Abstract
The widely found fungal iterative PKS-NRPS hybrid megasynthetases are highly programmed biosynthetic machines involved in the synthesis of 3- acyltetramic acids and related natural products. In vitro analysis of iterative PKS-NRPS have been hampered by the difficulties associated with obtaining pure and functional forms of these large enzymes (>400 kDa). We successfully expressed Aspergillus nidulans aspyridone synthetase (ApdA) from an engineered Saccharomyces cerevisiae strain. The complete functions of ApdA and its enoylreductase partner ApdC are reconstituted in vitro and in S. cerevisiae with the production of preaspyridone 7. The programming rules of both the PKS and NRPS modules were then examined in vitro. The key interaction between the PKS and the NRPS was dissected and reconstituted in trans by using standalone modules. Analogs of 7 were synthesized through heterologous combinations of PKS and NRPS modules from different sources. Our results represent the largest, multidomain enzyme reconstituted to date; and offer new opportunities for engineered biosynthesis of fungal natural products.
Filamentous fungi produce a diverse array of bioactive secondary metabolites. Among the small molecule natural products, polyketides and nonribosomal peptides are synthesized by multidomain enzymes such as polyketide synthases (PKSs) and nonribosomal peptide synthetases (NRPSs), respectively. The enzymology and biochemical properties of these mega-enzymes are highly complex and are different from the well-studied bacterial PKSs and NRPSs.1 In particular, the highly-reducing PKSs (HR-PKSs), in which individual domains are programmed to function in different permutations during the iterative process of chain elongation, are particularly enigmatic as exemplified by the lovastatin nonaketide synthase LovB.2 Notwithstanding the complexity of HR-PKSs, an even more impressive biosynthetic machinery that is found in nearly all filamentous fungi is the PKS-NRPS hybrid, in which a single module of NRPS is translationally fused to the C-terminus of a HR-PKS. A typical PKS-NRPS hybrid contains ~ 10 catalytic domains; exceeds 400 kDa as a standalone protein; and synthesizes acyltetramic acid (3-acyl-pyrrolidine- 2,4-dione) as the product. The tetramic acids are further tailored into complex natural products, such as fusarin C,3 equisetin,4 aspyridone A 1,5 pseurotin A,6 cyclopiazonic acid 2,7 chaetoglobosin,8 tenellin 3,9 etc (Figure 1A). The widespread presence of fungal PKS-NRPS machineries,10 along with their unique biosynthetic capabilities and highly evolved programming rules, make them attractive targets for biochemical investigations in vitro and in genetically tractable hosts.
Figure 1.
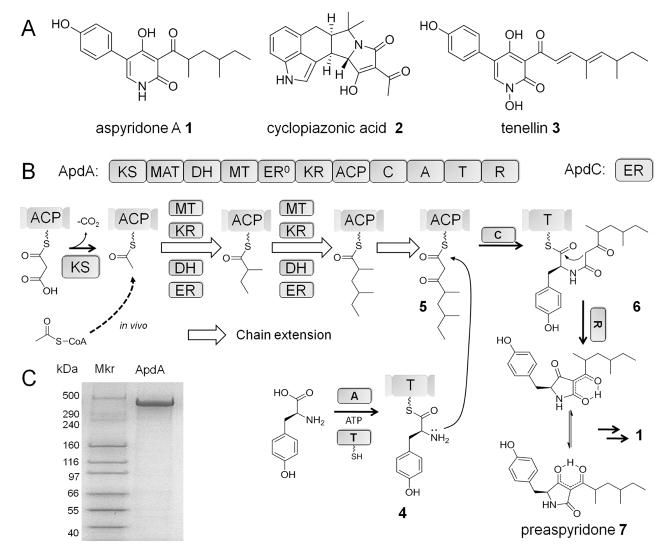
Tetramic acid synthesis by fungal PKS-NRPS. (A) Selected tetramic acid-derived natural products. (B) Proposed mechanism of preaspyridone 7 synthesis by ApdA (PKS-NRPS) and ApdC (ER). The phosphopantetheinyl arms of the ACP and T domains are shown as squiggle lines. Priming of the ACP by the acetyl group may be from the decarboxylative of malonyl-ACP by the KS domain (observed in vitro), or via the direct acetylation of KS by acetyl-CoA followed by transfer to the ACP domain (in vivo). The white arrow represents one round of chain elongation. 7 can be converted to the final natural product 1 after extra enzymatic oxidation and ring rearrangement.5 (C) SDS-PAGE of purified ApdA (439 kDa, tags included). The enzyme is purified from S. cerevisiae BJ5464-NpgA by an anti-FLAG antibody affinity resin.
ApdA is the PKS-NRPS that is involved in the synthesis of 1 (Figure 1A). The role of ApdA was discovered via artificial transcription activation of the cryptic apd gene cluster in Aspergillus nidulans.5 The HR-PKS module of the enzyme, which consists of ketosynthase (KS), malonyl–CoA:ACP transacylase (MAT), dehydratase (DH), methyltransferase (MT), ketoreductase (KR), and acyl carrier protein (ACP) domains, is proposed to synthesize the ACP-bound 4,6-dimethyl-3-oxooctanyl thioester 5. A dissociated enoylreductase (ER) ApdC is proposed to function in trans with ApdA, analogous to the role played by LovC during LovB-catalyzed synthesis of dihydromonacolin L.11 The downstream NRPS module contains the condensation (C), adenylation (A) and thiolation (T) domains, and catalyzes the formation of the L-tyrosinyl-thioester 4 and the amide linkage between 4 and 5 to yield 6 tethered to the T domain. The bimodular assembly line is terminated with a putative reductase (R) domain that facilitates formation and release of the tetramic acid product. Combined with ApdC, ApdA is proposed to synthesize a precursor of 1 via ~25 enzymatic steps. Although the PKS-NRPSs responsible for tenellin (TenS) and cyclopiazonic acid (CpaS) have both been reconstituted in the heterologous host Aspergillus oryzae,12 in vitro reconstitution of fungal PKS-NRPS activities has not been accomplished. Reconstitution of purified biosynthetic enzymes in vitro enables the most direct analysis of the functions and products of the megasynthases.13 Here we show that using the Saccharomyces cerevisiae strain BJ5464-NpgA as the expression host,2,14 the complete activities of the ApdA and ApdC can be reconstituted, which led to the identification of an acyltetramic acid 7 as the product.
The uninterrupted apdA and apdC genes were cloned from the sequenced A. nidulans strain FGSC A4 after re-annotation of introns (Figures S1–S2). A gene encoding N-terminal FLAG, C-terminal hexahistidine tagged ApdA was placed under the control of the ADH2 promoter15 in the 2-μm episomal vector pXW58. Full length ApdA (439 kDa) was then solubly expressed from BJ5464-NpgA transformed with pXW58 and purified to near homogeneity with an anti-FLAG antibody affinity resin to a final of yield of ~1.2 mg/L (Figure 1C). ApdC was solubly expressed from Escherichia coli BL21(DE3) when coexpressed with chaperone proteins GroEL and GroES (Figure S3A). To test the activities of the two enzymes and identify possible tetramic acid products, equimolar amounts (25 μM) of purified ApdA and ApdC were incubated at room temperature for 12 hours with all the cofactors and building blocks (2 mM NADPH, 1 mM SAM, 1 mM L-Tyr, 25 mM ATP, 10 mM MgCl2 and 2 mM malonyl-CoA). LC-MS analysis of the organic extract showed the production of a predominant compound 7 with a UV absorption maximum (λmax) at 279 nm with m/z [M+H]+ = 332 (see Figure 2A, trace i; Figure S4). Exclusion of any of the cofactors abolished the synthesis of 7. To assess the length of the polyketide portion of 7, the in vitro assay was performed in the presence of [2-13C] malonate (100 mM) and MatB (25 μM), which can generate [2-13C] malonyl-CoA in the presence of ATP, Mg2+ and CoA.16 The assay yielded the same product profile and the mass of 7 was increased to m/z [M+H]+ = 336 (Figure S4), suggesting the presence of a tetraketide chain which is consistent with the proposed function of AdpA as shown in Figure 1B.
Figure 2.
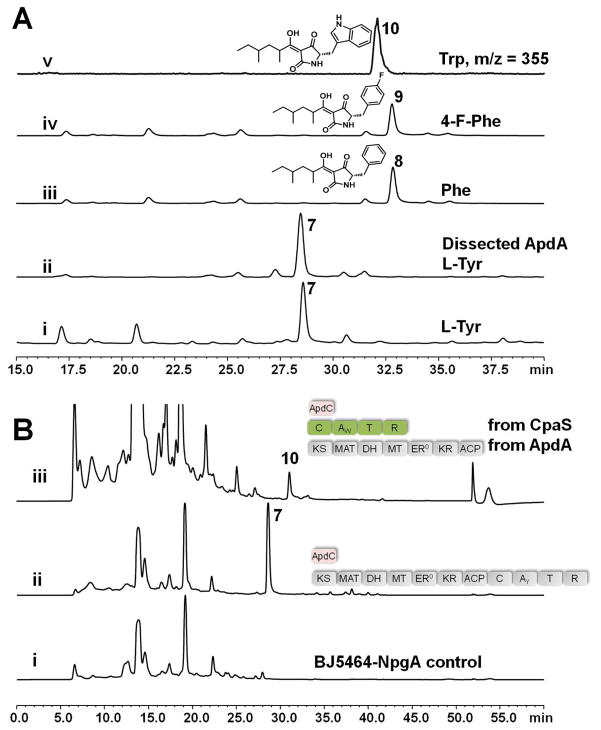
Reconstitution of ApdA in vitro and in S. cerevisiae. (A) In vitro reconstitution of ApdA using purified enzymes. In each trace, the supplied L-amino acid (1 mM) is shown. All experiments were performed with intact ApdA except trace ii in which dissected PKS and NRPS modules of ApdA were reassembled. Trace i–iv are HPLC traces monitored at UV λ=280 nm; trace v is a selected ion monitoring trace [M+H]+=355 of 10, which is produced at lower titers. More detailed traces can be found in Figure S9, S11–S14. (B) HPLC traces (280 nm) that shows the in vivo reconstitution of ApdA in BJ5464-NpgA. The cultures were extracted with ethyl acetate on the 5th day. Trace i: untransformed BJ5464 control; trace ii: BJ5464 transformed with ApdA. This culture was used to purify 7 for structural determination; trace iii: BJ5464 transformed with expression plasmids that separately encode the ApdA PKS and the CpaS NRPS modules. The synthesis of the expected analog 10 was observed in the extract (Trace iii is scaled differently to clearly show 10).
To obtain sufficient amounts of 7 for structural elucidation, BJ5464-NpgA was cotransformed with expression plasmids (pXW58 and pXW51) for both ApdA and ApdC and cultured as previously described.17,18 The RT, UV and mass spectra of 7 biosynthesized from yeast was identical to that of the in vitro sample (Figure 2B, trace ii). After 4 days, the culture was extracted and 7 was purified (final titer ~ 4 mg/L) and fully characterized by 1H, 13C and correlation NMR spectroscopy (Table S3; Figure S5–S8). Interestingly, both proton and carbon NMR spectra in CDCl3 showed two sets of signals with a relative ratio of 3:1 and these two sets collapsed into one single set when the NMR solvent was switched to CD3OD (Figure S9), which is characteristic for 3-acyltetramic acids. The two sets of signals in CDCl3 arise from the external tautomers, which slowly interconvert through the rotation around the acyl-tetramic acid linkage (Figure S10).19 The collapse can be explained by the inclination of external tautomeric equilibrium to one side as a result of solvent effect.20 The proton spectrum showed characteristics of a typical AA′BB′ spin system, which strongly indicated the presence of a para-substituted phenyl ring. COSY coupling between high field protons suggests a spin system of a branched aliphatic chain. Combining all the NMR information and in comparison to that of pretenellin-A21 and 1, 7 was determined as the 3-acyltetramic acid shown in Figure 1B and is named preaspyridone.
Synthesis of 7 using purified ApdA and ApdC confirms the complete reconstitution of the activities of the recombinant PKS-NRPS megasynthetase. Under in vitro conditions, the assembly of 7 can be initiated by loading of malonyl-CoA onto ApdA, followed by decarboxylation to yield the acetyl starter unit (Figure 1B). Under in vivo conditions, priming may also be initiated directly with an acetyl-CoA unit. The growing polyketide chain then elongates into the tetraketide 5 as shown in Figure 1B through differential tailoring of the α and β positions. None of the tailoring domains were used in the last PKS elongation step, which affords the β-keto thioester 5 required for formation of the acyltetramic acid. The acyl chain of 5 is then amidated with the α-NH2 of 4 to afford 6. No other tetramic acid or amidated polyketide product was observed in the in vitro reaction in detectable amounts, indicating the programming rules of AdpA are precise and only the correctly tailored polyketide acyl intermediates are transferred from the PKS to the NRPS. The in vitro synthesis of 7 in the absence of additional oxidative enzymes also confirms the release of 6 via a R-domain catalyzed Dieckmann cyclization instead of the originally proposed NADPH-dependent reductive release followed by cyclization and enzymatic oxidation.5 The Dieckmann cyclase property of ApdA R domain is therefore analogous to those of dissected R domains from the EqiS and CpaS fungal PKS-NRPSs.22
To probe the flexibility of the C and A domains towards different aromatic amino acids, we performed the in vitro reactions with various para-substituted phenylalanines and the synthesis of analogs of 7 was monitored by LC-MS (Figure 2A, trace iii, iv and v; Figure S11–S13). ApdA was able to synthesize the corresponding analogs 8 and 9 in the presence of L-phenylalanine and L-4-fluorophenylalanine, respectively, at ~50% efficiency compared to 7. Increasing the sizes of the para substituent (-NH2, -Cl, -Br) led to decreases in product turnover (Figure S14–S17). Surprisingly, adding L-tryptophan resulted in the synthesis of the indole-containing analog 10 (Figure 2A, trace v) as revealed by selected ion monitoring, albeit at significantly lower levels. Therefore, these results demonstrate while the NRPS module clearly prefers L-tyrosine in the in vitro assay, both the activation (A) and the condensing (C) domains have some flexibility in utilizing other aromatic amino acids and lead to formation of analogs of 7.
The in vitro assay developed for ApdA similarly allows probing of the programming rules of the HR-PKS portion of the megasynthetase. Towards this end, we first examined whether removal of SAM in the in vitro reaction can lead to unmethylated versions of 7. While no tetramic acid was observed, two unsaturated α-pyrones (13 and 14) were produced (Figure 3A, trace ii; Figure S18). 13 was previously recovered as a derailment product of CalE823 and 14 was isolated from an in vitro LovB assay in the absence of SAM, in which the ER LovC failed to function without C-methylation of a tetraketide intermediate.2 Therefore, the synthesis of 13 and 14 here indicates C-methylation is also a prerequisite modification for enoylreduction by the ER partner ApdC. The incorrectly tailored polyketide intermediates were then offloaded by ApdA as 13 and 14, analogous to that observed for LovB upon derailment in the tailoring steps. These convergent observations between ApdA/ApdC and LovB/LovC thus point to a conserved programming rule, in which a subtle α-methylation can modulate the interaction between HR-PKSs and their dissociated ER partners.
Figure 3.
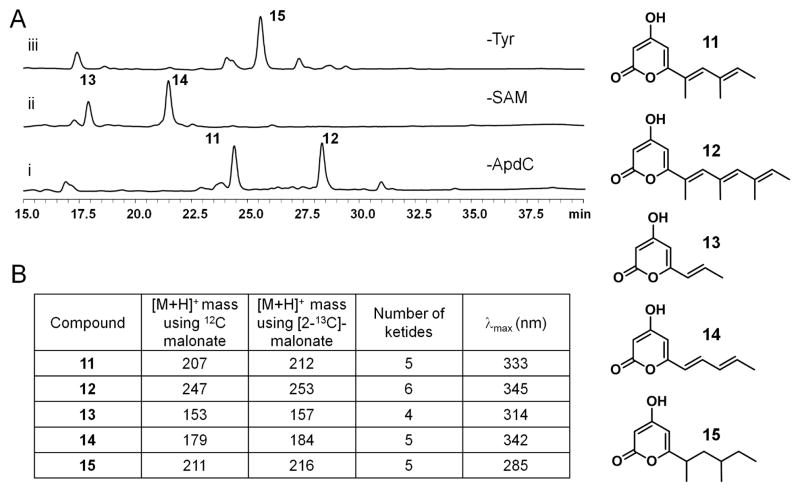
In vitro assays probing of the programming rules of the PKSNRPS system. The putative structures of these polyketides are listed on the right. (A) HPLC UV traces of that show the production of different polyketides through removal of the indicated components from the assay that synthesizes 7. Trace i and ii are monitored at λ=330 nm; trace iii is monitored at λ=280 nm. The relative retention times of compounds are in agreement with their relative polarities based on the proposed structures. (B) The masses of the mentioned polyketides. The increment in the mass when fed with [2-13C]-malonate indicates the number of ketides for the corresponding compound. Conjugation onto the pyrone ring will cause the λmax to shift a longer wavelength.
Similarly, removal of ApdC from the assay produced two α-pyrones 11 and 12 (Figure 3A, trace i, Figure S19). Repeating the assay with 2-13C malonate resulted in increase of 5 and 6 mu for 11 and 12, respectively (Figure 3B). We also observed increase in the λmax of the α-pyrones, which is indicative of increased levels of conjugation. These observations, combined with inference from biosynthetic logic and comparison to the LovB system,2,11,24 led us to propose the structures of 11 and 12 as shown in Figure 3, of which the two compounds contain two and three α-methyl modifications, respectively. The failure of ApdA to synthesize tetramic acids containing unsaturated polyketide portions is evident of the substrate specificity of the NRPS module towards incorrectly tailored polyketide acyl substrates. These stall products were instead offloaded as pyrones to afford 11 and 12. Intriguingly, this result is different from that of TenS reconstituted in A. oryzae in which the conjugated prototenellins were synthesized in the absence of the ER TenC.21 This is most likely due to the more relaxed substrate tolerance of the downstream C domain of TenS.
The formation of 11, 12 and 14 from the above assays points to an unexpected feature of ApdA, which is its ability to synthesize polyketide products longer than the tetraketide observed in 1 and 7. To probe the chain length specificity of the HR-PKS, we sought to assay the PKS portion independent of the activities of the NRPS module. To do so, we i) excluded L-tyrosine from the assay that produced 7; and ii) cloned and expressed the PKS portion as a standalone enzyme (Figure S3B) from BJ5464-NpgA (5 mg/L) and assayed with ApdC in the presence of malonyl-CoA, SAM and NADPH. In both assays, the same α-pyrone 15 was recovered from the reaction mixtures (Figure 3A, trace iii; Figure S20). The size of 15 was confirmed to be a pentaketide and λmax of 285 suggests that the linear portion of the molecule is not conjugated and is instead fully reduced, which is expected if all the tailoring domains functioned properly. Therefore the HRPKS module can indeed function correctly without the downstream NRPS module and synthesize a pentaketide. However, during the synthesis of 7, in order to prevent continual elongation of the polyketide beyond the β-keto tetraketide and eventual offloading as 15, the C domain of the NRPS must interact with the PKS accurately to capture 5 and condense with 4.
To examine if this critical interaction between the HRPKS and NRPS can be established in trans, we expressed and purified the NRPS module as a C-terminus hexahistidine tagged standalone enzyme from E. coli BAP125 (5 mg/L) (Figure S3C). Incubation of the dissected ApdA PKS and NRPS modules in equimolar amounts (25 μM), along with ApdC, cofactors and building blocks, produced 7 in comparable yield as the intact ApdA (Figure 2A, trace ii). This is the first demonstration of physically separating a fungal PKS-NRPS megasynthetase into the two independent, functional modules. Encouraged by the in trans complementation findings, we explored whether heterologous PKS and NRPS modules can be functionally matched, which was previously engineered for some bacterial PKS-NRPS hybrids.26 We cloned and expressed the NRPS module of Aspergillus flavus CpaS (Figure S3D), which incorporates L-tryptophan into the tetramic acid intermediate of 2.12b When BJ5464-NpgA was cotransformed with expression plasmids that expressed ApdA PKS, CpaS NRPS and ApdC, small amounts of 10 (~0.1 mg/L) was observed from the extracts of the yeast culture (Figure 2B, trace iii; Figure S21), indicating successful communication between the two modules. The lower titer of 10 may be attributed to the substrate specificity of the CpaS C domain donor site27 towards both the noncognate ApdA ACP and the bulkier 5 compared to the natural acetoacetyl thioester acyl donor. The lower turnover of 10 can also arise from the inability of the CpaS R domain to correctly process an unnatural substrate. Nevertheless, the result from the straightforward mix and match experiment shows the potential to use this dissociated platform to biosynthesize new tetramic acid compounds.
In conclusion, we reconstituted the complete activities of the PKS-NRPS ApdA through the synthesis of 7. To the best of our knowledge, this represents the largest, single biosynthetic megasynthase reconstituted in vitro. Using both in vitro assays and the yeast host, we were able to dissect the programming rules of ApdA, including the protein-protein interactions and the key acyl transfer step between the PKS and NRPS modules.
Supplementary Material
Acknowledgments
This work is supported by NIH grants 1R01GM085128 and 1R01GM092217, and an Alfred P. Sloan Fellowship to YT. We thank Prof. David Tirrell for providing all the unnatural phenylalanine analogs for our in vitro assays.
Footnotes
Supporting Information Available: Experimental details and spectroscopic information are available free of charge via the internet at http://pubs.acs.org.
References
- 1.(a) Cox RJ. Org Biomol Chem. 2007;5:2010. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]; (b) Staunton J, Weissman KJ. Nat Prod Rep. 2001;18:380. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 2.Ma SM, Li JW, Choi JW, Zhou H, Lee KK, Moorthie VA, Xie X, Kealey JT, Da Silva NA, Vederas JC, Tang Y. Science. 2009;326:589. doi: 10.1126/science.1175602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song Z, Cox RJ, Lazarus CM, Simpson TT. Chem Bio Chem. 2004;5:1196. doi: 10.1002/cbic.200400138. [DOI] [PubMed] [Google Scholar]
- 4.Sims JW, Fillmore JP, Warner DD, Schmidt EW. Chem Commun (Camb) 2005:186. doi: 10.1039/b413523g. [DOI] [PubMed] [Google Scholar]
- 5.Bergmann S, Schumann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Nat Chem Biol. 2007;3:213. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- 6.Maiya S, Grundmann A, Li X, Li SM, Turner G. Chem Bio Chem. 2007;8:1736. doi: 10.1002/cbic.200700202. [DOI] [PubMed] [Google Scholar]
- 7.(a) Tokuoka M, Seshime Y, Fujii I, Kitamoto K, Takahashi T, Koyama Y. Fungal Genet Biol. 2008;45:1608. doi: 10.1016/j.fgb.2008.09.006. [DOI] [PubMed] [Google Scholar]; (b) Chang PK, Horn BW, Dorner JW. Fungal Genet Biol. 2009;46:176. doi: 10.1016/j.fgb.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Schümann J, Hertweck C. J Am Chem Soc. 2007;129:9564. doi: 10.1021/ja072884t. [DOI] [PubMed] [Google Scholar]
- 9.Eley KL, Halo LM, Song Z, Powles H, Cox RJ, Bailey AM, Lazarus CM, Simpson TJ. Chem Bio Chem. 2007;8:289. doi: 10.1002/cbic.200600398. [DOI] [PubMed] [Google Scholar]
- 10.(a) von Dohren H. Fungal Genetics and Biology. 2009;46(Suppl 1):S45. doi: 10.1016/j.fgb.2008.08.008. [DOI] [PubMed] [Google Scholar]; (b) Collemare J, Billard A, Bohnert HU, Lebrun MH. Mycol Res. 2008;112:207. doi: 10.1016/j.mycres.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Kennedy J, Auclair K, Kendrew SG, Park C, Vederas JC, Hutchinson CR. Science. 1999;284:1368. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- 12.(a) Heneghan MN, Yakasai AA, Halo LM, Song Z, Bailey AM, Simpson TJ, Cox RJ, Lazarus CM. Chem Bio Chem. 2010 doi: 10.1002/cbic.201000259. [DOI] [PubMed] [Google Scholar]; b Seshime Y, Juvvadi PR, Tokuoka M, Koyama Y, Kitamoto K, Ebizuka Y, Fujii I. Bioorg Med Chem Lett. 2009;19:3288. doi: 10.1016/j.bmcl.2009.04.073. [DOI] [PubMed] [Google Scholar]
- 13.Sattely ES, Fischbach MA, Walsh CT. Nat Prod Rep. 2008;25:757. doi: 10.1039/b801747f. [DOI] [PubMed] [Google Scholar]
- 14.Lee KK, Da Silva NA, Kealey JT. Anal Biochem. 2009;394:75. doi: 10.1016/j.ab.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 15.Lee KM, DaSilva NA. Yeast. 2005;22:431. doi: 10.1002/yea.1221. [DOI] [PubMed] [Google Scholar]
- 16.An JH, Kim YS. Eur J Biochem. 1998;257:395. doi: 10.1046/j.1432-1327.1998.2570395.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhou H, Qiao K, Gao Z, Meehan MJ, Li JW, Zhao X, Dorrestein PC, Vederas JC, Tang Y. J Am Chem Soc. 2010;132:4530. doi: 10.1021/ja100060k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reeves CD, Hu Z, Reid R, Kealey JT. Appl Environ Microbiol. 2008;74:5121. doi: 10.1128/AEM.00478-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.(a) Royles BJL. Chem Rev. 1995;95:1981. [Google Scholar]; (b) Schobert R, Schlenk A. Bioorg Med Chem. 2008;16:4203. doi: 10.1016/j.bmc.2008.02.069. [DOI] [PubMed] [Google Scholar]
- 20.Saito K, Yamaguchi T. Bull Chem Soc Jpn. 1978;51:651. [Google Scholar]
- 21.Halo LM, Marshall JW, Yakasai AA, Song Z, Butts CP, Crump MP, Heneghan M, Bailey AM, Simpson TJ, Lazarus CM, Cox RJ. Chem Bio Chem. 2008;9:585. doi: 10.1002/cbic.200700390. [DOI] [PubMed] [Google Scholar]
- 22.(a) Sims JW, Schmidt EW. J Am Chem Soc. 2008;130:11149. doi: 10.1021/ja803078z. [DOI] [PubMed] [Google Scholar]; (b) Liu X, Walsh CT. Biochemistry. 2009;48:8746. doi: 10.1021/bi901123r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belecki K, Crawford JM, Townsend CA. J Am Chem Soc. 2009;131:12564. doi: 10.1021/ja904391r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burr DA, Chen XB, Vederas JC. Org Lett. 2007;9:161. doi: 10.1021/ol062763i. [DOI] [PubMed] [Google Scholar]
- 25.Pfeifer BA, Admiraal SJ, Gramajo H, Cane DE, Khosla C. Science. 2001;291:1790. doi: 10.1126/science.1058092. [DOI] [PubMed] [Google Scholar]
- 26.Liu F, Garneau S, Walsh CT. Chem Biol. 2004;11:1533. doi: 10.1016/j.chembiol.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 27.Samel SA, Schoenafinger G, Knappe TA, Marahiel MA, Essen LO. Structure. 2007;15:781. doi: 10.1016/j.str.2007.05.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.