recent review see Grewal and Elgin 2007). These characteristics include differences in DNA sequence organization, with high levels of repetitive elements in heterochromatin compared to low levels in euchromatin, and low gene density in heterochromatin compared to high gene density in euchromatin. Heterochromatin is generally considered to be more compact and the DNA less accessible than is the case for euchromatin. This difference in accessibility is invoked to explain the lower levels of transcription and low rates of recombination typical of heterochromatin as opposed to euchromatin. In addition, heterochromatin and euchromatin differ in certain biochemical marks that impact the nucleosome array and are likely fundamental to both packaging and silencing. Heterochromatin is associated with low levels of histone acetylation and high levels of histone 3 lysine 9 (H3K9) methylation, with concomitant association of the non-histone chromatin protein Heterochromatin Protein 1 (HP1); the latter features are so consistently associated with heterochromatin that they are referred to as “silent marks.” In contrast, euchromatin shows high levels of histone acetlyation as well as other “active marks” such as histone 3 lysine 4 (H3K4) methylation (Grewal and Elgin 2007). While the above characteristics, be they biochemical or genetic in nature, are commonly associated with heterochromatin or euchromatin, none are universal to all types of heterochromatin or euchromatin and exceptions do occur.
Oftentimes euchromatin and heterochromatin are portrayed as a simple dichotomy; however, this view is an oversimplification that ignores the highly dynamic nature of chromatin states during biological transitions. During the cell cycle, in particular during DNA synthesis, chromatin is unraveled and reestablished on the two sister chromatids. Thus, there are cell cycle dependent fluctuations in chromatin structure. In multi-cellular organisms, there is additional flux in chromatin structure due to the establishment of developmental differences in gene expression (for an example see Lu et al. 1996, Lu et al. 1998). The term constitutive heterochromatin applies to genomic regions such as the centromeres or telomeres that are heterochromatic in all cell types, while other regions of the genome fall under the term “facultative” heterochromatin. Facultative heterochromatin can include domains that are differentially package depending on the parent of origin (as in imprinting) and encompasses developmentally regulated genes or domains, which can exhibit alternative chromatin states in different cell types. The term also implies that such chromatin domains can switch between euchromatic and heterochromatic states, possibly regulated by the temporal pattern of gene expression. One prominent developmental system that can be considered to generate a type of facultative heterochromatin is the polycomb group of proteins (for a recent review see Trojer and Reinberg 2007). An example of such a switch between chromatin states involving polycomb components as well as a histone acetlytransferase is provided by the circadian clock system in mammalian cells. The circadian regulator CLOCK encodes a histone acetyltransferase and induces chromatin changes with a 24h periodicity (Doi et al. 2006, Nakahata et al. 2007), while the polycomb class protein EZH2, a methyltransferase modifying H3 K27, functions in the control of Period1 and Period2 (Etchegaray et al. 2006).
Besides the basic distinction between constitutive and facultative heterochromatin, further sub-categories of heterochromatin are supported by a variety of experimental evidence. Generally, there are at least three distinct regions of heterochromatin, centromeric, pericentric and telomeric heterochromatin. While the pathways controlling heterochromatin formation in these regions share common components, each is also affected by unique genetic factors as well. For examples, in Drosophila melanogaster, Su(var)205, the gene encoding HP1, acts as a suppressor of variegation in pericentric and fourth chromosome domains, but does not cause the release of silencing at telomeric reporters (Wallrath and Elgin 1995).
Chromosome four of Drosophila melanogaster exhibits interspersed heterochromatic and euchromatic domains
In addition to the “universal” types of heterochromatin, D. melanogaster has two unique heterochromatic regions, the Y chromosome and the small fourth chromosome (reviewed in Weiler and Wakimoto 1995). While little is known about regulation of Y chromosome heterochromatin, the fourth chromosome has been the focus of work in our laboratory and others for over a decade. The fourth chromosome, also called the F element (Müller 1940) or dot chromosome, is the smallest of the D. melanogaster chromosomes at approximately 4.2Mb (Locke and McDermid 1993). It consists of a small, entirely heterochromatic short arm, and a long arm, the distal 1.2Mb of which are polytenized in salivary gland nuclei. The chromosome as a whole is late replicating (Zhimulev et al. 2003) and exhibits no meiotic recombination (Sandler and Szauter 1978), characteristics of heterochromatin.
Further evidence for the heterochromatic nature of much of the fourth chromosome was gained from studies of position effect variegation (PEV). The earliest studies of PEV employed chromosomal rearrangements of loci such as white or yellow with easily scored phenotypes (for example see Muller 1930). However, PEV can also be studied using transgenic reporters inserted at various locations in the D. melanogaster genome; this case is illustrated in Figure 1 with a white transgene (Wallrath and Elgin 1995). PEV refers to the observation that a reporter gene can be either fully expressed in all units (ommatidia) of the compound eye, leading to a uniform red eye color in the case of a white transgene, or variably silenced in distinct units of the eye, leading to variegated eye color, depending its genomic location. (“Variably silenced” indicates that the gene is on in some cells but off in others in which it is normally expressed.) In theory, a reporter could be completely silenced due to its position in heterochromatin, but those cases are studied rarely as they are difficult to distinguish from accidental elimination of the reporter.
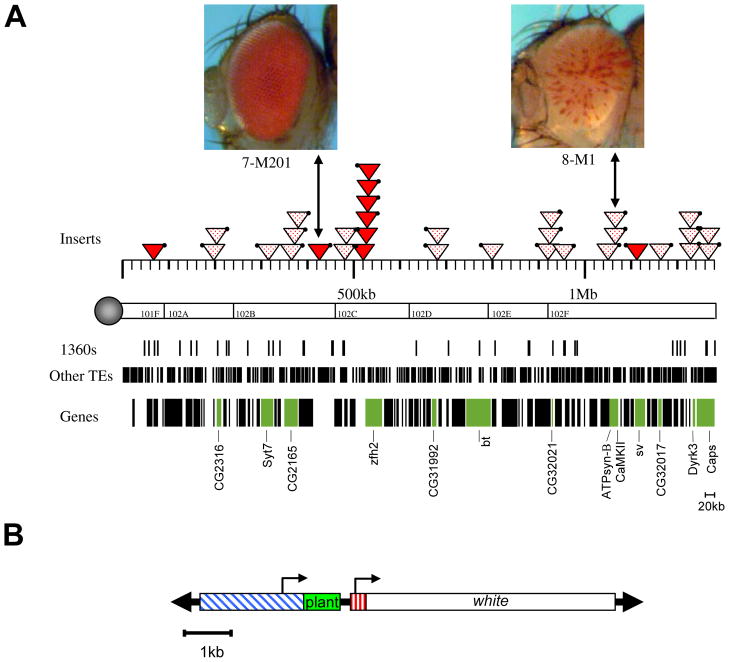
Figure 1.
Map positions of a collection of reporter transgenes on the 4th chromosome of D. melanogaster. A) Chromosome map. Above the chromosome map, representative pictures of a full red (7-M201) and a variegating (8-M1) eye are shown. Locations of these two inserts as well as the location of other inserts on the banded portion of the 4th chromosome are indicated by triangles; both variegating (mottled triangles) and non-variegating (red triangles) inserts are shown. The locations of 1360, other transposable elements as well as annotated genes are also shown. B). Map of the transgene construct. This transgene contains both an hsp26 promoter (blue) driving expression of a fragment of DNA derived from barley (green) and an hsp70 promoter (red) driving expression of the white gene.
Variegation is caused by stochastic silencing of the reporter gene in a subset of the cells in which it is normally expressed, and studies have shown that the level of silencing reflects the chromatin state at the reporter insertion site (Wallrath and Elgin 1995, Cryderman et al. 1999b, Sun et al. 2001). In the case of a white reporter, silencing is observed as a decrease in the number of ommatidia with red eye pigment. The more ommatidia lack pigment, the greater the probability of silencing and the greater percentage of nuclei with the reporter packaged as heterochromatin at the transgene insertion site as shown by chromatin digestion studies. Thus, the pigment level generally provides an easy readout of the local chromatin states.
However, the relationship between heterochromatin packaging and reporter phenotype is not simple. The heterochromatic state is thought to spread from either an initiation site or a larger heterochromatic mass via the action of HP1 and SU(VAR)3-9. HP1 can interact with histone H3 methylated at lysine 9, as well as with the methyltransferase SU(VAR)3-9, which generates this modification (Bannister et al. 2001, Jacobs et al. 2001, Lachner et al. 2001, Schotta et al. 2002). By doing so, HP1 is thought to mediate the spread of heterochromatin by recruiting SU(VAR)3-9, thus facilitating the modification of adjacent nucleosomes. (For more details on this and alternative models see the review by Talbert and Henikoff 2006.) Recently, chromatin immunoprecipitation (ChIP) studies have provided evidence for this spreading of the heterochromatic chromosome structure in a classical PEV system, the wm4 chromosome. The wm4 chromosome carries an inversion that places the white gene in close proximity to a heterochromatin block, thus causing PEV. Using ChIP followed by locus-specific quantitative PCR, Rudolph and colleagues discovered that H3K9 dimethlylation levels are highest at the white gene and gradually decrease with increasing distance from white and the heterochromatin breakpoint (Rudolph et al. 2007).
Several studies, including the ChIP analysis of the wm4 inversion described above (Rudolph et al. 2007), have demonstrated that while in general there is a correlation between proximity to heterochromatin and the level of variegation observed, the spreading of silencing usually is not linear, but can skip genes (Talbert and Henikoff 2000). While all genes tested for PEV in these cases show effects, the degree of impact can be gene-specific. In addition, there is evidence that the size and type of the most proximal heterochromatic block can influence the level of variegation (Wakimoto and Hearn 1990, Henikoff et al. 1995). These findings indicate that while reporter variegation (alternative expression levels) does reflect the chromatin structure at its insertion site, interpretation of PEV results must incorporate consideration of other aspects of chromosome organization as well.
Interestingly, many of the reporter lines with variegating eye color isolated in P element reporter screens in our laboratory map to the fourth chromosome (Figure 1; Riddle et al. 2008). The genome map in Figure 1 illustrates the composite results of several genetic screens, demonstrating that variegating reporters map to the entire length of the fourth chromosome, indicating that a heterochromatin-like state prevails on this chromosome at the developmental time assayed. However, the map also shows that euchromatic domains are interspersed with the heterochromatic domains (Sun et al. 2004, Riddle et al. 2008). In particular, with the P element white reporter system, four euchromatic domains have been identified, regions in which inserted transgenes show levels of red eye pigment similar to wildtype (Riddle et al. 2008). The domain structure of the fourth chromosome is very unusual, as the other chromosome arms show a transition from heterochromatin at the centromeres to euchromatin along the chromosome arms, but no reported interdigitation of these two types of chromatin (Yasuhara and Wakimoto 2008).
Subsequent screens with the white reporter construct shown in Figure 1 investigated the effect of local genomic deletions and duplications on the eye color phenotype associated with the reporter (Sun et al. 2004, Riddle et al. 2008). Changes in eye color phenotype, either from variegating to red, or from red to variegating, can be caused both by genomic deletions and duplications within the fourth chromosome. These duplications and deletions vary greatly in size, some as small as 4kb (a duplication adjacent to 39C-12), while others span hundreds of kb (for example 8M-192, which carries a deletion from nucleotide position 928,687 to 1,119408; Riddle et al. 2008). Based on these data, the size of some of the euchromatic domains can be estimated. For example, the euchromatic domain adjacent to hcf encompasses up to 50kb, while the domain including sv can be no larger than ~47kb (Riddle et al. 2008). How exactly genomic deletions and duplications cause a change in the reporter phenotype is under active investigation. Possible mechanisms explaining the effects include the deletion or introduction of boundary elements, or the deletion or introduction of heterochromatin initiator elements. The current data are more readily interpreted using the latter model. Alternatively, the domains on chromosome four supporting a more euchromatic chromatin structure and a red eye phenotype of white reporters might be under the control of a different system than the remaining portions of chromosome four, one that is more readily manipulated to maintain an active state. Together, the results of PEV studies demonstrate that events altering the sequence environment of the reporter can profoundly effect chromatin states and result in a switch from heterochromatin to euchromatin or euchromatin to heterochromatin.
The sequence composition of the dot chromosome is a unique mixture of euchromatic and heterochromatic components
A fundamental and important property of chromosome four is illustrated in Figure 1 beneath the chromosome map: its sequence composition. Compared to the other autosomes, the dot chromosome has a high density of repetitious elements (Wilson et al. 2008). The average density of repeats in the euchromatic regions of the autosomal chromosome arms is approximately 6% (Slawson et al. 2006). In contrast, the repeat content of the fourth chromosome is at least five times higher, 25% (Slawson et al. 2006), or as high as 35% in analyses using RepeatScout (W. Leung, personal communication; http://repeatscout.bioprojects.org). This high level of repeats resembles some of the pericentric regions (“beta heterochromatin”) of the other long chromosome arms, but in those regions the gene density is lower than that observed on chromosome four. The repeat types found interspersed with genes on the dot chromosome include simple sequence repeats, but the majority of the repetitive sequence on chromosome four consists of fragments derived from transposable elements, both DNA transposons and retroviral elements (Slawson et al. 2006). One simple sequence that is conspicuously absent in longer arrays on the fourth chromosome is (CA)n; the presence of this repeat is generally found to be characteristic of the euchromatic arms of D. melanogaster (Lowenhaupt et al. 1989, Slawson et al. 2006).
Curiously, the gene density on the fourth chromosome, 80 genes distributed over 1.2Mb, is almost identical to that of the euchromatic chromosome arms, which contain 74.6 genes/Mb based on recent estimates (Slawson et al. 2006, Wilson et al. 2008). Thus, with regard to repeat and gene density, the fourth chromosome exhibits characteristics of heterochromatin for one measure and those of euchromatin for the other. While one might initially anticipate blocks of repeats, packaged as heterochromatin, and blocks of genes, packaged as euchromatin, this model does not hold true. In reality, the interspersion of repetitive elements and genes is reminiscent of mammalian chromosomes, where repeat sequences are freely intermingled with genes along the chromosome arms. Gene and repeat density do not appear to change significantly between the chromatin domains delineated by the PEV studies. These findings point to the conclusion that at least some Drosophila genes have adapted to function in repeat-rich regions, even under circumstances where that characteristic has precipitated a heterochromatin-like state. The ability of the genes to function in a repeat-rich environment must be predominant in mammalian genomes.
Repetitive elements and chromatin structure on the dot chromosome
While no obvious correlation was detected between overall repeat density on the fourth chromosome and the character of the chromatin domains defined by the white reporter insertions (Riddle et al. 2008), individual elements can show a biased distribution either adjacent to heterochromatic/variegating sites or to euchromatic/red insertion sites. Such a correlation has been observed for the DNA element 1360 in a domain of approximately 200kb surrounding the Hcf locus – variegating insertions usually fall within 10kb of remnants of 1360 elements (Sun et al. 2004). Given that transposable elements in the genome of an organism need to be targeted for silencing (active mobile elements are mutagens), this finding led to the hypothesis that 1360 – and likely other repetitive elements – might serve as target sequences for the initiation of heterochromatin formation. This hypothesis was tested genetically by utilizing a P element reporter construct consisting of the hsp70-white reporter gene preceded by one copy of a 1360 element flanked by FRT sites. Out of a total of 22 insertion lines recovered, 19 showed a red eye phenotype, two lines exhibited orange eye color, and one line (T190-177) showed a variegating eye phenotype (Haynes et al. 2006). This result indicates that the presence of one copy of 1360 alone in a euchromatic domain is not sufficient to induce local heterochromatin formation.
Further analysis of the variegating line T190-177 demonstrated that it mapped to a domain at the base of chromosome 2L rich in transposable elements. When the 1360 element is removed from the transgene construct in this line by FLP-mediated recombination, the level of variegation is decreased, and the eye pigment level increases. However, wildtype red eye color is not recovered, and the line retains its variegating phenotype in the absence of 1360, albeit at a much higher level of pigmentation (Haynes et al. 2006). These results indicate that the insertion site can instill a variegating phenotype independent of 1360, but that the presence of 1360 is able to enhance the silencing, possibly recruiting additional silencing components. Thus, while one copy of 1360 is not sufficient to induce silencing at all genomic locations, it can indeed impinge on the degree of silencing observed at a subset of locations. At this point, given the single observation, it is unknown what determines the sensitivity of a genomic location to the presence of 1360 in regard to heterochromatin formation. The local density of repetitious elements, the nature of these elements, and proximity to a larger heterochromatic mass such as the centromere all seem likely to have an impact.
Subsequent analysis of an additional region of chromosome four (surrounding the sv locus) as well as a re-analysis of the data for the entire chromosome were carried out to examine the distribution of repetitive elements and their association with transgene states (Riddle et al. 2008). The genomic region surrounding sv (position 1,000,000–1,200,000 on the map in Figure 1) contains many fewer remnants of 1360 than the more proximal region surrounding Hcf (position 300,000–500,000) described above. Despite this dearth, variegating white reporter lines map to the vicinity of sv. Overall on the fourth chromosome, there are 9 transgenes with variegating phenotype that lack a 1360 element in their proximity. Conversely, in all but one case, if the transgene reporter is within 10kb of a 1360 remnant, a variegating phenotype is observed (Riddle et al. 2008). Thus, while 1360 is neither necessary nor sufficient to induce heterochromatin formation in general, it is able to “supplement” heterochromatin formation in appropriate genomic locations, perhaps in a manner common to many transposon sequences.
There are 65 additional types of repetitive elements present on the fourth chromosome (Wilson et al. 2008). When their correlation with the PEV phenotypes is considered, no single transgene is able to explain the chromatin domain structure on the fourth observed with the white reporter (Riddle et al. 2008). This type of analysis is hindered by the fact that it is unknown over what distance heterochromatin can spread from an initiation site, and thus, the correct window size to use in seeking a correlation is unknown. Thus, the results argue that repetitious elements do serve as targets to influence heterochromatin formation, but that no a single repetitive element serves as the sole heterochromatin initiator; most likely multiple sequence elements can serve this function. The key characteristics of such initiators remain to be elucidated.
The dot chromosome lacks recombination in females
In addition to its unique sequence organization and chromatin domain structure, the dot chromosome of D. melanogaster also is exceptional among the autosomes in that it does not undergo recombination in females under standard laboratory conditions (Sandler and Szauter 1978). In the classical Drosophila genetics literature, there are some reports indicating that recombination can be observed (Bridges 1921). Certain special conditions such heat-shock (Grell 1971) and abnormal chromosome configurations or certain mutations can induce recombination on chromosome four. For instance, the genetic map of chromosome four was constructed initially by Sturtevant using diplo-IV triploid females (triploid female flies lacking one copy of chromosome four; Sturtevant 1951).
Lack of recombination is a property usually associated with heterochromatic domains such as regions close to the centromeres or telomeres. The lack of recombination exhibited by the fourth chromosome might be due to several causes. One possibility is that the heterochromatic character of the entire fourth chromosome directly results in the lack of recombination. However, a different theory posits that the absence of recombination is caused by the “centromere effect” (Beadle 1932), i.e. its close proximity to the centromere and/or the pericentric heterochromatin. The entire fourth chromosome lies close to the centromere, pericentric heterochromatin, and the chromocenter due to its overall small size. In this sense, the lack of recombination on chromosome four would reflect a phenomenon that should be observed for other chromosomes as well, based entirely on the proximity of a particular genomic region to centromeric and/or pericentric chromatin. Evidence from translocation studies indicates that chromosome four sequences can undergo recombination if the banded portion is moved to a different location (Thompson 1963, Osborne 1998). The reciprocal translocation X chromosome fragment, which is of similar size as the banded portion of the dot chromosome, lacks recombination in its new genomic location (Osborne 1998). These findings argue that the sequence features of the distal fourth chromosome per se (high density of repetitious elements) do not themselves preclude recombination. This finding is supported by the observation that mutations in certain Su(var) genes, such was Su(var)205 (encoding HP1) can increase the recombination rate, with Su(var)2-2 and Su(var)2-14 increasing recombination rate even in the heterochromatic region on chromosome 2 containing light and rolled (Westphal and Reuter 2002).
However, other properties of the fourth chromosome also need to be considered. When inducing translocations to manipulate the genomic position of the white reporter from Figure 1, it was discovered that such translocations had a significant impact on the eye phenotype observed (Cryderman et al. 1999a). The 118E-15 transgene as well as approximately 1Mb of the adjacent chromosome four sequences were translocated from the telomeric region of chromosome four to the distal region of chromosome 2. This alteration in genomic location induces a change in the eye phenotype from variegating at the original location to red at the new chromosome 2 location (Cryderman et al. 1999a). These findings imply that in such a translocation, the heterochromatic character usually assumed by the chromosome four domain is diminished or absent. A possible cause is suggested by the observation that for one chromosome four associated protein, POF (painting-of-fourth), a site at the base of chromosome four is reported to be critical for its proper localization (Larsson et al. 2001); presumably its loss would impact proteins known to interact with POF, which include HP1. Removal of the fourth chromosome domain from the region of the nucleus where heterochromatin is concentrated could also impact the pool of heterochromatin proteins available to assemble the chromosome four chromatin domain. Consequently, removal of the distal chromosome four arm from its centromere could prevent proper formation of heterochromatin, due to the absence of an initiation site for the recruitment of POF and other chromatin components. In its entirety, the data available for chromosome four indicate that formation of its typical heterochromatic structure is essential for the lack of recombination observed. At this point, it remains ambiguous how the heterochromatic character of chromosome four is induced.
Comparative genomics of chromosome four
Further insights into the fourth chromosome of Drosophila melanogaster are possible by comparison with the same element from other species in the genus. Many species of Drosophila have a small pericentric chromosome, including 11 of the 12 species with published whole genome shotgun assemblies. While many of the properties of the F element are conserved among the different species, the chromosomes are not identical. Of particular interest is the F element in Drosophila virilis, which shows intriguing differences when compared to the D. melanogaster dot chromosome. For example, while the centromeric heterochromatin of both D. melanogaster and D. virilis contains HP1 and HP2 (see below), the banded portion of the F element in D. virilis does not associate with either HP1 or HP2 (Shaffer et al. 2002, Slawson et al. 2006). Yet POF appears to remain localized to this element in D. virilis (Larsson et al. 2004). In addition there is reported to be a much higher rate of recombination along the dot chromosome of D. virilis (Chino 1936/1937). These observations suggest a more euchromatic character to the D. virilis dot chromosome.
These differences are not due to a wholesale change in the genetic composition of the chromosomes, as a careful study of ~372kb of high quality finished sequences from the D. virilis dot chromosome found that 27 of 28 genes were found in common on both the D. melanogaster and D. virilis dot chromosomes, and that the overall levels of repetitious sequence are similar (Slawson et al. 2006). However, there are differences in the sequence composition of these two small chromosomes that could contribute to the observed differences in protein localization and recombination rates. Differences in repetitive DNA, both simple sequence repeats (SSRs) and types of middle repetitive elements, are noted (Slawson et al. 2006). Most striking of the SSRs is the dinucleotide repeat (dC-dA)·(dG-dT) commonly referred to as CA/GT. Long tandem copies of this repeat are found at significantly higher levels then would be predicted based on nucleotide composition in D. melanogaster long arms and are severely depleted on the D. melanogaster dot chromosome (Slawson et al. 2006). This sharp difference in CA/GT density between the euchromatic long arms and the D. melanogaster dot chromosome is not seen in D. virilis. Here both the dot chromosome sequences and the long arm sequences show long tandem CA/GT repeats at a higher than expected frequency, suggesting a euchromatic character.
There are also detectable differences in the type of repeats found on the D. virilis dot chromosome compared to the D. melanogaster dot chromosome. While both dot chromosomes are enriched for repetitive DNA, with at least one fourth of their sequence identified as repetitive by RepeatMasker as compared to about 6–8% for the long arms in both species, each appears to be enriched for different class of repeat type (Slawson et al. 2006). Thus for D. melanogaster, 18.4% of the dot chromosome is reported as DNA elements, while only 6.4% of the high quality sequence from the D. virilis dot chromosome is identified as such (Slawson et al. 2006). In contrast, the D. virilis dot chromosome appears to be enriched for retroelements, with 9.1% as compared to 4.2% found for the D. melanogaster dot chromosome (Slawson et al. 2006). Recent work in D. virilis which has extended the high quality sequence to a continuous section of 1.2 million bases of the D. virilis dot chromosome has found similar values, removing any concerns of bias in the original sample (C. Shaffer, W. Leung and S.C.R. Elgin; unpublished).
Heterochromatic proteins associate with chromosome four
Studies of polytene chromosomes have provided insights into the special chromatin character of chromosome four. When Heterochromatin Protein 1 was first described, it was noted that in addition to its association with the common regions of heterochromatin (centromeres and telomeres), HP1 also strongly associates with the 1.2Mb region of chromosome four amplified in polytene chromosomes (Figure 2; James et al. 1989). The HP1 distribution on the dot chromosome shows a banded pattern, in contrast to the diffuse distribution observed in the chromocenter (James et al. 1989).
Figure 2.
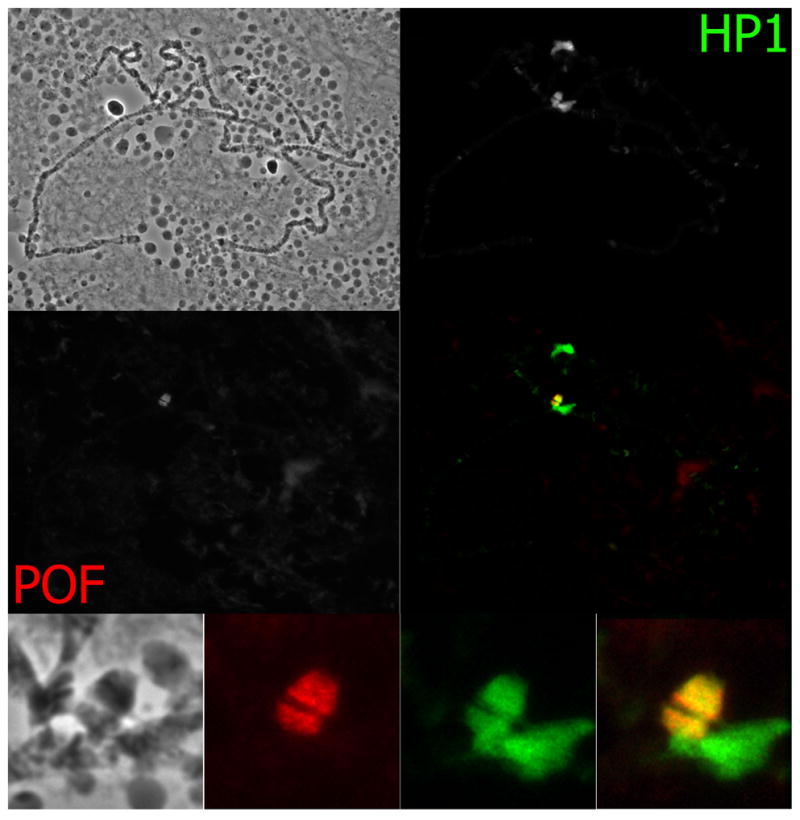
Immunofluorescent images of polytene chromosomes. Upper left: phase contrast. Staining with both HP1 (upper right) and POF (middle row, left) antibodies are shown separately. A merged image is shown in the middle row on the right. The bottom row shows a close up of the chromocenter. From left to right: phase contrast, POF signal, HP1 signal, merged signal.
The HP1 immunolocalization pattern on polytene chromosomes is mirrored by several other protein marks usually associated with heterochromatic regions of the genome. One of these proteins is Heterochromatin Protein 2 (HP2), which is a genetic and biochemical interactor with HP1 (Shaffer et al. 2002, Stephens et al. 2005). Like HP1, HP2 on polytene chromosomes is prominently localized to the chromocenter, telomeres, and chromosome four. The banding pattern on the dot chromosome displayed by HP2 is identical to that of HP1, providing additional evidence for their interaction (Shaffer et al. 2002). In addition to HP2, other chromatin proteins such as SU(VAR)3-7 (Cleard et al. 1997) and CAV (CARAVAGGIO/HOAP; Shareef et al. 2001) have been identified as interactors of HP1. They show a localization pattern on polytene chromosomes similar to HP1 and HP2, with SU(VAR)3-7 showing a clearly banded pattern on chromosome four (Cleard et al. 1997, Delattre et al. 2000). In addition, several other chromosomal proteins localize prominently to the fourth chromosome. These include POF (discussed below; Larsson et al. 2001), SuUR (Makunin et al. 2002), SU(VAR)3-9 (Schotta et al. 2002), and EGG (Stabell et al. 2006), among others.
In recent years, the study of histone modifications and their functions has led to an increased understanding of chromatin biology. These studies also have contributed to our understanding of fourth chromosome chromatin. H3K9 di- and tri-methylation is generally considered a heterochromatic mark, and these modified histones strongly associate with the dot chromosome of D. melanogaster in salivary gland nuclei (Jacobs et al. 2001, Cowell et al. 2002, Schotta et al. 2002). The localization of the H3K9 dimethyl mark on polytene chromosomes is similar to that of HP1, and the banded pattern observed on chromosome four is identical between HP1 and H3K9 dimethylation. In contrast, when a mark for active chromatin such as histone 4 lysine 8 acetylation is localized, the most prominent staining occurs in areas of chromosome four where HP1 is largely absent (Haynes et al. 2004). The findings on histone modifications and other heterochromatin proteins associated with chromosome four indicate that the genes resident on this chromosome have adapted to this unique environment, and are nonetheless developmentally controlled and oftentimes necessary for fly survival.
The dot chromosome binds Painting of Fourth, a chromosome-specific chromatin-associated protein
Further evidence for the notion that the dot chromosome represents a unique chromatin domain was brought to light by the discovery of painting of fourth (POF), a gene encoding a protein that specifically associates with the F element in several Drosophila species (Larsson et al. 2001). The chromosome-specific distribution of POF is shown in Figure 2 using polytene chromosomes from D. melanogaster salivary glands. While the function of POF is not well understood, it clearly associates exclusively with chromosome four in a banded pattern (Larsson et al. 2001). Amino acid sequence similarity suggests that POF is an RNA binding protein, with the strongest similarity to proteins in the polyadenylation system of flies (Larsson et al. 2001). POF provides a unique, chromosome-specific marking system; the only other such system known in D. melanogaster is the dosage compensation system, which operates on the X chromosome and is essential in males for proper gene expression. (For a review on connections between the chromosomes X and 4 see Larsson and Meller 2006)
The banding pattern of POF on polytene dot chromosomes is identical to that observed for the heterochromatic marks HP1 and H3K9 dimethylation, with one exception; POF does not spread into the most centromere-proximal regions adjacent to the chromocenter (Larsson et al. 2001). The interaction of HP1 and POF that is suggested by this colocalization is supported by genetic data as well (Johansson et al. 2007a). (Note however, that no physical interaction of HP1 with POF has been demonstrated to date.) Mutations in Su(var)205, the gene encoding HP1, result in the loss of binding of POF on the fourth chromosome (Johansson et al. 2007a). Conversely, in pof mutant larvae, HP1 binding to the fourth chromosome in polytene chromosomes is lacking as well. Recently published chromatin immunoprecipitation data for both POF and HP1 demonstrate that the two proteins bind to the same genomic locations, favoring exons (see Figure 3). This concomitant binding is strongly correlated with gene expression levels, with non-transcribed genes showing lower levels of both POF and HP1. Interestingly, transgene reporters showing a red eye phenotype are associated with regions that show low levels of POF and HP1, rather than correlating with the expression state of the fourth chromosome genes (Johansson et al. 2007b). This finding suggests that fourth chromosome genes can respond to HP1 levels in a way that the reporter genes cannot, perhaps through association with POF.
Figure 3.
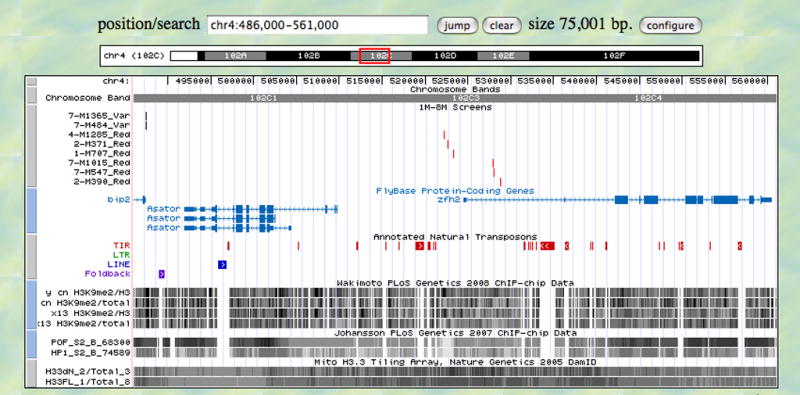
Region of the 4th chromosome with sample tracks. Shown is a 75 kb region of the 4th chromosome which contains both variegating (near the left edge) and non-variegating inserts (near the center). Below various information tracks are shown, displayed using the University of California at Santa Cruz genome browser software. Other tracks include genes from Flybase (version 5.3); annotated transposons (version 5.6); and various dense wiggle plots of selected genome scale experiments from Yasuhara et al. (2008) showing chromatin-IP/microarray analysis using antibodies to methylated H3K9, from Johansson et al. (2007) showing chromatin-IP/microarray analysis using antibodies to POF and HP1, and from Mito et al. (2005) showing results of genome-wide profiling of histone H3.3.
Despite the fact that HP1 and POF depend on each other for their localization to the fourth chromosome, their effects on gene expression are the opposite of each other; depletion of HP1 leads to an a small but consistent increase in gene expression of chromosome four genes, while depletion of POF leads to an small, but significant decrease in gene expression for the same set of genes (Johansson et al. 2007a). This finding is somewhat puzzling given that the results from polytene chromosome preparations indicate that loss of either HP1 or POF will also result in loss of the other. One model suggested to explain all of these data proposes that POF and HP1 have opposing influences on transcribed genes of chromosome four, providing a balanced control of gene expression (Johansson et al. 2007a).
D. melanogaster chromosome four has a dedicated histone 3 lysine 9 methyltransferase
As noted above, the dot chromosome shows enrichment for H3K9 dimethylation in addition to enrichment for HP1. Studies in several systems have demonstrated that HP1 can interact with both di- or tri-methylated H3K9 (Bannister et al. 2001, Jacobs et al. 2001, Lachner et al. 2001), and a histone methyltransferase imparting this modification (Schotta et al. 2002). In Drosophila, the latter interaction has been demonstrated for HP1 and the H3K9 methyltransferase SU(VAR)3-9 (Schotta et al. 2002). However, this enzyme is not responsible for histone methylation on chromosome four. Polytene chromosomes from Su(var)3-9 mutants lack prominent H3K9 dimethylation everywhere but on the fourth chromosome, which retains the modification in the usual banded staining pattern (Schotta et al. 2002). Recently, EGG (also known as dSETDB1) has been identified as the H3K9 methyltransferase responsible for maintaining histone methylation on the dot chromosome; however, it also carries out histone methylation on a more global scale (Clough et al. 2007, Seum et al. 2007, Tzeng et al. 2007). Immunoprecipitation studies have demonstrated that POF interacts with EGG (Tzeng et al. 2007). Lastly, binding of both POF and HP1 to the fourth chromosome appears to be dependent on EGG-mediated methylation of H3K9 residues (Tzeng et al. 2007). In egg mutants, HP1 and, to a greater extent, H3K9me2 levels observed on polytene chromosome squashes for chromosome four are diminished. In addition, POF staining in is almost completely absent from these chromosomes, with the exception of a small region at the base of chromosome four. Similarly, in POF deficient chromosomes, the fourth chromosome lacks H3K9m2, and to a large degree also HP1 (although some staining remains; (Tzeng et al. 2007)). In Su(var)205 mutants lacking HP1, no POF binding to the fourth chromosome is observed, and H3K9me2 is redistributed, loosing its preference for heterochromatin association (Schotta et al. 2002, Johansson et al. 2007a). Despite all this data, the interactions between EGG, POF, and HP1 currently are not well understood, but their interdependence suggests a complex relationship (for a model see Tzeng et al. 2007).
Genetic evidence for dot chromosome chromatin domain
In addition to the evidence provided above, ranging from the chromosome four-specific protein associations to its peculiar sequence composition, there is genetic evidence as well in support of the hypothesis that chromosome four represents a specialized chromatin domain. As noted earlier, some modifiers of PEV influence reporters in a domain-specific fashion, i.e. mutations in Su(var)205 will affect silencing of reporters in most heterochromatin domains with the exception of telomeres (Wallrath and Elgin 1995). When different reporter lines on the fourth chromosome were investigated systematically, two things were noted. One, for variegating reporters in the banded 1.2Mb of chromosome four, chromosome four itself, specifically the banded portion, acts as a suppressor of variegation (Haynes et al. 2007). Thus, adding additional copies of the banded portion of chromosome four (for example with an attached fourth chromosome) will result in increased pigment levels from a variegating reporter such as 39C-12 located in the banded portion of chromosome four (see Figure 1 for exact location). This Su(var) effect is not observed for variegating reporters that map to the pericentric region of chromosome four or to any other tested genomic location (Haynes et al. 2007). This finding argues that proteins (perhaps POF) critical for the fourth chromosome alone are titrated to low levels under such circumstances. Secondly, the effect of Su(var)3-9 mutations on variegating reporters on the dot chromosome is clearly distinct for insertions in the pericentric regions versus regions in the banded portion (Haynes et al. 2007). This finding is congruent with the identification of EGG as the main histone methyltransferase acting on the banded region of chromosome four (Tzeng et al. 2007) and the observation that the pericentric regions of all chromosomes, including chromosome four, are modified by SU(VAR)3-9 (Schotta et al. 2002). Thus, these data provide further support for the thesis that the dot chromosome represents a specialized chromatin domain. The fourth chromosome provides a model for those conditions necessary or sufficient to generate a heterochromatic domain that nonetheless allows gene function. This may provide an “intermediate” stage that will provide insights into the regulation of repetitive elements in mammalian genomes.
Concluding Remarks
Studies of the D. melanogaster dot chromosome over the last two decades have amassed a vast array of different data characterizing this small chromosome. In their totality, they provide support for the model of chromosome four as a specialized chromatin domain that shares characteristics with both heterochromatin and euchromatin. These data come from many subdisciplines, which have provided the necessary context and depth to understand the biology of this small but complex chromosome. Based on these different data types, the dot chromosome can be considered to represent a specialized chromatin domain, which is characterized by such unique features as HP1 binding in a region of high gene density, the use of the specialized histone methyltransferase EGG (Tzeng et al. 2007), and the chromosome-four-specific protein POF (Larsson et al. 2001).
While much progress has been made in understanding the peculiarities of the D. melanogaster fourth chromosome, one important aspect of its biology is still poorly understood. The relationship between HP1, POF, H3K9 methylation and EGG warrants further investigation and the mechanisms employed for targeting these proteins to the fourth chromosome needs to be identified. Two possible mechanisms have been suggested, both of which might operate on the dot chromosome. The first mechanism, discussed in detail by Johansson and colleagues, is based on the link between gene transcription and the localization of HP1 and POF (Johansson et al. 2007b). The sequence similarity of POF to RNA-binding proteins suggests a link to the transcription machinery that would initiate the recruitment of POF, EGG, and HP1. HP1 has also been associated with transcription at large puffs, such as the induced heat shock loci, where it is reported to play a role in transcript elongation (Piacentini et al. 2003). On the fourth chromosome, this mechanism might work in conjunction with a second mechanism that dictates the high basal level of HP1.
A second possible targeting mechanism is suggested by the finding that repetitive elements are enriched on chromosome four; that repetitive elements could potentially serve as targets for heterochromatin formation is supported by studies of RNAi and its role in transposon control and centromere function (based on data from other organisms as well as D. melanogaster; Riddle and Elgin 2008). In particular, in D. melanogaster the piRNA (PIWI-associated RNAs) pathway has been implicated in transposon control (Aravin et al. 2007). In the Drosophila germline, which for the purpose of this discussion also includes the somatic tissues of the ovaries and testes, specialized small RNA species were found to be associated with the three Argonaute proteins of the PIWI-clade, AGO3, AUB, and PIWI (Saito et al. 2006, Brennecke et al. 2007, Gunawardane et al. 2007). These short piRNAs are hypothesized to be generated from long transcripts derived from transposon-rich loci such as flamenco (Sarot et al. 2004, Brennecke et al. 2007). The piRNAs then could serve in targeting the heterochromatin machinery to loci complimentary to the piRNAs. Among the piRNA sequences that have been detected are many transposable elements and repetitive sequences also found on chromosome four, including sequences matching 1360 (Aravin et al. 2003, Brennecke et al. 2007, Nishida et al. 2007). Transposable elements are deregulated in mutants of piwi, demonstrating PIWI’s involvement in the control and silencing of these elements in the germline (Sarot et al. 2004, Kalmykova et al. 2005, Aravin et al. 2007). Similar results have been obtained for mutations in other RNAi pathway components (Aravin et al. 2001, Savitsky et al. 2006, Vagin et al. 2006). However, how transposon silencing is maintained and propagated outside of the germline is an area of active investigation.
Evidence supporting a role of the RNAi system, in particular the piRNA pathway, in the formation of heterochromatin is the finding that mutations in piwi can act as weak suppressors of variegation (Pal-Bhadra et al. 2004, Haynes et al. 2006). Eye pigment levels are increased in the presence of piwi mutations for a pericentric reporter (118-E10) and a chromosome four reporter (39-C12). The connection between the piRNA pathway and heterochromatin formation is further strengthened by yeast two-hybrid studies and immunoprecipitation experiments, which show that HP1 and PIWI physically interact with each other (Brower-Toland et al. 2007). Recent work from yeast has shed some light on the seemingly paradoxical situation where heterochromatin formation and silencing via the RNAi pathways actually depend on transcription of the heterochromatic sequences. HP1 and H3K9 dimethylation are lost from heterochromatic regions of the genome during mitosis, following phosphorylation of H3S10 (Fischle et al. 2003, Fischle et al. 2005, Dormann et al. 2006). Work in the yeast S. pombe has shown that this allows transcription of those regions, generating the needed rasiRNAs to reinstate silencing (Chen et al. 2008, Kloc et al. 2008). This finding holds great promise that investigating chromatin with its dynamic nature in mind will allow us to solve the mystery of heterochromatin formation and its inheritance.
Looking into the future, we see great promise in two areas of Drosophila research for contributing to our understanding of heterochromatin and the dot chromosome. One such area is the comparative genomics. With whole genome sequence data now available from the twelve Drosophila species – and more being planned – comparative work will be able to shed light on chromosome four biology by taking advantage of such species as D. willistoni, which has the F element fused to one of the long chromosome arms, or D. ananassae, which has a greatly expanded fourth chromosome. Another technique that is of immense value to understanding chromatin structure is chromatin immunoprecipitation, which is now being applied on a genome-wide scale to understand protein distributions and histone modification patterns (see Figure 3). With these studies being expanded by individual laboratories as well as by community service projects such as modENCODE (for a description of this project see www.modENCODE.org) to include a wide variety of proteins in a range of tissues, it will be possible to gain an understanding of chromatin and its dynamic nature in multiple dimensions.
Acknowledgments
We would like to acknowledge G. Volpi for the chromosome images shown in Figure 2. In addition, we would like to thank members of the Elgin laboratory, especially B. Brower-Toland and K. Huisinga, for helpful discussions and comments on the manuscript. Funding was provided by a grant from the National Institute of Health to S. C. R. Elgin (GM068388).
References
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318(5851):761–4. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5(2):337–50. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11(13):1017–27. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410(6824):120–4. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Beadle GW. A possible influence of the spindle fibre on crossing-over in Drosophila. Proc Natl Acad Sci U S A. 1932;18(2):160–5. doi: 10.1073/pnas.18.2.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128(6):1089–103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Bridges CB. Genetical and cytological proof of non-disjunction of the fourth chromosome of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1921;7(7):186–92. doi: 10.1073/pnas.7.7.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21(18):2300–11. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451(7179):734–7. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- Chino M. The genetics of Drosophila virilis. Japanese Journal of Genetics. 1936/1937;12(13):189–210. 257–277, 100–120. [Google Scholar]
- Cleard F, Delattre M, Spierer P. SU(VAR)3-7, a Drosophila heterochromatin-associated protein and companion of HP1 in the genomic silencing of position-effect variegation. Embo J. 1997;16(17):5280–8. doi: 10.1093/emboj/16.17.5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough E, Moon W, Wang S, Smith K, Hazelrigg T. Histone methylation is required for oogenesis in Drosophila. Development. 2007;134(1):157–65. doi: 10.1242/dev.02698. [DOI] [PubMed] [Google Scholar]
- Cowell IG, Aucott R, Mahadevaiah SK, Burgoyne PS, Huskisson N, Bongiorni S, Prantera G, Fanti L, Pimpinelli S, Wu R, Gilbert DM, Shi W, Fundele R, Morrison H, Jeppesen P, Singh PB. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma. 2002;111(1):22–36. doi: 10.1007/s00412-002-0182-8. [DOI] [PubMed] [Google Scholar]
- Cryderman DE, Morris EJ, Biessmann H, Elgin SC, Wallrath LL. Silencing at Drosophila telomeres: nuclear organization and chromatin structure play critical roles. Embo J. 1999a;18(13):3724–35. doi: 10.1093/emboj/18.13.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryderman DE, Tang H, Bell C, Gilmour DS, Wallrath LL. Heterochromatic silencing of Drosophila heat shock genes acts at the level of promoter potentiation. Nucleic Acids Res. 1999b;27(16):3364–70. doi: 10.1093/nar/27.16.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre M, Spierer A, Tonka CH, Spierer P. The genomic silencing ofposition-effect variegation in Drosophila melanogaster: interaction between the heterochromatin-associated proteins Su(var)3-7 and HP1. J Cell Sci. 2000;113(Pt 23):4253–61. doi: 10.1242/jcs.113.23.4253. [DOI] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125(3):497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Dormann HL, Tseng BS, Allis CD, Funabiki H, Fischle W. Dynamic regulation of effector protein binding to histone modifications: the biology of HP1 switching. Cell Cycle. 2006;5(24):2842–51. doi: 10.4161/cc.5.24.3540. [DOI] [PubMed] [Google Scholar]
- Etchegaray JP, Yang X, DeBruyne JP, Peters AH, Weaver DR, Jenuwein T, Reppert SM. The polycomb group protein EZH2 is required for mammalian circadian clock function. J Biol Chem. 2006;281(30):21209–15. doi: 10.1074/jbc.M603722200. [DOI] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438(7071):1116–22. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425(6957):475–9. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- Grell RF. Heat-induced exchange in the fourth chromosome of diploid females of Drosophila melanogaster. Genetics. 1971;69(4):523–7. doi: 10.1093/genetics/69.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447(7143):399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315(5818):1587–90. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Haynes KA, Caudy AA, Collins L, Elgin SC. Element 1360 and RNAi components contribute to HP1-dependent silencing of a pericentric reporter. Curr Biol. 2006;16(22):2222–7. doi: 10.1016/j.cub.2006.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes KA, Gracheva E, Elgin SC. A distinct type of heterochromatin within Drosophila melanogaster chromosome 4. Genetics. 2007;175(3):1539–42. doi: 10.1534/genetics.106.066407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes KA, Leibovitch BA, Rangwala SH, Craig C, Elgin SC. Analyzing heterochromatin formation using chromosome 4 of Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 2004;69:267–72. doi: 10.1101/sqb.2004.69.267. [DOI] [PubMed] [Google Scholar]
- Heitz E. Das Heterochromatin der Moose. Jb Wiss Bot. 1928;69:728–818. [Google Scholar]
- Henikoff S, Jackson JM, Talbert PB. Distance and pairing effects on the brownDominant heterochromatic element in Drosophila. Genetics. 1995;140(3):1007–17. doi: 10.1093/genetics/140.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Li J, Eissenberg JC, Allis CD, Khorasanizadeh S. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. Embo J. 2001;20(18):5232–41. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TC, Eissenberg JC, Craig C, Dietrich V, Hobson A, Elgin SC. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur J Cell Biol. 1989;50(1):170–80. [PubMed] [Google Scholar]
- Johansson AM, Stenberg P, Bernhardsson C, Larsson J. Painting of fourth and chromosome-wide regulation of the 4th chromosome in Drosophila melanogaster. Embo J. 2007a;26(9):2307–16. doi: 10.1038/sj.emboj.7601604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson AM, Stenberg P, Pettersson F, Larsson J. POF and HP1 bind expressed exons, suggesting a balancing mechanism for gene regulation. PLoS Genet. 2007b;3 (11):e209. doi: 10.1371/journal.pgen.0030209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmykova AI, Klenov MS, Gvozdev VA. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res. 2005;33(6):2052–9. doi: 10.1093/nar/gki323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol. 2008;18(7):490–5. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410(6824):116–20. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Larsson J, Chen JD, Rasheva V, Rasmuson-Lestander A, Pirrotta V. Painting of fourth, a chromosome-specific protein in Drosophila. Proc Natl Acad Sci U S A. 2001;98(11):6273–8. doi: 10.1073/pnas.111581298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J, Meller VH. Dosage compensation, the origin and the afterlife of sex chromosomes. Chromosome Res. 2006;14(4):417–31. doi: 10.1007/s10577-006-1064-3. [DOI] [PubMed] [Google Scholar]
- Larsson J, Svensson MJ, Stenberg P, Makitalo M. Painting of fourth in genus Drosophila suggests autosome-specific gene regulation. Proc Natl Acad Sci U S A. 2004;101(26):9728–33. doi: 10.1073/pnas.0400978101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke J, McDermid HE. Analysis of Drosophila chromosome 4 using pulsed field gel electrophoresis. Chromosoma. 1993;102(10):718–23. doi: 10.1007/BF00650898. [DOI] [PubMed] [Google Scholar]
- Lowenhaupt K, Rich A, Pardue ML. Nonrandom distribution of long mono-and dinucleotide repeats in Drosophila chromosomes: correlations with dosage compensation, heterochromatin, and recombination. Mol Cell Biol. 1989;9(3):1173–82. doi: 10.1128/mcb.9.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu BY, Bishop CP, Eissenberg JC. Developmental timing and tissue specificity of heterochromatin-mediated silencing. Embo J. 1996;15(6):1323–32. [PMC free article] [PubMed] [Google Scholar]
- Lu BY, Ma J, Eissenberg JC. Developmental regulation of heterochromatin-mediated gene silencing in Drosophila. Development. 1998;125(12):2223–34. doi: 10.1242/dev.125.12.2223. [DOI] [PubMed] [Google Scholar]
- Makunin IV, Volkova EI, Belyaeva ES, Nabirochkina EN, Pirrotta V, Zhimulev IF. The Drosophila suppressor of underreplication protein binds to late-replicating regions of polytene chromosomes. Genetics. 2002;160(3):1023–34. doi: 10.1093/genetics/160.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37(10):1090–7. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- Muller HJ. Types of visible variations induced by X-rays in Drosophila. J Genet. 1930;22:299–334. [Google Scholar]
- Nakahata Y, Grimaldi B, Sahar S, Hirayama J, Sassone-Corsi P. Signaling to the circadian clock: plasticity by chromatin remodeling. Curr Opin Cell Biol. 2007;19(2):230–7. doi: 10.1016/j.ceb.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada T, Inagaki S, Siomi H, Siomi MC. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. Rna. 2007;13(11):1911–22. doi: 10.1261/rna.744307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne JD. Crossing over in a T(1;4) Translocation in Drosophila melanogaster. University of Alberta; Edmonton, Alberta: 1998. [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303(5658):669–72. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- Piacentini L, Fanti L, Berloco M, Perrini B, Pimpinelli S. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J Cell Biol. 2003;161(4):707–14. doi: 10.1083/jcb.200303012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle NC, Elgin SC. A role for RNAi in heterochromatin formation in Drosophila. Curr Top Microbiol Immunol. 2008;320:185–209. doi: 10.1007/978-3-540-75157-1_9. [DOI] [PubMed] [Google Scholar]
- Riddle NC, Leung W, Haynes KA, Granok H, Wuller J, Elgin SC. An investigation of heterochromatin domains on the fourth chromosome of Drosophila melanogaster. Genetics. 2008;178(3):1177–91. doi: 10.1534/genetics.107.081828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schafer C, Phalke S, Walther M, Schmidt A, Jenuwein T, Reuter G. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell. 2007;26(1):103–15. doi: 10.1016/j.molcel.2007.02.025. [DOI] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20 (16):2214–22. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler L, Szauter P. The effect of recombination-defective meiotic mutants on fourth-chromosome crossing over in Drosophila melanogaster. Genetics. 1978;90(4):699–712. doi: 10.1093/genetics/90.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarot E, Payen-Groschene G, Bucheton A, Pelisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166(3):1313–21. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006;20(3):345–54. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. Embo J. 2002;21(5):1121–31. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seum C, Reo E, Peng H, Rauscher FJ, 3rd, Spierer P, Bontron S. Drosophila SETDB1 is required for chromosome 4 silencing. PLoS Genet. 2007;3(5):e76. doi: 10.1371/journal.pgen.0030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer CD, Stephens GE, Thompson BA, Funches L, Bernat JA, Craig CA, Elgin SC. Heterochromatin protein 2 (HP2), a partner of HP1 in Drosophila heterochromatin. Proc Natl Acad Sci U S A. 2002;99(22):14332–7. doi: 10.1073/pnas.212458899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shareef MM, King C, Damaj M, Badagu R, Huang DW, Kellum R. Drosophila heterochromatin protein 1 (HP1)/origin recognition complex (ORC) protein is associated with HP1 and ORC and functions in heterochromatin-induced silencing. Mol Biol Cell. 2001;12(6):1671–85. doi: 10.1091/mbc.12.6.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawson EE, Shaffer CD, Malone CD, Leung W, Kellman E, Shevchek RB, Craig CA, Bloom SM, Bogenpohl JI, Dee J, Morimoto ETA, Myoung J, Nett AS, Ozsolak F, Tittiger ME, Zeug A, Pardue ML, Buhler J, Mardis ER, Elgin SCR. Comparison of dot chromosome sequences from D. melanogaster and D. virilis reveals an enrichment of DNA transposon sequences in heterochromatic domains. Genome Biol. 2006;7(2):R15. doi: 10.1186/gb-2006-7-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabell M, Bjorkmo M, Aalen RB, Lambertsson A. The Drosophila SET domain encoding gene dEset is essential for proper development. Hereditas. 2006;143(2006):177–88. doi: 10.1111/j.2006.0018-0661.01970.x. [DOI] [PubMed] [Google Scholar]
- Stephens GE, Slawson EE, Craig CA, Elgin SC. Interaction of heterochromatin protein 2 with HP1 defines a novel HP1-binding domain. Biochemistry. 2005;44(40):13394–403. doi: 10.1021/bi051006+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH. A map of the fourth chromosome of Drosophila melanogaster, based on crossing over in triploid females. Proc Natl Acad Sci U S A. 1951;37(7):405–7. doi: 10.1073/pnas.37.7.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun FL, Cuaycong MH, Elgin SC. Long-range nucleosome ordering is associated with gene silencing in Drosophila melanogaster pericentric heterochromatin. Mol Cell Biol. 2001;21(8):2867–79. doi: 10.1128/MCB.21.8.2867-2879.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun FL, Haynes K, Simpson CL, Lee SD, Collins L, Wuller J, Eissenberg JC, Elgin SC. cis-Acting determinants of heterochromatin formation on Drosophila melanogaster chromosome four. Mol Cell Biol. 2004;24(18):8210–20. doi: 10.1128/MCB.24.18.8210-8220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. A reexamination of spreading of position-effect variegation in the white-roughest region of Drosophila melanogaster. Genetics. 2000;154(1):259–72. doi: 10.1093/genetics/154.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. Spreading of silent chromatin: inaction at a distance. Nat Rev Genet. 2006;7(10):793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- Thompson PE. Centric pairing and crossing-over in Drosophila melanogaster. Genetics. 1963;48:697–701. doi: 10.1093/genetics/48.5.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P, Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature? Mol Cell. 2007;28(1):1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Tzeng TY, Lee CH, Chan LW, Shen CK. Epigenetic regulation of the Drosophila chromosome 4 by the histone H3K9 methyltransferase dSETDB1. Proc Natl Acad Sci U S A. 2007;104(31):12691–6. doi: 10.1073/pnas.0705534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313(5785):320–4. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Wakimoto BT, Hearn MG. The effects of chromosome rearrangements on the expression of heterochromatic genes in chromosome 2L of Drosophila melanogaster. Genetics. 1990;125(1):141–54. doi: 10.1093/genetics/125.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrath LL, Elgin SC. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 1995;9(10):1263–77. doi: 10.1101/gad.9.10.1263. [DOI] [PubMed] [Google Scholar]
- Weiler KS, Wakimoto BT. Heterochromatin and gene expression in Drosophila. Annu Rev Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- Westphal T, Reuter G. Recombinogenic effects of suppressors of position-effect variegation in Drosophila. Genetics. 2002;160(2):609–21. doi: 10.1093/genetics/160.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RJ, Goodman JL, Strelets VB. FlyBase: integration and improvements to query tools. Nucleic Acids Res. 2008;36(Database issue):D588–93. doi: 10.1093/nar/gkm930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara JC, Wakimoto BT. Molecular landscape of modified histones in Drosophila heterochromatic genes and euchromatin-heterochromatin transition zones. PLoS Genet. 2008;4(1):e16. doi: 10.1371/journal.pgen.0040016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhimulev IF, Belyaeva ES, Makunin IV, Pirrotta V, Volkova EI, Alekseyenko AA, Andreyeva EN, Makarevich GF, Boldyreva LV, Nanayev RA, Demakova OV. Influence of the SuUR gene on intercalary heterochromatin in Drosophila melanogaster polytene chromosomes. Chromosoma. 2003;111(6):377–98. doi: 10.1007/s00412-002-0218-0. [DOI] [PubMed] [Google Scholar]