Abstract
An accurate reconstruction of the eukaryotic tree of life is essential to identify the innovations underlying the diversity of microbial and macroscopic (e.g., plants and animals) eukaryotes. Previous work has divided eukaryotic diversity into a small number of high-level “supergroups,” many of which receive strong support in phylogenomic analyses. However, the abundance of data in phylogenomic analyses can lead to highly supported but incorrect relationships due to systematic phylogenetic error. Furthermore, the paucity of major eukaryotic lineages (19 or fewer) included in these genomic studies may exaggerate systematic error and reduce power to evaluate hypotheses. Here, we use a taxon-rich strategy to assess eukaryotic relationships. We show that analyses emphasizing broad taxonomic sampling (up to 451 taxa representing 72 major lineages) combined with a moderate number of genes yield a well-resolved eukaryotic tree of life. The consistency across analyses with varying numbers of taxa (88–451) and levels of missing data (17–69%) supports the accuracy of the resulting topologies. The resulting stable topology emerges without the removal of rapidly evolving genes or taxa, a practice common to phylogenomic analyses. Several major groups are stable and strongly supported in these analyses (e.g., SAR, Rhizaria, Excavata), whereas the proposed supergroup “Chromalveolata” is rejected. Furthermore, extensive instability among photosynthetic lineages suggests the presence of systematic biases including endosymbiotic gene transfer from symbiont (nucleus or plastid) to host. Our analyses demonstrate that stable topologies of ancient evolutionary relationships can be achieved with broad taxonomic sampling and a moderate number of genes. Finally, taxon-rich analyses such as presented here provide a method for testing the accuracy of relationships that receive high bootstrap support (BS) in phylogenomic analyses and enable placement of the multitude of lineages that lack genome scale data.
Keywords: Excavata, microbial eukaryotes, Rhizaria, supergroups, systematic error, taxon sampling
Perspectives on the structure of the eukaryotic tree of life have shifted in the past decade as molecular analyses provide hypotheses for relationships among the approximately 75 robust lineages of eukaryotes. These lineages are defined by ultrastructural identities (Patterson 1999)—patterns of cellular and subcellular organization revealed by electron microscopy—and are strongly supported in molecular analyses (Parfrey et al. 2006; Yoon et al. 2008). Most of these lineages now fall within a small number of higher level clades, the supergroups of eukaryotes (Simpson and Roger 2004; Adl et al. 2005; Keeling et al. 2005). Several of these clades—Opisthokonta, Rhizaria, and Amoebozoa—are increasingly well supported by phylogenomic (Rodríguez-Ezpeleta et al. 2007a; Burki et al. 2008; Hampl et al. 2009) and phylogenetic (Parfrey et al. 2006; Pawlowski and Burki 2009), analyses, whereas support for “Archaeplastida” predominantly comes from some phylogenomic studies (Rodríguez-Ezpeleta et al. 2005; Burki et al. 2007) or analyses of plastid genes (Yoon et al. 2002; Parfrey et al. 2006). In contrast, support for “Chromalveolata” and Excavata is mixed, often dependent on the selection of taxa included in analyses (Rodríguez-Ezpeleta et al. 2005; Parfrey et al. 2006; Rodríguez-Ezpeleta et al. 2007a; Burki et al. 2008; Hampl et al. 2009). We use quotation marks throughout to note groups where uncertainties remain. Moreover, it is difficult to evaluate the overall stability of major clades of eukaryotes because phylogenomic analyses have 19 or fewer of the major lineages and hence do not sufficiently sample eukaryotic diversity (Rodríguez-Ezpeleta et al. 2007b; Burki et al. 2008; Hampl et al. 2009), whereas taxon-rich analyses with 4 or fewer genes yield topologies with poor support at deep nodes (Cavalier-Smith 2004; Parfrey et al. 2006; Yoon et al. 2008).
Estimating the relationships of the major lineages of eukaryotes is difficult because of both the ancient age of eukaryotes (1.2–1.8 billion years; Knoll et al. 2006) and complex gene histories that include heterogeneous rates of molecular evolution and paralogy (Maddison 1997; Gribaldo and Philippe 2002; Tekle et al. 2009). A further issue obscuring eukaryotic relationships is the chimeric nature of the eukaryotic genome—not all genes are vertically inherited due to lateral gene transfer (LGT) and endosymbiotic gene transfer (EGT)—that can also mislead efforts to reconstruct phylogenetic relationships (Andersson 2005; Rannala and Yang 2008; Tekle et al. 2009). This is especially true among photosynthetic lineages that comprise “Chromalveolata” and “Archaeplastida” where a large portion of the host genome (approximately 8–18%) is derived from the plastid through EGT (Martin and Schnarrenberger 1997; Martin et al. 2002; Lane and Archibald 2008; Moustafa et al. 2009; Tekle et al. 2009).
There is a long-standing debate among systematists as to the relative benefits of increasing gene or taxon sampling (Hillis et al. 2003; Cummings and Meyer 2005; Rokas and Carroll 2005). Both approaches improve phylogenetic reconstruction by alleviating either stochastic or systematic phylogenetic error (e.g., Rokas and Carroll 2005; Hedtke et al. 2006). Stochastic error results from too little signal in the data (e.g., single to few gene trees) to estimate relationships and results in poorly resolved trees with low support, especially at deep levels (Swofford et al. 1996; Rokas and Carroll 2005). The problems of stochastic error are amplified for deep relationships, such as relationships among major clades of eukaryotes (Roger and Hug 2006). Many researchers opt to increase the number of genes, exemplified by phylogenomic studies, which alleviates stochastic error and yields well-resolved trees that are highly supported (Rokas and Carroll 2005; Burki et al. 2007; Hampl et al. 2009). However, analyses of many genes are still vulnerable to systematic error and often include very few lineages.
Systematic error results from biases in the data that mislead phylogenetic reconstruction, yielding incorrect sister group relationships that do not reflect historical relationships; the most well known of these is long-branch attraction (Felsenstein 1978). Incongruence can also arise from conflicts between gene trees and species trees resulting from population genetic processes or the chimeric nature of eukaryotic genomes (Maddison 1997; Rannala and Yang 2008). Systematic errors can be detected and eliminated by several methods that are often combined, including using more realistic models of sequence evolution (e.g., Rodríguez-Ezpeleta et al. 2007b), removing rapidly evolving genes and/or taxa that cause errors (Brinkmann et al. 2005), and by increasing taxonomic sampling (Zwickl and Hillis 2002; Hedtke et al. 2006). Increased taxon sampling has been shown to improve phylogenetic accuracy even when the additional taxa contain large amounts of missing data (Philippe et al. 2004; Wiens 2005; Wiens and Moen 2008). In contrast, the abundance of data in phylogenomic studies can yield highly supported, but incorrect relationships caused by these systematic biases (Philippe et al. 2004; Hedtke et al. 2006; Jeffroy et al. 2006; Rokas and Chatzimanolis 2008). Taxon-rich analyses provide a method for testing the accuracy of relationships that receive high BS support in phylogenomic analyses (Zwickl and Hillis 2002; Heath et al. 2008).
Here, we assess the eukaryotic tree of life by analyzing 16 genes from a broadly sampled data set that includes 451 diverse taxa from 72 lineages. We aim to overcome both stochastic and systematic phylogenetic error by assessing two measures of clade robustness: (i) statistical support (bootstrap), and (ii) the stability of clades across analyses with varying numbers of taxa and levels of missing data. We demonstrate that extensive taxon sampling coupled with selection of a modest number of well-sampled genes counteracts systematic error and correctly places many rapidly evolving lineages without the removal of genes or taxa. Furthermore, this approach enables us to place the numerous lineages that have only a few genes sequenced, and to assess support for the hypothesized clades of eukaryotes with a more inclusive sampling of diverse lineages.
METHODS
Gene Sequencing
Ovammina opaca and Ammonia sp. T7 were collected from a salt marsh on Cabretta Island, Georgia with assistance from Susan T. Goldstein (University of Georgia). DNA was isolated from 60 cells each that were individually picked, washed, and purged of food items overnight using a plant DNeasy kit (Qiagen). Gromia sp. Antarctica DNA was isolated from one cell undergoing gametogenesis and generously provided by Sam Bowser and Andrea Habura (Wadsworth Center). DNA for all other taxa was obtained from American Type Culture Collection (ATCC; Table S1, available from http://www.sysbio.oxfordjournals.org/) and accessions have been photodocumented (http://eutree.lifedesks.org/). Small subunit ribosomal DNA (SSU-rDNA) was amplified with previously described primers (Medlin et al. 1988) and 3 additional primers were used to generate overlapping sequences from each clone (Snoeyenbos-West et al. 2002). Hsp90 was amplified with CAC CTG ATG TCT YTN ATH ATH AAY and CTG GCG AGA NAN RTT NAR NGG, and reamplified with nested primers TCT CTG ATC ATC AAY RCN TTY TAY and AGA GAT GTT NAR NGG NAN RTC. Primers for actin, alpha-tubulin and beta-tubulin are from Tekle et al. (2008). Phusion DNA Polymerase (Finnzymes Inc.), a strict proofreading enzyme, was used to amplify the genes of interest and Invitrogen Zero Blunt Topo cloning kits were used for cloning. Sequencing of cloned plasmid DNA was accomplished using vector- or gene-specific primers and the BigDye terminator kit (Applied Biosystems). Sequences were run on an ABI 3100 automated sequencer. We have fully sequenced 1–4 clones of each gene and surveyed up to 10 clones per taxon in order to detect paralogs. Stephanopogon apogon SSU-rDNA is extremely large and we were unable to amplify it using standard methods. Instead, we amplified 3 overlapping fragments that were then combined for use in our analyses. All new sequences, including any paralogs identified, have been deposited in GenBank (GQ377645–GQ377715 and HM244866–HM244878).
Cultures of microbial eukaryotes for expressed sequence tag (EST) sequencing were obtained from ATCC or the Culture Collection of Algae and Protozoa (Table S1) and grown in Corning culture flasks according to supplier's recommended protocols. Cultures of Heteromita sp. were kindly provided by Linda Amaral Zettler and subsequently deposited at ATCC (ATCC PRA-74). Cultures were harvested and pooled as needed to obtain approximately 2 × 107 cells. Cells were pelleted and messenger RNA (mRNA) was extracted using the Qiagen Oligotex direct mRNA protocol. The resultant mRNA was quantitated by NanoDrop and/or Agilent Bioanalyzer RNA chip. Complementary DNA was generated using the ClonTech SMART cDNA construction protocol and ligated into the Lucigen pSMART vector (Diplonema papillatum) or the ClonTech pDNRlib vector (all others). Electrocompetent cells were transformed using the ligation products and plated on Luria broth-kanamycin agar. Clones were grown in 96-well polypropylene 2.0 mL deep well growth blocks containing 1.2 mL superbroth (with 30μL/mL kanamycin) per well and plasmid DNA was prepared using a modified alkaline lysis procedure adapted for automation (GenomicSolutions RevPrep Orbit or Beckman BiomekFX). Approximately 10,000 clones from each library were sequenced bidirectionally with vector primers using Sanger cycle sequencing (Applied Biosystems BigDye Terminator chemistry). Paired reads from the same clone were trimmed using custom Perl scripts and assembled based on sequence overlap using phrap (www.phrap.org). Clustering was done after assembly of paired reads, by TGICL (Pertea et al. 2003), and was used to group highly similar sequences that were extremely likely to be copies of the same gene. The size of a cluster thus reflects number of transcripts of a particular gene (gene copy number and expression level).
Data set Assembly
Taxa and genes were selected to maximize taxonomic diversity and evenness given the availability of molecular data. This strategy was used to improve phylogenetic accuracy by breaking up long branches with dense sampling across the eukaryotic tree (Hillis 1998). The classifications systems of Patterson (1999) and Adl et al. (2005) were used as guides as we aimed to sample eukaryotic diversity by including representatives of as many lineages defined by ultrastructural identities as possible (Table S2). These lineages have generally proven to be robust as they are well supported in molecular analyses (e.g., Adl et al. 2005; Parfrey et al. 2006; Yoon et al. 2008), including the current study, and they represent monophyletic groups that serve as a proxy for taxonomic diversity. Our data set has representatives from 72 lineages, including 53 of the 71 lineages plus 7 of 200 unplaced genera as defined in Patterson (1999). Additionally, we include 3 unplaced lineages isolated more recently, Malawimonas jakobiformis (O'Kelly and Nerad 1999), Breviata anathema (Walker et al. 2006), and ATCC strain 50646 (an isolate given the candidate name “Soginia anisocystis” that has yet to be described formally). We use an updated classification (Adl et al. 2005) to designate lineages in Amoebozoa and Rhizaria that belonged to the single unsupported clade (Ramicristate) from Patterson 1999 (Table S2). In order to maximize taxon evenness along with breadth, we chose limited but diverse members from within lineages where possible (e.g., we included 15 phylogenetically distant animals).
To maximize gene sampling for diverse taxa, we include markers historically targeted by polymerase chain reaction–based analyses (e.g., SSU-rDNA, actin, elongation factor 1α; Table S3) plus commonly sequenced ESTs (e.g., ribosomal proteins, 14-3-3; Table S3). The comprehensively sampled SSU-rDNA and the historical markers facilitate inclusion of many additional taxa for which only these genes have been characterized (Table S4). The minimum sequence data required for inclusion were nearly full-length SSU-rDNA, which provided the core of information necessary for phylogenetic placement with large amounts of missing data (Wiens and Moen 2008).
SSU-rDNA sequences were hand curated for target taxa by removing introns, unalignable regions, non-nuclear rDNAs, and misannotated sequences. This alignment was crucial to overall accuracy because nearly half of the target taxa are represented only by SSU-rDNA, thus several alignment and masking methods were assessed to ensure the robustness of the SSU-rDNA alignment. SSU-rDNA sequences were aligned by HMMER (Eddy 2001), version 2.1.4 with default settings, taking secondary structure into account. HMMER used a set of previously aligned sequences to model the secondary structure of a sequence. The training alignment for building the model, consisting of all available SSU-rDNA eukaryote sequences (as of December 2008) aligned according to their secondary structure, was downloaded from the European Ribosomal Database (Wuyts et al. 2002). An additional SSU-rDNA alignment was constructed in MAFFT 6 implemented in SeaView (Galtier et al. 1996) with the E-INS-i algorithm (Katoh and Toh 2008). Both alignments were further edited manually in MacClade v4.08 (Maddison D.R. and Maddison W.P. 2005). To assess the effect of rate heterogeneity on the SSU-rDNA topologies, we partitioned the data matrices into 8 rate classes using the general time-reversible (GTR) model with invariable sites and rate variation among sites following a discrete gamma distribution, as implemented in HyPhy version .99b package (Kosakovsky Pond et al. 2005). We then ran analyses without the fastest and two fastest rate classes, resulting in 1197 and 1019 characters, respectively. However, the reduced data sets resulted in less resolution in the backbone without improving apparent the long-branch attraction. Thus, we used the alignment generated in MAFFT and masked with GBlocks (Talavera and Castresana 2007) and by eye in MacClade, resulting in 867 unambiguously aligned characters.
Assembly of the protein data set relied on a custom-built pipeline and database that combined Perl and Python scripts to identify homologs from diverse eukaryotes. Our goal in developing this pipeline was to ensure that we captured the broadest possible set of sequences given the tremendous heterogeneity among microbial eukaryotes. All available protein and EST data from our target taxa (Table S4) were downloaded from GenBank in January 2009 and ESTs were analyzed in all 6 translated frames to identify correct sequences for our alignment. A fasta file of 6 sequences representing the six “supergroups” was created for each target gene and used to query our database of target taxa by BLASTp. Results were limited by length, e-value, and identity, and all sequences with greater than 1% divergence within each taxon were retained for assessment of paralogy. The resulting sequences were aligned with ClustalW (Thompson et al. 1994) and the resulting single gene alignments were assessed by eye to remove nonhomologous sequences.
The inferred amino acid sequences for each of the protein genes from our data pipeline were combined with the new sequences generated for this study and again aligned in Clustal W (Thompson et al. 1994). The alignment was adjusted by eye in MacClade (Maddison D.R. and Maddison W.P. 2005). As these alignments included all paralogs extracted from the pipeline, individual gene trees were examined to choose appropriate orthologs. For example, in cases where paralogs formed a monophyletic group, the shortest branch sequence was retained. When paralogs fell into multiple locations on the tree, we aimed to maintain orthologous groups that included the greatest taxonomic representation. The individual gene alignments were then concatenated to build a 16 gene, 451-taxon matrix with 6578 unambiguously aligned characters, including SSU-rDNA. All other data sets were constructed by removing taxa and/or genes from this matrix. All data matrices are available at TreeBASE (submission ID S10552).
Creation of Subdata Matrices
We created an array of data matrices by subsampling our full data matrix of 16 genes (15 protein-coding genes plus SSU-rDNA) and 451 taxa (denoted all:16) in order to assess the impact of taxon sampling, missing data, and gene sampling. First, seven data sets were created to assess the impact of missing data and taxon sampling (summarized in Table 1). The least inclusive of these contained 16 genes and all 88 taxa that had at least 10 of the 16 genes (10:16), which resulted in 17% missing data. Similarly, the 6:16 and 4:16 matrices include all taxa with at least 6 and 4 of the targeted 16 genes, respectively. SSU-rDNA is ubiquitously sampled in our data set and many phylogenetic hypotheses are based on SSU-rDNA genealogies. To address the concern that SSU-rDNA was driving our results, we deleted it from each of the 16 gene data sets resulting in 9:15, 5:15, 3:15, and all:15 matrices.
TABLE 1.
Support for major clades of eukaryotes in analyses containing varying levels of taxon inclusion and missing data
| 10:16 | 6:16 | 4: 16 | all:16 | 9:15 | 5:15 | 3:15 | all:15 | |
| Supported clades | ||||||||
| Opisthokonta | 99a | 97a | 97a | 69 | 100a | 99a | 85 | 19 |
| Rhizaria | 100a | 99a | 94a | 82 | 100a | 100a | 47 | 29 |
| SAR | 97a | 98a | 63 | 22 | 100a | 100a | 32 | 19 |
| Rhizaria + stramenopiles | 94a | 94a | 57 | 26 | 92a | 96a | 29 | 18 |
| Excavata | 83 | 77 | 65 | 6 | 84 | 76 | 44 | 19 |
| Amoebozoa | 59 | 46 | 49 | nm | 68 | 56 | 44 | 5 |
| “Unikonta” | 63 | 39 | 21 | nm | 54 | 50 | 15 | 3 |
| Weak/unsupported hypotheses | ||||||||
| “Archaeplastida” | nm | nm | nm | nm | nm | nm | nm | nm |
| “Chromalveloata” | nm | nm | nm | nm | nm | nm | nm | nm |
| Cryptomonads + haptophytes | 33 | 50 | nm | 29 | 38 | 56 | 22 | 25 |
| Haptophytes + SAR | nm | nm | 15 | nm | nm | nm | nm | nm |
| Alveolates + stramenopiles | nm | nm | nm | nm | nm | nm | nm | nm |
| Red algae + green algae | nm | nm | nm | nm | nm | nm | nm | nm |
| Red, Green, Glauco, Hapto, Crypt | 47 | 32 | nm | 9 | 39 | 27 | 16 | 8 |
| Data set statistics | ||||||||
| Number of taxa | 88 | 111 | 160 | 451 | 88 | 111 | 160 | 240 |
| Number of lineages | 26 | 30 | 45 | 72 | 26 | 30 | 45 | 54 |
| % Missing data (characters) | 17 | 25 | 38 | 69 | 19 | 28 | 43 | 59 |
Note: Supported clades are stable across analyses, albeit with decreasing support as the percentage of missing data increases. Bootstrap support values from RAxML analyses. Support values greater than 75 are indicated by bold text and greater than 85 are indicated with a. nm = nonmonophyletic. Column headings describe the data sets. For example, “10:16” includes all taxa that have at least 10 of the 16 genes, with a total of 88 taxa representing 26 lineages and containing 17% missing data. The “all:15” includes the protein-coding genes from all taxa and contains 59% missing data. See Table S2 for lineages and Figure S1a–h for individual trees.
Support values greater than 85.
To assess the relative importance of gene versus taxon sampling, we compared our full analysis to data sets with taxon sampling based a recent phylogenomic analysis (Hampl et al. 2009; Table S5, Hampl:16 gene) and phylogenetic analysis (Yoon et al. 2008; Table S5, Yoon: 16 gene). We also analyzed a data set of the 4 genes used by Yoon et al. (2008) (actin, alpha tubulin, beta tubulin, and SSU-rDNA) with our taxon sampling (Table S5; all:4 gene). Although a thorough test of the impact of gene sampling would require a large number of analyses of data sets with genes systematically deleted, we feel that this approach provides insight into the contributions of genes and taxa.
Photosynthetic lineages have chimeric genomes that are composed of genes originating both from the host eukaryote, the endosymbiotic plastid (through EGT), and, in cases of secondary or greater endosymbiosis, from the symbiont nucleus. If genes of multiple origins were retained in our concatenated data set, the resulting conflicting signal between host, symbiont, and plastid could mislead phylogenetic reconstruction. This chimerism may contribute to the instability observed for photosynthetic lineages without clear sister groups (red algae, green algae, glaucocystophytes, cryptomonads, and haptophytes). Thus, we used 2 methods to detect discordance among loci that could indicate EGT. First, the 16 genes from representatives of each of these photosynthetic lineages were analyzed by top BLASTp hit. We scored the first 2 lineages hit, with red algae, green algae, plants, or glaucophytes taken as evidence for EGT. Nine genes showed some evidence of EGT, and these were removed to create non-EGT data sets (5:non-EGT and 3:non-EGT; Table S6). The second approach was to use Concaterpillar to identify protein-coding genes with discordant histories (Leigh et al. 2008), which could be caused by EGT or LGT. Repeated runs yielded different results, indicating an absence of supported discordances. Nevertheless, we analyzed several gene sets identified by Concaterpillar as concordant, including (i) the largest set of concordant genes plus SSU-rDNA (3: cater 7 gene; Table S6), (ii) a 13-gene data set that excluded the 3 genes that were not concordant with any others (5: cater 13 gene; Table S6). To target discordance caused by EGT, we ran Concaterpillar on photosynthetic lineages alone and analyzed the largest concordant gene set (5: cater 9 gene; Table S6).
Phylogenetic Analyses
Genealogies for this study were constructed almost exclusively in RaxML. The MPI version of RaxML 7.0.4 with rapid bootstrapping was used (Stamatakis et al. 2008). The SSU-rDNA partition was analyzed with GTR+gamma as this was the best fitting model available in RAxML, according to MrModelTest (Nylander 2004). ProtTest (Abascal et al. 2005) was used to select the appropriate model of sequence evolution for the amino acid data using the 9:15 data set. The WAG amino acid replacement matrix was found to be the best-fitting model for the concatenated data, but the rtREV amino acid replacement matrix was the best for some of the individual partitions and both WAG and rtREV were among the top 3 models for all but 1 gene (and with similar likelihood scores). We ran our data under both WAG and rtREV models and found consistent results, indicating that our interpretations are robust to at least this level of model choice. The results presented are from the WAG analyses and the rtREV analyses differed only in level of BS for key nodes (usually ± 5 points). In initial analyses, the appropriate number of independent bootstrap replicates was determined for each data set using bootstopping criteria in RAxML 7.0.4 as implemented on Cyberinfrastructure for Phylogenetic Research (CIPRES) portal 2 (Miller et al. 2009). All analyses stopped after 200 or fewer replicates, except all:16, which stopped after 400 replicates. In later analyses, using the MPI version of RAxML, which does not implement a bootstopping criterion, 200 rapid bootstrap replicates followed by a full maximum-likelihood search was used for all analyses except all:16, for which 600 bootstrap replicates were run. Because of the computational cost of the all:16 analysis, this was run as 6 separate analyses: 100 bootstraps followed by a full maximum likelihood search and 5 other runs of 100 bootstraps each. These data were combined in RAxML to complete the analysis. We found no significant difference in comparisons between fast and slow RAxML bootstrap methods (Fig. S1i), which we tested because the fast bootstrapping method in RAxML can produce misleading results particularly for long-branch taxa (Leigh 2008). The results of rapid bootstrapping are shown.
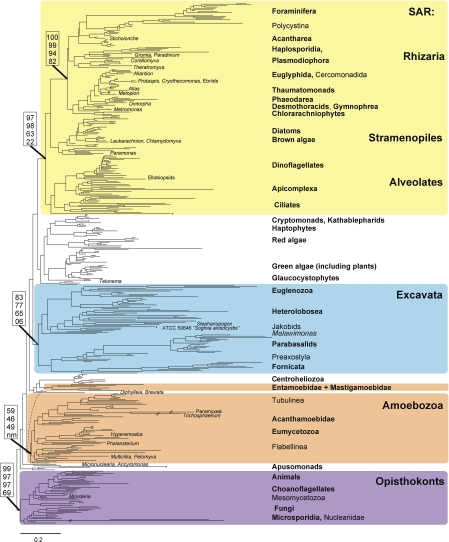
To investigate the stability of our tree topology under different analytic methods, select data sets were analyzed with Bayesian approaches and Parsimony (Fig. S1s–v). Parsimony analysis of 10:16, implemented in Paup* (Shimodaira 2002), yielded a less resolved version of the RAxML topology (i.e. Excavata as a polytomy) that is generally concordant with the more resolved tree obtained by maximum-likelihood methods. The one exception was the misplacement of some rapidly evolving lineages (including Giardia, Microsporidia, Foraminifera, and Entamoeba). PhyloBayes was run on the 9:15 data set using the CAT model with recoded amino acids. The amino acids were recoded using the Dayhoff (6) model, based on the chemical properties of the amino acids. PhyloBayes was stopped after building 2 chains of > 13,000 trees with a maxdiff of 0.26, which indicates weak convergence, but that the chains disagreed on at least one clade 26% of the time. A burn-in of 100 trees was removed and the posterior probabilities were calculated after sampling every other tree. The topology of the consensus tree is consistent with, though less well resolved than the results from RAxML. The parallel version of MrBayes 3.1.4 was used to analyze the 10:16 data matrix using the GTR+I+γ (for nucleotide partition) and WAG (for amino acid partition) models of sequence evolution (Ronquist and Huelsenbeck 2003). Six simultaneous MCMCMC chains were run for 5,600,000 generations, sampling every 1000 generations. An average standard deviation of split frequencies of <0.1 indicated weak convergence. Stationarity was determined by plotting the maximum-likelihood values of the 2 runs, and 10,756 trees were retained. The resulting topology is the same as shown in Figure 2, except that Breviata nests within Amoebozoa sister to Mastigamoeba + Entamoeba. Most nodes are strongly supported: posterior probability equals 1.00 for Amoebozoa, Opisthokonta, Rhizaria, and SAR, and 0.66 for Excavata and “Unikonta.”
FIGURE 2.
Most likely eukaryotic tree of life reconstructed with 10:16, which includes 88 taxa (each with 10 or more of the genes analyzed in this study) and 16 genes (SSU-rDNA plus 15 protein genes). Thickened lines receive >95% bootstrap support. Other notes as in Methods section and Figure 1.
Topology Testing
We performed the approximately unbiased (AU) test (Shimodaira 2002) as well as the more conventional Kishono-Hasegawa and Shimodaira-Hasegawa tests, as implemented in Consel 0.1j (Shimodaira and Hasegawa 2001) to test the monophyly of “Chromalveolata,” “Archaeplastida,” and “Chromista.” The most likely trees with these groups constrained to be monophyletic were built, and the site likelihood values for each constrained topology and the unconstrained topology were estimated using RAxML 7.0.4 (Table S7). In addition, we explored in Paup* v4.08b (Shimodaira 2002) the number of Bayesian trees that were consistent with these hypotheses (Table S7).
RESULTS AND DISCUSSION
Robust Topology of the Eukaryotic Tree of Life
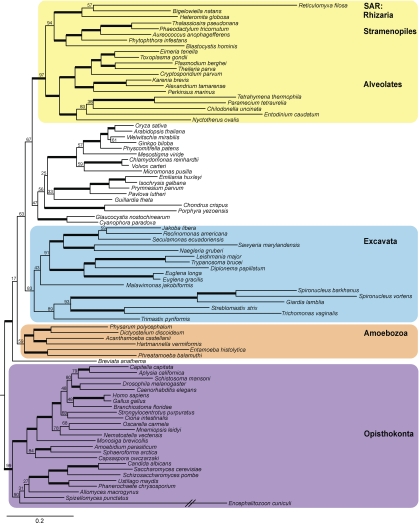
Many major clades were consistently recovered across our analyses (Fig. 1 and Table 1). These stable groups receive moderate to strong support in analyses with limited missing data (Fig. 2) and less support as missing data increases. The Opisthokonta, which includes animals and fungi, and the heterogeneous clade Rhizaria are recovered in all analyses with strong support (Fig. 1 and Table 1). Excavata are recovered in all analyses with moderate support (Fig. 1 and Table 1). Amoebozoa receives low to moderate support in all but our most inclusive analysis (all:16) where most members form a clade with the exception of Mastigamoebidae + Entamoeba that form a separate clade with Breviata, DIphylleia and Centroheliozoa (Fig. 1 and Table 1). Both Rhizaria and Amoebozoa are heterogeneous assemblages of organisms with diverse body plans (Pawlowski and Burki 2009; Tekle et al. 2009) that were created based on molecular analyses (Parfrey et al. 2006). There are no defining morphological features or molecular signatures for Rhizaria, which now encompasses nearly 30 of the 75 lineages with ultrastructural identities (Pawlowski and Burki 2009). Excavata was hypothesized in part on the basis of ultrastructural characters associated with the ventral feeding groove (Simpson 2003), but is generally polyphyletic in phylogenetic (Parfrey et al. 2006; Simpson et al. 2006) and phylogenomic analyses unless rapidly evolving taxa and characters are removed from the analyses (Rodríguez-Ezpeleta et al. 2007a; Hampl et al. 2009). We also find strong support for the clade of stramenopiles, alveolates, plus Rhizaria (SAR; Burki et al. 2007; Hackett et al. 2007; Burki et al. 2008) and a sister relationship between stramenopiles and Rhizaria (Fig. 2 and Table 1). This latter finding is at odds with many phylogenomic analyses (Rodríguez-Ezpeleta et al. 2007a; Burki et al. 2008; Hampl et al. 2009) that find stramenopiles and alveolates are sister to one another.
FIGURE 1.
Most likely eukaryotic tree of life reconstructed using all 451 taxa and all 16 genes (SSU-rDNA plus 15 protein genes). Major nodes in this topology are robust to analyses of subsets of taxa and genes, which include varying levels of missing data (Table 1). Clades in bold are monophyletic in analyses with 2 or more members except in all:15 in which taxa represented by a single gene were sometimes misplaced. Numbers in boxes represent support at key nodes in analyses with increasing amounts of missing data (10:16, 6:16, 4:16, and all:16 analyses; see Table 1 for more details). Given uncertainties around the root of the eukaryotic tree of life (see text), we have chosen to draw the tree rooted with the well-supported clade Opisthokonta. Dashed line indicates alternate branching pattern seen for Amoebozoa in other analyses. Long branches, indicated by //, have been reduced by half. The 6 lineages labeled by * represent taxa that are misplaced, probably due to LBA, listed from top to bottom with expected clade in parentheses. These are Protoopalina japonica (Stramenopiles), Aggregata octopiana (Apicomplexa), Mikrocytos mackini (Haplosporidia), Centropyxis laevigata (Tubulinea), Marteilioides chungmuensis (unplaced), and Cochliopodium spiniferum (Amoebozoa).
In contrast, the relationships among photosynthetic lineages and the position of most orphan lineages (e.g., Breviata and Centroheliozoa) remain unresolved, as discussed below. Furthermore, the root of the eukaryotic tree of life has been hypothesized to be between a clade containing Amoebozoa and Opisthokonta (“Unikonta”) and all remaining eukaryotes (Stechmann and Cavalier-Smith 2003), although there is conflict among evidence (reviewed in Roger and Simpson 2009; Tekle et al. 2009). In our analyses, we find at best moderate support for “Unikonta” (Table 1), but concatenated analyses such as these cannot resolve the root.
In exploring the tradeoffs between increasing taxonomic sampling and decreasing missing data, we analyzed varying combinations of genes and taxa using almost exclusively a maximum-likelihood approach implemented in the software RAxML 7.0.4 (Stamatakis et al. 2005). Node support was highest when we included taxa with 10 or more of our targeted 16 genes (10:16, with 17% missing data and 88 taxa; Fig. 2 and Table 1). As taxa are added, node support decreases (Table 1, BS in Fig. 1) due to the diminishing amount of character data available to estimate a growing number of relationships (i.e., 211 of 451 taxa are represented by SSU-rDNA only). Put another way, stochastic error increases with increasing missing data because the signal-to-noise ratio is decreasing. The mosaic structure of missing data in phylogenomic studies using ESTs is known to decrease phylogenetic accuracy (Hartmann and Vision 2008). However, Wiens and Moen (2008) found that taxa with large amounts of missing data (up to 90%) could be accurately placed so long as there is a shared core of informative data. The ubiquitous SSU-rDNA plus a few well-sampled protein genes likely provide such a core of informativeness in this study.
In addition to allowing assessment of the phylogenetic diversity of eukaryotes, a strength of this taxon-rich analysis is that it enables us to assess clade stability by comparing tree topologies across analyses that vary in numbers of taxa and genes included. Much of the topology remains consistent across all analyses: supported clades (Table 1) and most clades with ultrastructural identities (bold lineages Fig. 1; Table S2) are recovered regardless of the number of genes/level of missing data included. We argue that this is strong evidence that these clades are accurately reconstructed—they reflect true relationships. The ability to accurately place so many lineages that are represented only by SSU-rDNA demonstrates the robustness of these analyses.
We tested the hypothesis that SSU-rDNA was driving our results, as this gene is ubiquitously sampled but is not present in phylogenomic analyses. However, the 15-protein data sets yielded similar topologies that were again robust to varying taxonomic representation (Table 1). We also looked for supported incongruences among loci using Concaterpillar (Leigh et al. 2008) on the 15 protein-coding genes. Repeated runs yielded varying gene sets, suggesting there are no well-supported incongruences. Analyses of several of these gene sets yielded a topology consistent with that depicted in Figures 1 and 2, although support was low in analyses with few genes (Table S6). Here again, the placement of photosynthetic lineages was unstable, suggesting that they may be responsible for discordance among loci.
We also assessed the extent to which choice of these particular 16 genes versus the breadth of our taxon sampling impacted the generation of stable topologies by comparing with previously published studies. Using our 16 genes and a taxon set comparable with Hampl et al. (2009) that included only 48 taxa representing 19 lineages, we generated a highly supported tree similar to what we find using broader taxon sampling (Table S5). Indeed, with our 16 genes and this Hampl-like data set, we recover monophyletic Excavata with 82% BS, whereas this clade is only monophyletic after removal of rapidly evolving lineages in the phylogenomic analysis (Hampl et al. 2009). In contrast, using the broader taxon set of Yoon et al. (2008) (101 taxa representing 26 lineages) generates a topology that is less well supported at many nodes, and Excavata is polyphyletic (Table S5). Finally, using all our taxa and the 4 genes from Yoon et al. generates poorly supported topologies (Table S5). Together, these analyses demonstrate that it is an interaction of gene choice and taxon sampling that yields well-resolved trees.
The ability of our taxon-rich approach to place lineages known to be problematic for phylogenetic reconstruction into correct territories, including Microsporidia, Giardia and ciliates (e.g., Hirt et al. 1999; Zufall et al. 2006; Yoon et al. 2008; Hampl et al. 2009), is a testament to the role of sufficient gene and taxon sampling in accurately reconstructing relationships. Other analyses with fewer taxa and/or genes routinely remove rapidly evolving taxa and/or sites so that these clades “behave” (Hackett et al. 2007; Rodríguez-Ezpeleta et al. 2007b; Burki et al. 2008; Yoon et al. 2008; Hampl et al. 2009). However, removal of taxa weakens the credibility of the process and support for taxonomic hypotheses while also decreasing the power of interpretation of the resulting phylogenetic trees (Hillis 1998).
Orphan Lineages
Our taxon-rich analyses enable inclusion of numerous unplaced lineages that have only limited molecular data. Some of these remain orphans (i.e., without clear sister taxa) including Breviata, Centroheliozoa, Ancyromonas, and Micronuclearia, as their position is unstable and support values are very low (Table S8). These taxa may be either independent lineages or their sister taxa may not yet be sequenced. Consistent with other analyses, we find support for the sister relationships of Apusomonadida with Opisthokonta (Cavalier-Smith and Chao 2003; 85–100%; Table S8), and the nonphotosynthetic kathablepharids with cryptomonads (Okamoto and Inouye 2005; 65–88%; Table S8). Telonema is consistently basal to green algae (including plants), albeit with low support (Table S8), which is in contrast to the hypothesis that this lineage is sister to cryptomonads (Shalchian-Tabrizi et al. 2006). Several unplaced lineages represented only by SSU-rDNA are placed within robust groups, but often on long branches and with low support (Paramyxea, Mikrocytos; Table S8). We believe that their placement is artifactual, either due to long-branch attraction or the lack of a sequenced sister lineage. In support of this hypothesis, these taxa also bounce around in analyses of SSU-rDNA alone with and without rapidly evolving sites (as described in Methods section).
Photosynthetic Lineages
Our analyses do not resolve the placement of many lineages with photosynthetic ancestry including the green algae, red algae (rhodophytes), glaucocystophytes, haptophytes, and cryptomonads. Notably, there is no support in any analysis for “Archaeplastida” (“Plantae”) or “Chromalveolata” (Tables 1 and S6) or the nested hypothesis “Chromista” (stramenopiles, cryptomonads, and haptophytes). These hypothesized clades rest on the assertion that plastid acquisition is a rare event, happening once in the “Archaeplastida” (primary acquisition of a cyanobacterium in the ancestor of red algae, green algae and glaucocystophtes; Cavalier-Smith 1981) and once in “Chromalveolata” (secondary acquisition of a red algal plastid in the ancestor of stramenopiles, alveolates, haptophytes, and cryptomonads; Cavalier-Smith 1999). We hypothesize that the lack of resolution among the photosynthetic lineages (e.g., cryptomonads, haptophytes, glaucocystophytes, rhodophytes, and green algae) is due to conflicting signal following endosymbiotic gene transfer from plastid genomes or from the nuclei of secondary (or tertiary) eukaryotic endosymbionts (Martin and Schnarrenberger 1997; Lane and Archibald 2008; Tekle et al. 2009). We discuss this hypothesis and alternatives below.
Our analyses, like many others (Cavalier-Smith 2004; Parfrey et al. 2006; Rodríguez-Ezpeleta et al. 2007b; Kim and Graham 2008; Yoon et al. 2008; Hampl et al. 2009) find polyphyletic “Chromalveolata” and thus falsify the chromalveolate hypothesis as it was originally proposed. Furthermore, “Chromalveolata” and the nested hypothesis “Chromista” (stramenopiles, cryptomonads, and haptophytes) are rejected by the AU test (P = 0.007 and P < 0.001, respectively) and other statistical methods, and this topology was not found among the 10,756 trees in Bayesian analyses (Table S7). A single endosymbiotic event at the base of the chromalveoate lineages necessitates that the descendant lineages be monophyletic, although not everyone agrees with this interpretation (Keeling 2009). Instead, our analyses are consistent with alternative hypotheses that postulate multiple secondary endosymbioses of red algal plastids in the ancestors of “Chromalveoata” (Grzebyk et al. 2003, Howe et al. 2008, Bodył et al. 2009).
Recent findings indicate that plastid acquisition is not as rare as once assumed, challenging the central tenet that plastid acquisition is much more difficult than loss. Two independent primary endosymbioses that may be first steps toward organelles have been detailed in the testate amoeba Paulinella chromatophora (Nakayama and Ishida 2009) and the diatom Rhopalodia gibba (Kneip et al. 2008). Further, numerous secondary endosymbiotic events are also known in lineages such as euglenids, chlorarachniophytes, and kathablepharids (Archibald 2009), and there is evidence for tertiary endosymbiosis in diatoms (Moustafa et al. 2009) and dinoflagellates (Archibald 2009). Thus, plastid acquisition is more common across the eukaryotic tree of life than previously believed. The possibility that plastid acquisition may have occurred multiple times will make a stable resolution of photosynthetic lineages difficult (Lane and Archibald 2008; Bodył et al. 2009).
As the stramenopiles and alveolates (2 putative members of the “Chromalveolata”) form a well-supported clade including Rhizaria (SAR), we suggest it is time to abandon the chromalveolate hypothesis. Although some argue for expanding the chromalveolate concept to include Rhizaria and other heterotrophic assemblages of eukaryotes as descendants of an ancestor with a red algal symbiont (Keeling 2009), we do not think this revision is warranted due to the large number of losses and replacement of plastids that this would necessitate. Instead, multiple endosymbioses are a much more parsimonious scenario and are consistent with the monophyly of former chromalveolate lineages in analyses of plastid genes (Yoon et al. 2008; Bodył2005; Parfrey et al. 2006). Similarly, the mere handful of genes that are potentially of photosynthetic origin in heterotrophic lineages such as ciliates (16 genes from a total of 27,446 in the complete genome; Reyes-Prieto et al. 2008) or the basal dinoflagellate Oxyrrhis marina (8 genes from 9876 ESTs; Slamovits and Keeling 2008) are more consistent with the “you are what you eat” hypothesis (Doolittle 1998) than the chromalveolate hypothesis.
A single primary plastid acquisition at the base of “Archaeplastida” is the prevailing view (Gould et al. 2008; Archibald 2009; Keeling 2009). The Archaeplastida hypothesis is supported by many shared features of plastids and their integration into the host cell, including plastid protein import machinery, conserved gene order, and metabolic pathways (Mcfadden 2001; Larkum et al. 2007; Gould et al. 2008). Although analyses of few genes do not generally support “Archaeplastida” (Parfrey et al. 2006; Kim and Graham 2008), support is strong in some phylogenomic analyses (Rodríguez-Ezpeleta et al. 2005; Rodríguez-Ezpeleta et al. 2007a, 2007b; Burki et al. 2008, though see Hampl et al. 2009). It has been suggested that 100+ genes are necessary to recover “Archaeplastida” with strong support (Rodríguez-Ezpeleta et al. 2005).
The Archaeplastida hypothesis is not supported in our analyses (Tables 1 and S6 and Figs. 1 and 2) or those of others (Parfrey et al. 2006; Kim and Graham 2008; Yoon et al. 2008; Hampl et al. 2009). Here, the “Archaeplastida” lineages red algae, green algae, and glaucocystophytes are never monophyletic, but instead generally form a poorly supported cluster with the secondarily photosynthetic haptophytes and cryptomonads plus other nonphotosynthetic lineages (Table 1 and Figs. 1 and 2). This lack of resolution is not simply a by-product of our overall approach as the same analyses yield relatively well-supported nodes for much of the rest of the tree (Table 1 and Figs. 1 and 2), and recover groups with ultrastructural identities with strong support, including photosynthetic lineages (e.g., green algae including land plants; Fig. 2). The confounding effects of EGT (from plastid or nucleus of secondary endosymbiont) may explain the lack of resolution and failure to recover “Archaeplastida”. Being aware of these issues, we attempted to identify conflicting signal and remove genes impacted by EGT both by inspection of individual genes using BLAST analyses and by assessing concordant data sets identified by Concaterpillar (Table S6 and Fig. S1m–r). These approaches failed to yield robust placement of the problematic photosynthetic lineages (Table S6). For example, we hypothesized that the secondarily photosynthetic haptophytes and cryptomonads were branching within “Archaeplastida” due to EGT; however, “Archaeplastida” remains polyphyletic in analyses without haptophytes and cryptomonads (Table S6). In contrast to the “Archaeplastida”, other lineages with photosynthetic ancestry are robustly placed in clades containing both photosynthetic and heterotrophic lineages (e.g., dinoflagellates within alveolates, diatoms within stramenopiles, and euglenids as sister to kinetoplastids). This may reflect differential timing of endosymbiotic events as ancient events will be more difficult to reconstruct than recent secondary transfers because (i) more genes in the plastid were available for transfer early and (ii) more time for subsequent confounding events will have elapsed.
Alternatively, nonmonophyly of “Archaeplastida” may be reflective of the true host histories if there were multiple endosymbiotic events in the ancestors of red algae, green algae, and glaucocystophytes. Many scenarios are consistent with both the nonmonophyly of “Archaeplastida” and the similarities of the plastids of these lineages (Palmer 2003; Stiller 2003; Larkum et al. 2007). Two of these are (i) multiple primary endosymbioses of closely related cyanobacteria followed by a convergent path of plastid reduction plus extinction of intervening cyano bacterial lineages and (ii) a single primary endosymbiosis into one lineage followed by ancient secondary endosymbioses into the remaining “Archaeplastida” lineages. Such scenarios, as well as a single primary acquisition, are also consistent with the well-supported monophyly of plastid genes with respect to cyanobacteria (Rodríguez-Ezpeleta et al. 2005; Parfrey et al. 2006) plus possibly the confounding data on the divergent Rubisco genes in red and green algae (Delwiche and Palmer 1996). Furthermore, the phylogenetic position of “Archaeplastida” lineages may be difficult to resolve because their sister groups have not yet been sequenced, or are extinct. The unstable position of these lineages across our analyses mimics the patterns observed in orphan lineages (Table S8) in support of this hypothesis. Under these scenarios, phylogenomic analyses that recover “Archaeplastida” may be picking up misleading EGT signal of genes independently transferred from the plastid to the host nucleus of these three lineages.
We suspect that resolving relationships among photosynthetic groups will require more intensive taxon and more careful gene sampling to disentangle signals from host and symbiont genomes, coupled with the recognition that plastid genes may be derived from several sources (Larkum et al. 2007). These data, combined with methods that distinguish between conflicting phylogenetic signal (Ahmadinejad et al. 2007; Leigh et al. 2008) or gene-tree species-tree reconciliation (Wehe et al. 2008; Akerborg et al. 2009), are likely required to elucidate the history of photosynthetic lineages.
Relationships Within the Well-Sampled Rhizaria and Excavata
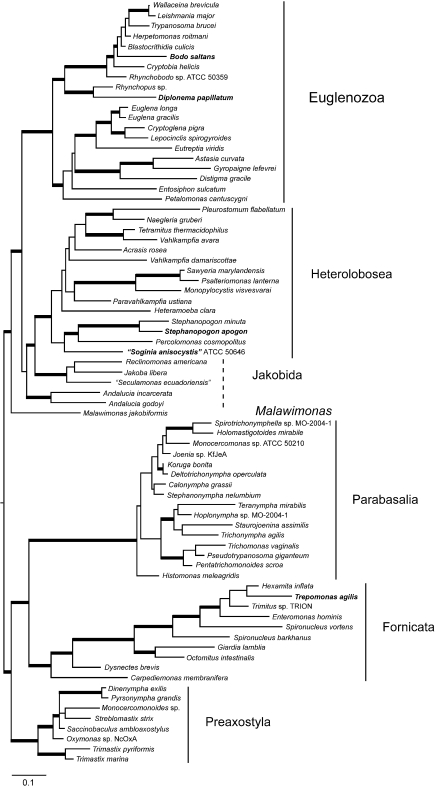
We subsampled the data set to estimate relationships within 2 diverse clades, Excavata and Rhizaria, for which we had large numbers of taxa. We analyzed a 97-taxon data set of Rhizaria that included all lineages with previously published data plus additional multigene data for 12 taxa added for this study (Table S1). Three major clades are strongly supported, though the relationships among them are unresolved: i) Cercozoa, ii) Foraminifera plus Polycystinea and Acantharea (formerly classified with Phaeodarea as radiolarians), and (iii) the parasitic Haplosporidia and Plasmodiophorida with Gromia and vampyrellids (Fig. 3; Bass et al. 2009). We show that Theratromyxa, a nematode-eating soil amoeba, is related to vampyrellid amoebae (Fig. 3; 100% BS), and together they are sister to the plant parasites plasmodiophorids (100% BS). The SSU-rDNA sequence for Theratromyxa is identical to an amoeba isolated from Siberia where it was identified as Arachnula impatiens (EU567294; Bass et al. 2009).
FIGURE 3.
Maximum likelihood tree of Rhizaria reconstructed with 103 Rhizaria taxa and 16 genes. The SSU-rDNA partition was analyzed with GTR+gamma and proteins with rtREV. Thickened lines receive >80% bootstrap support in all analyses. Node support in boxes from Rhizaria:4-gene, Rhizaria:16-gene, all:16 analyses. Taxa with new data are in bold. Dashed lines indicate nonmonophyly.
The topology within the Excavata is consistent with previous hypotheses and clades with ultrastructural identities (Simpson 2003; Fig. 4), when contaminant EST data originally mislabeled as Streblomastix strix are excluded (Slamovits and Keeling 2006). Excavata is often polyphyletic in other analyses because Malawimonas branches outside the other clades of Excavata (Rodríguez-Ezpeleta et al. 2007a; Hampl et al. 2009), whereas in analyses of fewer genes Excavata members fall into 2 or 3 clades (Parfrey et al. 2006; Simpson et al. 2006). Although Malawimonas nests robustly within Excavata in our analyses, it does not have a stable sister group and may represent an independent lineage (Fig. 4). Our analyses confirm that Stephanopogon (unplaced in Patterson 1999) branches within Heterolobosea (Cavalier-Smith and Nikolaev 2008; Yubuki and Leander 2008) and suggests that another enigmatic flagellate, ATCC 50646 (tentatively named Soginia anisocystis) is a basal member of Heterolobosea.
FIGURE 4.
Maximum-likelihood tree of Excavata with 75 taxa and 16 genes. The SSU-rDNA partition was analyzed with GTR+gamma and proteins with rtREV. See Figure 3 for other notes.
CONCLUSIONS
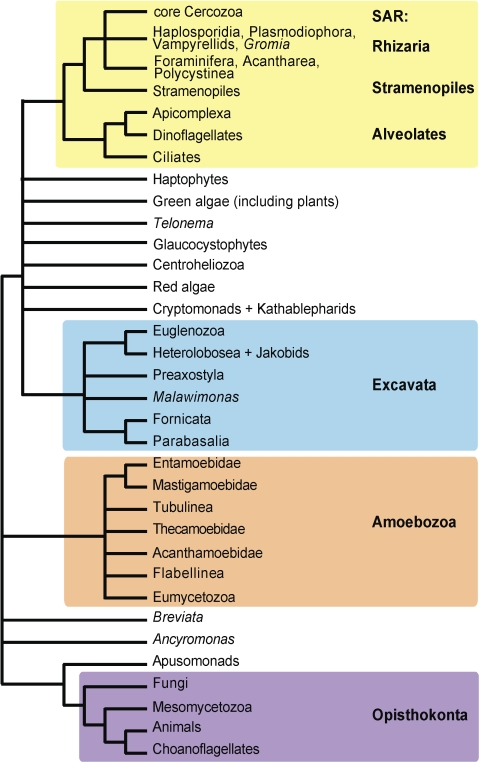
The robust tree of life emerging from this study demonstrates the benefits of improved taxon sampling for reconstructing deep phylogeny as our analyses produce stable topologies that include a broad representation of eukaryotes. The current study, combined with insights from other studies referenced herein, has refined the eukaryotic tree of life from over 70 major lineages (Patterson 1999) to ∼ 16 major groups (Fig. 5, http://eutree.lifedesks.org/). Most significantly, we attribute the stability of major clades (e.g., Excavata, Amoebozoa, Opisthokonta, and SAR) to broader taxonomic sampling combined with analyses of sufficient characters (16 genes or 6578 characters). In our view, inclusion of more taxa coupled with carefully chosen genes is necessary to further resolve the 16 or so major lineages of microbial eukaryotes for which sister group relationships remain uncertain.
FIGURE 5.
Summary of major findings—the evolutionary relationships among major lineages of eukaryotes. Clades have been collapsed into those that we view to be strongly supported. The many polytomies represent uncertainties that remain.
SUPPLEMENTARY MATERIAL
Supplementary material can be found at http://www.sysbio.oxfordjournals.org/.
FUNDING
This work was made possible by the US National Science Foundation Assembling the Tree of Life grant to L.A.K. and D.J.P. (043115) and US National Institutes of Health 5R01AI058054-05 to M.L.S. Funding to collect Foraminifera was provided by a Society of Systematic Biologists MiniPEET grant to L.W.P.
Supplementary Material
Acknowledgments
We are grateful to Robert Molestina at ATCC who provided DNAs through a collaborative National Science Foundation grant. We acknowledge the assistance of Kasia Hammar, Leslie Murphy, and Jillian Ward in preparation and sequencing of EST libraries. Our manuscript was improved following detailed comments from the editors, Alastair Simpson, and 1 anonymous reviewer. Thanks to David Hillis for conversations on early versions of the manuscript. Many thanks also to Daniel J. G. Lahr for comments and discussions, and to Wayne Pfeiffer and Mark Miller at CIPRES plus Tony Caldanaro at Smith College for technical help in running the analyses.
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Adl SM, Simpson AGB, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, James TY, Karpov S, Kugrens P, Krug J, Lane CE, Lewis LA, Lodge J, Lynn DH, Mann DG, McCourt RM, Mendoza L, Moestrup O, Mozley-Standridge SE, Nerad TA, Shearer CA, Smirnov AV, Spiegel FW, Taylor M. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Euk. Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- Ahmadinejad N, Dagan T, Martin W. Genome history in the symbiotic hybrid Euglena gracilis. Gene. 2007;402:35–39. doi: 10.1016/j.gene.2007.07.023. [DOI] [PubMed] [Google Scholar]
- Akerborg O, Sennblad B, Arvestad L, Lagergren J. Simultaneous Bayesian gene tree reconstruction and reconciliation analysis. Proc. Natl. Acad. Sci. USA. 2009;106:5714–5719. doi: 10.1073/pnas.0806251106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson JO. Lateral gene transfer in eukaryotes. Cell. Mol. Life Sci. 2005;62:1182–1197. doi: 10.1007/s00018-005-4539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald JM. The puzzle of plastid evolution. Curr. Biol. 2009;19:R81–R88. doi: 10.1016/j.cub.2008.11.067. [DOI] [PubMed] [Google Scholar]
- Bass D, Chao EEY, Nikolaev S, Yabuki A, Ishida KI, Berney C, Pakzad U, Wylezich C, Cavalier-Smith T. Phylogeny of novel naked filose and reticulose Cercozoa: Granofilosea cl. n. and Proteomyxidea revised. Protist. 2009;160:75–109. doi: 10.1016/j.protis.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Bodył A. Do plastid-related characters support the chromalveolate hypothesis? J. Phycol. 2005;41:712–719. [Google Scholar]
- Bodył A, Stiller JW, Mackiewicz P. Chromalveolate plastids: direct descent or multiple endosymbioses? Trends Ecol. Evol. 2009;24:119–121. doi: 10.1016/j.tree.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Brinkmann H, Van der Giezen M, Zhou Y, De Raucourt GP, Philippe H. An empirical assessment of long-branch attraction artefacts in deep eukaryotic phylogenomics. Syst. Biol. 2005;54:743–757. doi: 10.1080/10635150500234609. [DOI] [PubMed] [Google Scholar]
- Burki F, Inagaki Y, Brate J, Archibald JM, Keeling PJ, Cavalier-Smith T, Sakaguchi M, Hashimoto T, Horak A, Kumar S, Klaveness D, Jakobsen KS, Pawlowski J, Shalchian-Tabrizi K. Large-scale phylogenomic analyses reveal that two enigmatic protist lineages, Telonemia and Centroheliozoa, are related to photosynthetic chromalveolates. Genome Biol. Evol. 2009;1:231–238. doi: 10.1093/gbe/evp022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Shalchian-Tabrizi K, Minge M, Skjæveland Å, Nikolaev SI, Jakobsen KS, Pawlowski J. Phylogenomics reshuffles the eukaryotic supergroups. PLoS ONE. 2007;2:e790. doi: 10.1371/journal.pone.0000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Shalchian-Tabrizi K, Pawlowski J. Phylogenomics reveals a new 'megagroup' including most photosynthetic eukaryotes. Biol. Letters. 2008;4:366–369. doi: 10.1098/rsbl.2008.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Eukaryote kingdoms–seven or nine. Biosystems. 1981;14:461–481. doi: 10.1016/0303-2647(81)90050-2. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Principles of protein and lipid targeting in secondary symbiogenesis: Euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J. Euk. Microbiol. 1999;46:347–366. doi: 10.1111/j.1550-7408.1999.tb04614.x. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Only six kingdoms of life. Proc. R. Soc. B Biol. Sci. 2004;271:1251–1262. doi: 10.1098/rspb.2004.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EEY. Phylogeny of Choanozoa, Apusozoa, and other Protozoa and early eukaryote megaevolution. J. Mol. Evol. 2003;56:540–563. doi: 10.1007/s00239-002-2424-z. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Nikolaev S. The zooflagellates Stephanopogon and Percolomonas are a clade (Class Percolatea: Phylum Percolozoa) J. Euk. Microbiol. 2008;55:501–509. doi: 10.1111/j.1550-7408.2008.00356.x. [DOI] [PubMed] [Google Scholar]
- Cummings MP, Meyer A. Magic bullets and golden rules: data sampling in molecular phylogenetics. Zoology. 2005;108:329–336. doi: 10.1016/j.zool.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Delwiche CF, Palmer JD. Rampant horizontal transfer and duplication of rubisco genes in eubacteria and plastids. Mol. Biol. Evol. 1996;13:873–882. doi: 10.1093/oxfordjournals.molbev.a025647. [DOI] [PubMed] [Google Scholar]
- Doolittle WF. You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 1998;14:307–311. doi: 10.1016/s0168-9525(98)01494-2. [DOI] [PubMed] [Google Scholar]
- Eddy SR. HMMER: Profile hidden markov models for biological sequence analysis. 2001. Available from: http://hmmer.janelia.org/ [Google Scholar]
- Felsenstein J. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 1978;27:401–410. [Google Scholar]
- Galtier N, Gouy M, Gautier C. Seaview and Phylo_win, two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Gould SB, Waller RR, McFadden GI. Plastid evolution. Annu. Rev. Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- Gribaldo S, Philippe H. Ancient phylogenetic relationships. Theor. Popul. Biol. 2002;61:391–408. doi: 10.1006/tpbi.2002.1593. [DOI] [PubMed] [Google Scholar]
- Grzebyk D, Schofield O, Vetriani C, Falkowski PG. The mesozoic radiation of eukaryotic algae: The portable plastid hypothesis. J. Phycol. 2003;39:259–267. [Google Scholar]
- Hackett JD, Yoon HS, Li S, Reyes-Prieto A, Rummele SE, Bhattacharya D. Phylogenomic analysis supports the monophyly of cryptophytes and haptophytes and the association of 'Rhizaria' with chromalveolates. Mol. Biol. Evol. 2007;8:1702–1713. doi: 10.1093/molbev/msm089. [DOI] [PubMed] [Google Scholar]
- Hampl V, Hug L, Leigh JW, Dacks JB, Lang BF, Simpson AGB, Roger AJ. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc. Natl. Acad. Sci. USA. 2009;106:3859–3864. doi: 10.1073/pnas.0807880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann S, Vision TJ. Using ESTs for phylogenomics: Can one accurately infer a phylogenetic tree from a gappy alignment? BMC Evol. Biol. 2008;8:95. doi: 10.1186/1471-2148-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath TA, Hedtke SM, Hillis DM. Taxon sampling and the accuracy of phylogenetic analyses. J. Syst. Evol. 2008;46:239–257. [Google Scholar]
- Hedtke SM, Townsend TM, Hillis DM. Resolution of phylogenetic conflict in large data sets by increased taxon sampling. Syst. Biol. 2006;55:522–529. doi: 10.1080/10635150600697358. [DOI] [PubMed] [Google Scholar]
- Hillis DM. Taxonomic sampling, phylogenetic accuracy, and investigator bias. Syst. Biol. 1998;47:3–8. doi: 10.1080/106351598260987. [DOI] [PubMed] [Google Scholar]
- Hillis DM, Pollock DD, McGuire JA, Zwickl DJ. Is sparse taxon sampling a problem for phylogenetic inference? Syst. Biol. 2003;52:124–126. doi: 10.1080/10635150390132911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt RP, Logsdon JM, Healy B, Dorey MW, Doolittle WF, Embley TM. Microsporidia are related to Fungi: Evidence from the largest subunit of RNA polymerase II and other proteins. Proc. Natl. Acad. Sci. USA. 1999;96:580–585. doi: 10.1073/pnas.96.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe CJ, Barbrook AC, Nisbet RER, Lockhart PJ, Larkum AWD. The origin of plastids. Philos. T. Roy. Soc. B. 2008;363:2675–2685. doi: 10.1098/rstb.2008.0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffroy O, Brinkmann H, Delsuc F, Philippe H. Phylogenomics: the beginning of incongruence? Trends Genet. 2006;22:225–231. doi: 10.1016/j.tig.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Keeling PJ. Chromalveolates and the evolution of plastids by secondary endosymbiosis. J. Euk. Microbiol. 2009;56:1–8. doi: 10.1111/j.1550-7408.2008.00371.x. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, Pearlman RE, Roger AJ, Gray MW. The tree of eukaryotes. Trends Ecol. Evol. 2005;20:670–676. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Kim E, Graham LE. EEF2 analysis challenges the monophyly of Archaeplastida and Chromalveolata. PLoS ONE. 2008;3:e2621. doi: 10.1371/journal.pone.0002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneip C, Voss C, Lockhart PJ, Maier UG. The cyanobacterial endosymbiont of the unicellular algae Rhopalodia gibba shows reductive genome evolution. BMC Evol. Biol. 2008;8:30. doi: 10.1186/1471-2148-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll AH, Javaux EJ, Hewitt D, Cohen P. Eukaryotic organisms in proterozoic oceans. Philos. Trans. R. Soc. B Biol. Sci. 2006;361:1023–1038. doi: 10.1098/rstb.2006.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Lane CE, Archibald JM. The eukaryotic tree of life: endosymbiosis takes its TOL. Trends Ecol. Evol. 2008;23:268–275. doi: 10.1016/j.tree.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Larkum AWD, Lockhart PJ, Howe CJ. Shopping for plastids. Trends Plant Sci. 2007;12:189–195. doi: 10.1016/j.tplants.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Leigh JW. Congruence in phylogenomic data: exploring artifacts in deep eukaryotic phylogeny [Ph.D. Thesis] [Halifax (Nova Scotia)]: Dalhousie University; 2008. [Google Scholar]
- Leigh JW, Susko E, Baumgartner M, Roger AJ. Testing congruence in phylogenomic analysis. Syst. Biol. 2008;57:104–115. doi: 10.1080/10635150801910436. [DOI] [PubMed] [Google Scholar]
- Maddison DR, Maddison WP. MacClade version 4.08: an analysis of phylogeny and character evolution. Sunderland (MA): Sinauer Associates; 2005. [Google Scholar]
- Maddison WP. Gene trees in species trees. Syst. Biol. 1997;46:523–536. [Google Scholar]
- Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M, Penny D. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. USA. 2002;99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Schnarrenberger C. The evolution of the Calvin cycle from prokaryotic to eukaryotic chromosomes: a case study of functional redundancy in ancient pathways through endosymbiosis. Curr. Genet. 1997;32:1–18. doi: 10.1007/s002940050241. [DOI] [PubMed] [Google Scholar]
- McFadden GI. Primary and secondary endosymbiosis and the origin of plastids. J. Phycol. 2001;37:951–959. [Google Scholar]
- Medlin L, Elwood HJ, Stickel S, Sogin ML. The characterization of enzymatically amplified eukaryotes 16s-like ribosomal RNA coding regions. Gene. 1988;71:491–500. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- Miller MA, Holder MT, Vos R, Midford PE, Liebowitz T, Chan L, Hoover P, Warnow T. The CIPRES portals. 2009 Available from: http://www.phylo.org/sub_sections/portal. [Google Scholar]
- Moustafa A, Beszteri B, Maier UG, Bowler C, Valentin K, Bhattacharya D. Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science. 2009;324:1724–1726. doi: 10.1126/science.1172983. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Ishida K. Another acquisition of a primary photosynthetic organelle is underway in Paulinella chromatophora. Curr. Biol. 2009;19:R284–R285. doi: 10.1016/j.cub.2009.02.043. [DOI] [PubMed] [Google Scholar]
- Nylander JA. MrModelTest. 2004 Uppsala. Distributed by the author. Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- O'Kelly CJ, Nerad TA. Malawimonas jakobiformis n. Gen., n. Sp (Malawimonadidae n. Fam.): a Jakoba-like heterotrophic nanoflagellate with discoidal mitochondrial cristae. J. Euk. Microbiol. 1999;46:522–531. [Google Scholar]
- Okamoto N, Inouye I. The katablepharids are a distant sister group of the Cryptophyta: A proposal for Katablepharidophyta divisio nova/Kathablepharida phylum novum based on ssu-rDNA and beta-tubulin phylogeny. Protist. 2005;156:163–179. doi: 10.1016/j.protis.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Palmer JD. The symbiotic birth and spread of plastids: how many times and whodunit? J. Phycol. 2003;39:4–11. [Google Scholar]
- Parfrey LW, Barbero E, Lasser E, Dunthorn M, Bhattacharya D, Patterson DJ, Katz LA. Evaluating support for the current classification of eukaryotic diversity. PLoS Genet. 2006;2:2062–2073. doi: 10.1371/journal.pgen.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson DJ. The diversity of eukaryotes. Am. Nat. 1999;154:S96–S124. doi: 10.1086/303287. [DOI] [PubMed] [Google Scholar]
- Pawlowski J, Burki F. Untangling the phylogeny of amoeboid protists. J. Euk. Microbiol. 2009;56:16–25. doi: 10.1111/j.1550-7408.2008.00379.x. [DOI] [PubMed] [Google Scholar]
- Pertea G, Huang XQ, Liang F, Antonescu V, Sultana R, Karamycheva S, Lee Y, White J, Cheung F, Parvizi B, Tsai J, Quackenbush J. TIGR gene indices clustering tools (tgicl): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19:651–652. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- Philippe H, Snell EA, Bapteste E, Lopez P, Holland PWH, Casane D. Phylogenomics of eukaryotes: Impact of missing data on large alignments. Mol. Biol. Evol. 2004;21:1740–1752. doi: 10.1093/molbev/msh182. [DOI] [PubMed] [Google Scholar]
- Phillips MJ, Delsuc F, Penny D. Genome-scale phylogeny and the detection of systematic biases. Mol. Biol. Evol. 2004;21:1455–1458. doi: 10.1093/molbev/msh137. [DOI] [PubMed] [Google Scholar]
- Rannala B, Yang ZH. Phylogenetic inference using whole genomes. Annu. Rev. Genomics Hum. Genet. 2008;9:217–231. doi: 10.1146/annurev.genom.9.081307.164407. [DOI] [PubMed] [Google Scholar]
- Reyes-Prieto A, Moustafa A, Bhattacharya D. Multiple genes of apparent algal origin suggest ciliates may once have been photosynthetic. Curr. Biol. 2008;18:956–962. doi: 10.1016/j.cub.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, Brinkmann H, Burey SC, Roure B, Burger G, Loffelhardt W, Bohnert HJ, Philippe H, Lang BF. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr. Biol. 2005;15:1325–1330. doi: 10.1016/j.cub.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, Brinkmann H, Burger G, Roger AJ, Gray MW, Philippe H, Lang BF. Toward resolving the eukaryotic tree: The phylogenetic positions of jakobids and cercozoans. Curr. Biol. 2007a;17:1420–1425. doi: 10.1016/j.cub.2007.07.036. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, Brinkmann B, Roure N, Lartillot BF, LangPhilippe H. Detecting and overcoming systematic errors in genome-scale phylogenies. Syst. Biol. 2007b;56:389–399. doi: 10.1080/10635150701397643. [DOI] [PubMed] [Google Scholar]
- Roger AJ, Hug LA. The origin and diversification of eukaryotes: Problems with molecular phylogenetics and molecular clock estimation. Phil. Trans. R. Soc. B Biol. Sci. 2006;361:1039–1054. doi: 10.1098/rstb.2006.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger AJ, Simpson AGB. Evolution: revisiting the root of the eukaryote tree. Curr. Biol. 2009;19:R165–R167. doi: 10.1016/j.cub.2008.12.032. [DOI] [PubMed] [Google Scholar]
- Rokas A, Carroll SB. More genes or more taxa? The relative contribution of gene number and taxon number to phylogenetic accuracy. Mol. Biol. Evol. 2005;22:1337–1344. doi: 10.1093/molbev/msi121. [DOI] [PubMed] [Google Scholar]
- Rokas A, Chatzimanolis S. From gene-scale to genome-scale phylogenetics: the data flood in, but the challenges remain. In: Murphy WJ, editor. Methods in Molecular Biology: Phylogenomics. Totowa (NJ): Humana Press Inc; 2008. pp. 1–12. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Shalchian-Tabrizi K, Eikrem W, Klaveness D, Vaulot D, Minge MA, Le Gall F, Romari K, Throndsen J, Botnen A, Massana R, Thomsen HA, Jakobsen KS. Telonemia, a new protist phylum with affinity to chromist lineages. Proc. R. Soc. B Biol. Sci. 2006;273:1833–1842. doi: 10.1098/rspb.2006.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. Consel: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Simpson AGB. Cytoskeletal organization, phylogenetic affinities and systematics in the contentious taxon excavata (eukaryota) Int. J. Syst. Evol. Microbiol. 2003;53:1759–1777. doi: 10.1099/ijs.0.02578-0. [DOI] [PubMed] [Google Scholar]
- Simpson AGB, Inagaki Y, Roger AJ. Comprehensive multigene phylogenies of excavate protists reveal the evolutionary positions of “primitive” eukaryotes. Mol. Biol. Evol. 2006;23:615–625. doi: 10.1093/molbev/msj068. [DOI] [PubMed] [Google Scholar]
- Simpson AGB, Roger AJ. The real 'kingdoms' of eukaryotes. Curr. Biol. 2004;14:R693–R696. doi: 10.1016/j.cub.2004.08.038. [DOI] [PubMed] [Google Scholar]
- Slamovits CH, Keeling PJ. A high density of ancient spliceosomal introns in oxymonad excavates. BMC Evol. Biol. 2006;6:8. doi: 10.1186/1471-2148-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamovits CH, Keeling PJ. Plastid-derived genes in the nonphotosynthetic alveolate Oxyrrhis marina. Mol. Biol. Evol. 2008;25:1297–1306. doi: 10.1093/molbev/msn075. [DOI] [PubMed] [Google Scholar]
- Snoeyenbos-West OLO, Salcedo T, McManus GB, Katz LA. Insights into the diversity of choreotrich and oligotrich ciliates (class: Spirotrichea) based on genealogical analyses of multiple loci. Int. J. Syst. Evol. Microbiol. 2002;52:1901–1913. doi: 10.1099/00207713-52-5-1901. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web-servers. Syst. Biol. 2008;57(5):758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T. The root of the eukaryote tree pinpointed. Curr. Biol. 2003;13:R665–R666. doi: 10.1016/s0960-9822(03)00602-x. [DOI] [PubMed] [Google Scholar]
- Stiller JW. Weighing the evidence for a single origin of plastids. J. Phycol. 2003;39:1283–1285. [Google Scholar]
- Swofford D. Paup*. Phylogenetic analysis using parsimony (*and other methods). version 4.0b8. Sunderland (MA): Sinauer Associates; 2002. [Google Scholar]
- Swofford DL, Olsen GJ, Waddell PJ, Hillis DM. Phylogenetic inference. In: Hillis DM, Moritz C, Mable BK, editors. Molecular systematics. Sunderland (MA): Sinauer Associates; 1996. pp. 407–514. [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Tekle YI, Parfrey LW, Katz LA. Molecular data are transforming hypotheses on the origin and diversification of eukaryotes. Bioscience. 2009;59:471–481. doi: 10.1525/bio.2009.59.6.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G, Dacks JB, Embley TM. Ultrastructural description of Breviata anathema, n. Gen., n. Sp., the organism previously studied as “Mastigamoeba invertens”. J. Euk. Microbiol. 2006;53:65–78. doi: 10.1111/j.1550-7408.2005.00087.x. [DOI] [PubMed] [Google Scholar]
- Wehe A, Bansal MS, Burleigh JG, Eulenstein O. DupTree: a program for large-scale phylogenetic analyses using gene tree parsimony. Bioinformatics. 2008;24:1540–1541. doi: 10.1093/bioinformatics/btn230. [DOI] [PubMed] [Google Scholar]
- Wiens JJ. Can incomplete taxa rescue phylogenetic analyses from long-branch attraction? Syst. Biol. 2005;54:731–742. doi: 10.1080/10635150500234583. [DOI] [PubMed] [Google Scholar]
- Wiens JJ, Moen DS. Missing data and the accuracy of Bayesian phylogenetics. J Syst. Evol. 2008;46:307–314. [Google Scholar]
- Wuyts J, Van de Peer Y, Winkelmans T, De Wachter R. The European database on small subunit ribosomal RNA. Nucleic Acids Res. 2002;30:183–185. doi: 10.1093/nar/30.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Grant J, Tekle YI, Wu M, Chaon BC, Cole JC, Logsdon JM, Patterson DJ, Bhattacharya D, Katz LA. Broadly sampled multigene trees of eukaryotes. BMC Evol. Biol. 2008;8:14. doi: 10.1186/1471-2148-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Pinto G, Bhattacharya D. The single, ancient origin of chromist plastids. Proc. Natl. Acad. Sci. USA. 2002;99:15507–15512. doi: 10.1073/pnas.242379899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yubuki N, Leander BS. Ultrastructure and molecular phylogeny of Stephanopogon minuta: An enigmatic microeukaryote from marine interstitial environments. Eur. J. Protistol. 2008;44:241–253. doi: 10.1016/j.ejop.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Zufall RA, McGrath CL, Muse SV, Katz LA. Genome architecture drives protein evolution in ciliates. Mol. Biol. Evol. 2006;23:1681–1687. doi: 10.1093/molbev/msl032. [DOI] [PubMed] [Google Scholar]
- Zwickl DJ, Hillis DM. Increased taxon sampling greatly reduces phylogenetic error. Syst. Biol. 2002;51:588–598. doi: 10.1080/10635150290102339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.