Abstract
Long QT Syndrome (LQTS) is a cardiac channelopathy associated with syncope, seizures, and sudden death. Approximately 75% of LQTS is due to mutations in genes encoding for three cardiac ion channel alpha-subunits (LQT1-3). However, traditional mutational analyses have limited detection capabilities for atypical mutations such as large gene rearrangements. Here, we set out to determine the prevalence and spectrum of large deletions/duplications in the major LQTS-susceptibility genes among unrelated patients who were mutation-negative following point mutation analysis of LQT1-12-susceptibility genes. Forty-two unrelated clinically strong LQTS patients were analyzed using multiplex ligation-dependent probe amplification (MLPA), a quantitative fluorescent technique for detecting multiple exon deletions and duplications. The SALSA-MLPA LQTS Kit from MRC-Holland was used to analyze the three major LQTS-associated genes: KCNQ1, KCNH2, and SCN5A and the two minor genes: KCNE1 and KCNE2. Overall, 2 gene rearrangements were found in 2/42 (4.8%, CI, 1.7–11%) unrelated patients. A deletion of KCNQ1 exon 3 was identified in a 10 year-old Caucasian boy with a QTc of 660 milliseconds (ms), a personal history of exercise-induced syncope, and a family history of syncope. A deletion of KCNQ1 exon 7 was identified in a 17 year-old Caucasian girl with a QTc of 480 ms, a personal history of exercise-induced syncope, and a family history of sudden cardiac death. In conclusion, since nearly 5% of patients with genetically elusive LQTS had large genomic rearrangements involving the canonical LQTS-susceptibility genes, reflex genetic testing to investigate genomic rearrangements may be of clinical value.
Keywords: Long QT syndrome, Genetic Testing, Sudden Cardiac Death, Gene Rearrangements
INTRODUCTION
Recently, two studies suggest that genomic rearrangements (large deletion/duplication mutations) involving KCNQ1 and KCNH2 may explain 5–12% of long QT syndrome (LQTS) among patients who were previously considered genotype negative following traditional open reading frame mutational analysis of the 3 canonical LQTS-causing genes: KCNQ1 (LQT1), KCNH2 (LQT2), and SCN5A (LQT3)1,2. Additionally, Caselli and colleagues identified a 5.27 Mb deletion of chromosome 7 (7q36.1-q36.2) encompassing a whole gene deletion of KCNH2 in a 9-year-old with significant QT prolongation (QTc of 490 msec), renal hypoplasia, and mental retardation3. Given these reports, we sought to determine the spectrum and prevalence of genomic rearrangements among our cohort of LQTS patients who remained genetically elusive following conventional open reading frame mutation analysis of all 12 currently known LQTS-susceptibility genes (i.e. genotype negative/phenotype positive LQTS).
MATERIALS AND METHODS
Our cohort consisted of 42 unrelated patients with robust clinical evidence for LQTS (QTc ≥ 480 ms and/or Schwartz-Moss score ≥ 4, Table 1)4. All of the patients were mutation negative following LQTS mutation analysis (DHPLC and sequencing) of the three canonical LQTS genes: KCNQ1, KCNH2, and SCN5A and the nine minor LQTS genes: ANKB, KCNE1, KCNE2, KCNJ2, CACN1AC, CAV3, SCN4B, AKAP9, and SNTA15–15. The study was approved by the Mayo Foundation Institutional Review Board.
Table 1.
Patient Demographics (n=42)
| Male/female | 14/28 |
| Average Age (Years) | 22.5 ± 15 |
| Average QTc (ms) | 530 ± 66 |
| QTc > 480ms | 38 (90%) |
| Schwarz-Moss Score > 4 | 20 (48%) |
| Syncope or Cardiac Arrest | 21 (50%) |
| Family History of SCD | 10 (24%) |
Genomic DNA was subjected to multiplex ligation-dependent probe amplification (MLPA) analysis, a quantitative technique for detecting large deletions and duplications otherwise missed by traditional mutation detection methods, using the SALSA P114 LQT kit (MRC-Holland) according to manufacturer’s instructions. Data was collected and analyzed with GeneMarker v.1.75 software (Soft Genetics). Significantly (>35%) decreased or increased signal relative to control probes were scored as exon deletions or duplications, respectively.
In order to confirm and decipher the deletion breakpoints in KCNQ1, long range PCR amplification using the Expand Long Template PCR System (Roche, Mannheim, Germany) and PCR primers flanking the suspected boundaries of the deletion was performed. The amplicon was isolated on agarose gel electrophoresis, excised from the gel, purified using the Qiaquick gel extraction kit (Qiagen Sciences, Maryland) and DNA sequenced. Primer sequences and PCR conditions are available upon request.
RESULTS
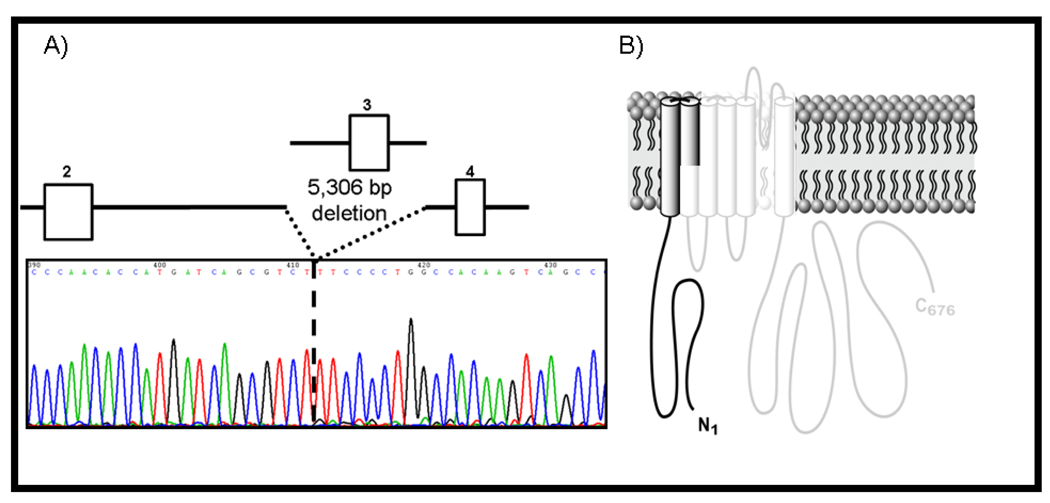
Following MLPA analysis, 2 unique KCNQ1 gene rearrangements were identified in 2/42 (4.8%, CI, 1.7–11%) unrelated patients with genotype negative/phenotype positive LQTS. A 5,306 base pair (bp) deletion (c.478-5001_604+178del, Figure 1A) of KCNQ1 (NM_000218.2), involving a portion of intron 2 (5001 bp), all of exon 3 (127 bp), and a portion of intron 3 (178 bp) that results in the complete deletion of exon 3 and a subsequent frameshift of the KCNQ1 transcript leading to a premature stop codon (p.E160fs34X, Figure 1B) was identified in a 10- year-old Caucasian boy. This patient was diagnosed clinically at age 5 years with extreme QT prolongation (QTc, 660 ms), recurrent seizures with activity and flashing lights since 2 years of age (normal EEG), a personal history of 5 syncopal episodes during physical activity, and although atypical of LQTS, a family history of syncope associated with the onset of pain in both his mother and maternal grandmother. The patient has been deaf since infancy which may be related to viral meningitis occurring at age 2 months. At 10 years of age, he received an implantable cardioverter defibrillator (ICD) following breakthrough syncope during exertion while on propranolol beta blocker therapy. Six weeks following device implantation, the patient received 16 appropriate shocks on one day following multiple episodes of torsade de pointes (TdP). Samples from the affected relatives were unavailable to confirm proper pedigree co-segregation of the mutation.
Figure 1. Molecular characterization of a large KCNQ1 exon 3 deletion identified in a 10-year-old male LQTS case.
Depicted is the A) sequence chromatogram (shown is the reverse sequence) and schematic representation of a large 5,306 bp deletion involving the complete loss of exon 3 and B) the subsequent predicted result of an early truncation mutation of the KCNQ1 alpha subunit occurring in the S2 transmembrane domain. The “washed-out” area of the channel is predicted to be missing.
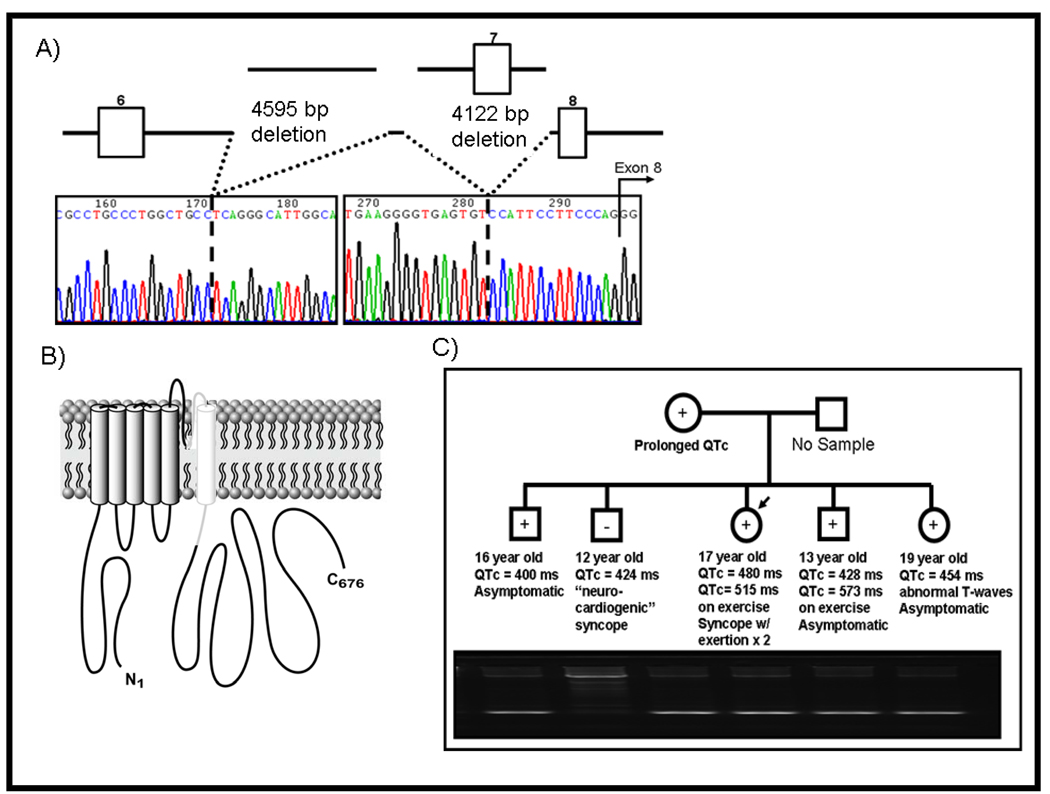
Deletions of 4595 bp (portion of intron 6, c.922-7066 _ 922-2471del, Figure 2A) and 4122 bp (c.922-2359_1032+1652del, Figure 2A) of KCNQ1 involving a portion of intron (2359 bp), all of exon 7 (111 bp), and all of intron 7 (1652 bp) except for 14 bp leading up to exon 8 were identified in a 17-year-old Caucasian girl. These deletions presumably result in a complete deletion of exon 7 and exon skipping of exon 8 due to abnormal splicing resulting from loss of the putative branch point sequence of intron 7 (Figure 2B). This patient had a QTc of 480 ms with lengthening to 515 ms during exercise stress testing, a personal history of 2 syncopal episodes during physical activity at 6 years of age, and a family history of documented QT prolongation during exercise stress testing in a 13-year-old brother (QTc of 428 ms at rest lengthening to 573 ms during exercise), abnormal T-waves in a 19-year-old sister (QTc = 454 ms), and a clinical diagnosis of LQTS in the mother.
Figure 2. Molecular characterization of a large KCNQ1 exon 7 deletion identified in a 17-year-old female LQTS case.
Depicted is the A) sequence chromatogram and schematic representation of a two large deletions (4595 bp deletion in intron 6 and a 4122 bp deletion encompassing all of exon 7 and most of intron 7 and B) the subsequent predicted result of an inframe deletion of exon 7 and exon skipping of exon 8 of the KCNQ1 alpha subunit involving the PORE and S6 transmembrane domain of the channel. The “washed-out” area of the channel is predicted to be missing. Panel C) shows the pedigree analysis revealing co-segregation with incomplete penetrance and variable expressivity of this maternally derived mutation. The upper band on the gel represents the wild-type allele and the lower band represents the smaller mutant allele secondary to the deletion.
All three family members with clinically suspected LQTS are currently asymptomatic and being treated with nadolol. Additionally, two brothers (16 year-old with a QTc of 400 ms and asymptomatic and a 12 year-old with a QTc of 424 ms and a history of “neurocardiogenic” syncope: once while going to the rest room and twice while standing in the heat have been diagnosed clinically as normal and are currently untreated. Pedigree mutational analysis (Figure 2C) confirmed that all clinically diagnosed relatives indeed hosted the mutation. In addition, one of the siblings dismissed as “normal” is genotype positive and preventative measures (QT drug avoidance) for his concealed LQT1 have been implemented.
DISCUSSION
To date, nearly 7% (6/89, Table 2) of patients, with a clinically strong diagnosis of LQTS that are mutation-negative following a coding (exonic) and splice-site region point mutation analysis of the major LQTS-susceptibility genes, host large genomic rearrangements that would elude detection by traditional mutational analysis techniques, but are easily detected by MLPA. In 2006, Koopmann and colleagues provided the first report for any cardiovascular disease of MLPA-exposed mutations demonstrating that large gene rearrangements in the LQT2- susceptibility gene KCNH2 may serve as a pathogenic basis for cases of seemingly genotype-negative LQTS1. The authors analyzed 21 clinically strong LQTS patients (Schwartz-Moss score ≥ 4) and found that 1 patient had a tandem duplication of 3,682 bp (involving 2.5 kb of intron 5, all of exon 6, and 121 bp of exon 7) in KCNH2 (LQT2). In 2008, Eddy and colleagues used MLPA to identify large multiple-exon deletions in KCNQ1 (deletion of exons 13–14) and KCNH2 (deletion of exons 6–14), and a large intragenic duplication in KCNH2 (duplication of exons 9–14) in 3 of 26 (11.5%) patients with genotype negative/phenotype positive LQTS2.
Table 2.
Summary of Long QT Syndrome-Associated Large Gene Rearrangements Discovered to Date.
| Gene | Deletion/ Duplication |
Exon(s) | Age (years) |
Gender | QTc* | Personal History | Family History | Reference |
|---|---|---|---|---|---|---|---|---|
| KCNQ1 | Deletion | 3 | 10 | M | 660 ms | Syncope / Seizures (Exercise)/ICD Shocks | Syncope | This study |
| Deletion | 7 | 17 | F | 480 ms | Syncope (Exercise) | SCD | This study | |
| Deletion | 13–14 | 11 | M | 580 ms | Syncope (Exercise) | Seizures | Eddy2 | |
| KCNH2 | Duplication | 6–7 | 17 | F | 480 ms | Syncope (Auditory) | Syncope and SCD | Koopman1 |
| Deletion | 6–14 | 22 | F | 560 ms | Seizures | Seizures and SCD | Eddy2 | |
| Duplication | 9–14 | 12 | M | 550 ms | Asymptomatic | SCD | Eddy2 |
The longest specified QTc; M = male, F=female; SCD = sudden cardiac death
Here, we report the identification of two large KCNQ1 deletions involving a deletion of exon 3 and a deletion of exon 7 in 2 patients (5%) with seemingly genotype negative/phenotype positive LQTS using MLPA technology. While Eddy and colleagues limited their cohort to patients with a Schwartz-Moss score ≥4, we included patients with either a QTc ≥ 480 ms and/or a Schwartz-Moss score ≥ 4. While 90% of our cohort had a QTc greater than 480 ms, only 48% of our patients had a measurable Schwartz-Moss score of ≥4, suggesting that the Eddy cohort may represent a collection of patients with a more severe phenotype. This may account for the disparity in yield of large gene rearrangements between our cohorts and may suggest that a higher expected diagnostic rate for this modality among patients with a more severe phenotype may exist. In fact, if we limit the denominator of our cohort to the 20 cases with a Schwartz- Moss score ≥4; our yield is similar at 10%. Expectantly, no large gene rearrangements have been reported in SCN5A from analysis of these LQTS cohorts as such rearrangements in SCN5A would predictably result in a “loss-of-function” Brugada syndrome-like phenotype. Whether large SCN5A gene rearrangements provide a molecular substrate for some of Brugada syndrome (BrS) is largely unknown. However, Koopman et al. recently reported the absence of large SCN5A rearrangements among their Dutch cohort of 38 SCN5A mutation negative BrS patients16.
Large gene rearrangements in KCNQ1 and KCNH2 may confer more “torsadogenic” substrates then other LQT1- and LQT2-causative mutations. Here, we report a 5,306 bp deletion encompassing all of KCNQ1 exon 3, resulting in a frameshift and early truncation of the channel, that was identified in a young boy with a personal history of five syncopal episodes during physical activity (consistent with LQT1), including a breakthrough event during exertion while on the beta blocker propanolol; leading to an ICD. However, at least for the KCNQ1-encoded IKs channel, the subunit assembly domain that facilitates formation of the tetrameric channel resides in the C-terminus. As such, both KCNQ1 gene rearrangements identified in our study yield a truncated protein that lacks this domain. Therefore, this likely yields haploinsufficiency rather than a dominant negative mechanism where prior studies have demonstrated that dominant negative LQT1 is associated with a two-fold greater probability of an LQT1-triggered cardiac event when compared to haploinsufficiency-mediated LQT117. In addition, the three previously described large KCNH2 gene rearrangements were identified in families with a remarkable history of autopsy negative sudden unexplained death in the young, totaling six cases between the three families1,2. While the six sudden deaths involved individuals over the age of one year, it is plausible that such large gene rearrangements in these cardiac channel genes could provide a channelopathic mechanism of some sudden infant death syndrome as well18,19.
Taken together, these reports suggest that genetic testing for genomic rearrangements should be considered as a possible reflex test when the primary LQTS genetic test is negative. This additional molecular scrutiny for genomic rearrangements in KCNQ1 and KCNH2 may increase the maximal anticipated yield of the current clinically available genetic test by 1 – 3%. Although minute in magnitude, this incremental increase is similar to that derived by inclusion of all 9 minor LQTS-susceptibility genes and more importantly, may have an important clinical and life saving impact in those families found to be positive for such mutations. For example, in our current study, family pedigree analysis has exposed the potentially lethal molecular substrate of a large KCNQ1 gene rearrangement in an untreated family member with apparently concealed LQT1 (normal QTc and asymptomatic). Although at very low risk for sudden death, simple preventative measures such as QT drug avoidance and perhaps even prophylactic beta blocker therapy can now be considered because of the patient’s correct classification as someone with genetically concealed LQT1.
ACKNOWLEDGMENTS
This work was supported by the Mayo Clinic Windland Smith Rice Comprehensive Sudden Cardiac Death Program (MJA) and the National Institutes of Health (HD42569, MJA).
Funding: Mayo Clinic Windland Smith Rice Comprehensive Sudden Cardiac Death Program (MJA).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: MJA is a consultant for PGxHealth. Intellectual property derived from MJA’s research program resulted in license agreements in 2004 between Mayo Clinic Health Solutions (formerly Mayo Medical Ventures) and PGxHealth (formerly Genaissance Pharmaceuticals).
REFERENCES
- 1.Koopmann TT, Alders M, Jongbloed RJ, Guerrero S, Mannens M, Wilde AAM, Bezzina CR. Long QT syndrome caused by a large duplication in the KCNH2 (HERG) gene undetectable by current polymerase chain reaction-based exon-scanning methodologies. Heart Rhythm. 2006;3:52–55. doi: 10.1016/j.hrthm.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 2.Eddy C-A, MacCormick JM, Chung S-K, Crawford JR, Love DR, Rees MI, Skinner JR, Shelling AN. Identification of large gene deletions and duplications in KCNQ1 and KCNH2 in patients with long QT syndrome.[see comment] Heart Rhythm. 2008;5:1275–1281. doi: 10.1016/j.hrthm.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 3.Caselli R, Mencarelli MA, Papa FT, Ariani F, Longo I, Meloni I, Vonella G, Acampa M, Auteri A, Vicari S, Orsi A, Hayek G, Renieri A, Mari F. Delineation of the phenotype associated with 7q36.1q36.2 deletion: long QT syndrome, renal hypoplasia and mental retardation. American Journal of Medical Genetics Part A. 2008;146:1195–1199. doi: 10.1002/ajmg.a.32197. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz PJ, Moss AJ, Vincent GM, Crampton RS. Diagnostic criteria for the long QT syndrome. An update. Circulation. 1993;88:782–784. doi: 10.1161/01.cir.88.2.782. [DOI] [PubMed] [Google Scholar]
- 5.Splawski I, Shen J, Timothy K, Vincent GM, Lehmann MH, Keating MT. Genomic structure of three long QT syndrome genes: KVLQT1, HERG, and KCNE1. Genomics. 1998;51:86–97. doi: 10.1006/geno.1998.5361. [DOI] [PubMed] [Google Scholar]
- 6.Curran ME, Splawski I, Timothy KW, Vincent GM, Green ED, Keating MT. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995 80;:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 7.Wang Q, Shen J, Splawski I, Atkinson D, Li Z, Robinson JL, Moss AJ, Towbin JA, Keating MT. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80:805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 8.Mohler PJ, Schott J-J, Gramolini AO, Dilly KW, Guatimosim S, duBell WH, Song LS, Haurogne K, Kyndt F, Ali ME, Rogers TB, Lederer WJ, Escande D, Le Marec H, Bennett V. Ankyrin-B mutation causes type 4 long-QT cardiac arrhythmia and sudden cardiac death. Nature. 2003;421:634–639. doi: 10.1038/nature01335. [DOI] [PubMed] [Google Scholar]
- 9.Abbott GW, Sesti F, Splawski I, Buck ME, Lehmann MH, Timothy KW, Keating MT, Goldstein SA. MiRP1 forms IKr potassium channels with HERG and is associated with cardiac arrhythmia. Cell. 1999;97:175–187. doi: 10.1016/s0092-8674(00)80728-x. [DOI] [PubMed] [Google Scholar]
- 10.Andelfinger G, Tapper AR, Welch RC, Vanoye CG, George AL, Jr, Benson DW. KCNJ2 mutation results in Andersen syndrome with sex-specific cardiac and skeletal muscle phenotypes. American Journal of Human Genetics. 2002;71:663–668. doi: 10.1086/342360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, Napolitano C, Schwartz PJ, Joseph RM, Condouris K, Tager-Flusberg H, Priori SG, Sanguinetti MC, Keating MT. Cav1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 12.Vatta M, Ackerman MJ, Ye B, Makielski JC, Ughanze EE, Taylor EW, Tester DJ, Balijepalli RC, Foell JD, Li Z, Kamp TJ, Towbin JA. Mutant caveolin-3 induces persistent late sodium current and is associated with long-QT syndrome. Circulation. 2006;114:2104–2112. doi: 10.1161/CIRCULATIONAHA.106.635268. [DOI] [PubMed] [Google Scholar]
- 13.Medeiros-Domingo A, Kaku T, Tester DJ, Iturralde-Torres P, Itty A, Ye B, Valdivia C, Ueda K, Canizales-Quinteros S, Tusie-Luna MT, Makielski JC, Ackerman MJ. SCN4B-encoded sodium channel beta4 subunit in congenital long-QT syndrome. Circulation. 2007;116:134–142. doi: 10.1161/CIRCULATIONAHA.106.659086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L, Marquardt ML, Tester DJ, Sampson KJ, Ackerman MJ, Kass RS. Mutation of an A-kinase-anchoring protein causes long-QT syndrome. Proceedings of the National Academy of Sciences. 2007;104:20990–20995. doi: 10.1073/pnas.0710527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ueda K, Valdivia CR, Medeiros-Domingo A, Tester DJ, Vatta M, Farrugia G, Ackerman MJ, Makielski JC. Syntrophin mutation associated with long QT syndrome through activation of the nNOS-SCN5A macromolecular complex. Proceedings of the National Academy of Sciences. 2008;105:9355–9360. doi: 10.1073/pnas.0801294105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koopmann TT, Beekman L, Alders M, Meregalli PG, Mannens MMAM, Moorman AFM, Wilde AAM, Bezzina CR. Exclusion of multiple candidate genes and large genomic rearrangements in SCN5A in a Dutch Brugada syndrome cohort. Heart Rhythm. 2007;4:752–755. doi: 10.1016/j.hrthm.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 17.Moss AJ, Shimizu W, Wilde AAM, Towbin JA, Zareba W, Robinson JL, Qi M, Vincent GM, Ackerman MJ, Kaufman ES, Hofman N, Seth R, Kamakura S, Miyamoto Y, Goldenberg I, Andrews ML, McNitt S. Clinical aspects of type-1 long-QT syndrome by location, coding type, and biophysical function of mutations involving the KCNQ1 gene. Circulation. 2007;115:2481–2489. doi: 10.1161/CIRCULATIONAHA.106.665406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tester DJ, Ackerman MJ. Sudden infant death syndrome: How significant are the cardiac channelopathies? Cardiovasc Res. 2005;67:388–396. doi: 10.1016/j.cardiores.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Arnestad M, Crotti L, Rognum TO, Insolia R, Pedrazzini M, Ferrandi C, Vege A, Wang DW, Rhodes TE, George AL, Jr, Schwartz PJ. Prevalence of long-QT syndrome gene variants in sudden infant death syndrome. Circulation. 2007;115:361–367. doi: 10.1161/CIRCULATIONAHA.106.658021. [DOI] [PubMed] [Google Scholar]