Abstract
Spermiogenesis represents the process through which haploid male germ cells differentiate from round spermatids into elongated spermatids and eventually the male gametes called spermatozoa. Haploid cell differentiation is unique to male germ cell development and many unique genes/proteins essential for this process have been discovered. SPEM1 is one of these spermiogenesis-essential proteins encoded by a testis-specific gene exclusively expressed in the developing spermatids. Inactivation of Spem1 in mice results in deformed spermatozoa characterized by “head-bent-back” abnormalities with 100% penetrance. Using yeast two-hybrid screening assays, we identified UBQLN1 as one of the SPEM1-interacting partners. UBQLN1 and SPEM1 were colocalized to the manchette of elongating spermatids. Since UBQLN1 functions through binding and directing poly-ubiquitinated proteins to the proteasome for degradation, interactions between UBQLN1and SPEM1 suggest a role in the regulation of protein ubiquitination during spermiogenesis.
Keywords: Spermatogenesis, ubiquitination, ubiquitin, sperm, infertility, manchette, protein turnover
1. Introduction
Male gametes are formed through a complex process called spermatogenesis. Spermatogenesis starts with male germline stem cells called spermatogonia, some of which are committed to differentiation and become differentiated spermatogonia (type A, intermediate and type B spermatogonia) (Clermont, 1972). Type B spermatogonia then enter meiosis after DNA synthesis and become sequentially preleptotene, leptotene, zygotene, pachytene and diplotene spermatocytes, during which numerous meiotic events including tetrad formation, crossover (homologous recombination), and meiotic sex chromosome inactivation occur (Clermont, 1972; Wolgemuth et al., 1995). Following two consecutive meiotic cell divisions, spermatocytes become round spermatids, which are male haploid germ cells (Esponda, 1985). Round spermatids then undergo a dynamic morphogenetic process called spermiogenesis, during which the nuclei containing the haploid paternal genome are heavily condensed and packed, apparatuses essential for motility (flagellum), for penetrating zona pellucida (acrosome and its enzymatic contents), and for sensing environment in the female reproductive tracts (surface ion channels, receptors, etc.) are developed (Esponda, 1985). Since spermiogenesis is unique to male gamete formation, numerous spermatid-specific genes/proteins have been identified (Schultz et al., 2003; Shima et al., 2004) and many of them are essential for spermiogenesis (Cooke and Saunders, 2002; Matzuk and Lamb, 2002, 2008; Yan, 2009). Sperm maturation 1 (Spem1) encodes a 330 a.a. protein without known functional domains (Zheng et al., 2007). SPEM1 is exclusively expressed in the cytoplasm of elongating and elongated spermatids. Spem1 knockout mice displayed a “head-bent-back” deformation in their sperm with 100% penetrance (Zheng et al., 2007). Spem1-null sperm possess cytoplasmic droplet-like remnants translocated to the junction between the head and the neck, suggesting the process of cytoplasm removal may have been impaired either directly or indirectly. unwanted or misfolded proteins through the ubiquitin-proteasome pathway. We therefore further validated our Y2H results, and examined the expression and localization of UBQLN1 in the mouse testis. Our results indeed suggest that SPEM1-UBQLN1 interaction is involved in the regulation of spermiogenesis.
2. Methods and materials
2.1. Animals
Wild-type and Spem1-null mice (on a C57Bl/6:129SvEv hybrid background) were maintained at the University of Nevada School of Medicine in a temperature- and humidity-controlled animal facility with free access to water and food. Testes were collected from male mice at the ages of postnatal days 7, 14, 21, 28, 35 and adult. One of the paired testes from each mouse was snap-frozen in liquid nitrogen for subsequent RNA and protein analyses. The other one was cryoprotected and cryosections were prepared for immunofluorescent staining. The animal use protocol was approved by the Institutional Animal Care and Use Committee of the University of Nevada, Reno.
2.2. Yeast 2-hybrid (Y2H) assays
Y2H screening of an adult mouse testis library using the full-length SPEM1 as bait was performed as described previously (Wang et al., 2006). Briefly, the open reading frame (ORF) of Spem1 cDNA was subcloned into the EcoRI-Sal I site of pGBKT7 vector as the bait construct (pGBKT7/SPEM1). Sequencing analyses were performed to verify the lack of mutation and in-frame of the ORF in the pGBKT7. The pGBKT7/SPEM1 was transformed into Y187 competent yeast cells followed by mating with AH109 yeast cells transformed with an adult mouse testis cDNA library cloned into the pGDT7 vector. Clones grown on the selective medium (SD/-Leu/-Trp/-Ade/-His) were picked and the pGDT7 plasmid DNA contained was sequenced. Co-transformation assays were performed on non-selective (SD/-Leu/-Trp) and selective (SD/-Leu/-Ade/-His) plates by co-transforming pGDT7/Ubquilin1 with an empty bait vector pGBKT7, or with a pGBKT7/SPEM1.
2.3. Mammalian expression constructs
The PCR fragment of the ORF of Spem1 cDNA was subcloned into the EcoRI-KpnI sites of the pGEM-T easy vector (Promega). Sequencing analyses confirmed that the amplified ORF of Spem1 was mutation-free. After double digestion by EcoRI and Kpn1, the Spem1 ORF fragment was subcloned into p3XFLAG-myc-CMV™-26 expression vector (Sigma). The ORF of Spem1 was in frame and thus the full-length SPEM1 was expected to be expressed after transfection into mammalian cell lines. We named this vector p3XFLAG-Spem1. The full-length cDNA clone for Ubqln1 was purchased from Invitrogen (clone ID#4935292). A pair of primers was used to amplify the ORF with a 6-nucleotide Kozak sequence (GCCACC) addition to the 5’ primer before the start codon ATG to facilitate the expression of UBQLN1 protein. The PCR fragment was then cloned into pcDNA3.1/ V5-TOPO vector (Invitrogen) in frame with the deletion of the stop codon. This vector was named pcDNA3.1/ V5-Ubqln1. Sequencing analyses were performed on all final expression vectors to confirm that the inserts were in frame and the sequences were mutation-free.
2.4. Cell culture and transfection
COS-7 cells (ATCC™ Cat#: CRL-1651) were cultured and transfected according to the manufacturer's instructions (PolyFect Transfection Reagent, QiaGen). Briefly, two million cells were seeded in a culture dish (100mm diameter) in DMEM medium supplemented with 10% FCS and the cells usually reached 40%~80% confluence the next day. The transfection complex contains 2μg p3XFLAG-Spem1 and 2μg pcDNA3.1/ V5-Ubqln1 mixed with OPT-MEM in a total volume of 300μl. An aliquot of 25μl PolyFect reagent (QiaGen) was added to the DNA mixture and mixed well by pipetting up and down several times. During the incubation at RT for 5-10min, the old culture medium was removed and the cells were washed once with 5ml D-PBS (Invitrogen) followed by addition of 7ml complete growth medium (containing serum and antibiotics). The transfection complex was then added directly into the culture medium followed by gentle swirling to ensure uniformed distribution of the complexes. Cells were cultured at 37°C with 5% CO2 and harvested for analyses 48h after transfection.
2.5. Co-immunoprecipitation assays
A co-immunoprecipitation kit (Roche, Cat. #: 11719386001) was used and all the procedures were carried out according to the manufacturer's protocols. Briefly, after 48h of culture, COS-7 cells transfected with p3XFLAG-Spem1 and pcDNA3.1/ V5-Ubqln1 were lysed using a buffer containing 50mM Tris-HCl, pH 7.4, 150mM NaCl; 1% sodium deoxycholate, 1% NP-40 and 100mM EDTA supplemented with one tablet of protease inhibitor cocktail (Roche). Anti-V5 or anti-FLAG monoclonal antibodies (Sigma) and protein-G agarose slurry were added into the lysates and the incubation was performed overnight at 4°C with gentle shaking. After sequential washes with high salt and low salt washing buffers, the immuno-complex was subsequently analyzed by SDS-PAGE and standard Western blot analyses as described (Wang et al., 2006).
2.6. Northern Blot Analyses
Total RNA was isolated from heart, liver, spleen, lung, kidney, brain, stomach, small intestine, colon, testis, ovary, uterus tissues as described (Wang et al., 2006). RNA samples (15μg/lane) were fractioned in a 1.2% agarose/formaldehyde gel and transferred to nylon membrane (Hybond –XL, Amersham Biosciences) followed by UV cross-linking (Spectrolinker XL-1500, Spectronics Corp.). A 352bp-long probe corresponding to part of the last exon of Ubiquilin1 was amplified from mouse testicular cDNAs using primers (forward: 5′-TGTGACTGGCTCTGGTTCAG-3′; reverse: 5′-TGCATATTGCCATTCACCAG-3′). The probe was labeled with (α-32P)-dCTP using a Rediprime II Labeling Kit (Amersham). Hybridization, washing, and autoradiography were performed as described (Wang et al., 2006). Gel images of the 28S and 18S bands were used as loading and RNA quality controls.
2.7. Semi-quantitative PCR
Spermatogenic cell populations were purified as described (Song et al., 2009). RNA isolation and cDNA synthesis were performed as described (Song et al., 2009). RT-PCR primers for mouse Ubqln1 were the same as those for Northern blot analyses. PCR was performed at 25-28 cycles, which were tested to be in the exponential range. Mouse house-keeping gene Hprt was used as a loading control, as described (Wang et al., 2006).
2.8. Immunofluorescent staining and confocal microscopy
Testes of adult mice were dissected and fixed with 4% paraformaldehyde for 3 hours at 4°C. The fixed testes were then cryoprotected in serial sucrose solutions with increasing concentrations by mixing 5% and 20% sucrose in ratios at 2:1, 1:1 and 1:2, respectively. The testis samples were incubated for 1/2 hour in each of the sucrose mixtures at room temperature followed by a final incubation in 20% sucrose at 4°C overnight. The testes were then embedded into sucrose: OCT of 1:1. Cryosections of 10-μm thickness were prepared for immunofluorescent staining. The slides were blocked in normal goat and fetal bovine sera at room temperature for 1 hour in a humidity box. A rabbit anti-UBQLN1 antibody (ABGENT, Cat#AP2176c, diluted at 1:200) was added onto the sections and the slides were placed into a humidity box for overnight incubation at 4°C. The slides were washed with 1XPBS for 3 times prior to incubation with an AlexaFLuo488-conjugated goat-anti-rabbit secondary antibody (Molecular Probes, at a dilution of 1:500) for 1 hour. After three times of washing with 1XPBS, aqueous mounting medium containing propidium iodide was applied onto the sections. Imaging analyses was conducted using a confocal microscope (Carl Zeiss, LSM510).
Squashed seminiferous tubules were prepared as described with a minor modification (Yan et al., 2000). After fixation with 4% freshly-prepared paraformaldehyde at RT for 15min, slides with the squashed seminiferous tubules were permeabilized by microwaving 3 times soaked in 1×citrate buffer (pH6.0). Washed with 1XPBS for 3 times (5min/wash), the slides were then blocked with 100μl Blocking Buffer (30μl FBS+30μl normal goat serum+960μl 0.1M PBS) at RT for 1hr. Primary antibody incubation was performed at 4°C overnight. The rabbit-anti-UBQLN1 polyclonal antibody and a mouse anti-β-Tubulin monoclonal antibody (Developmental Studies Hybridoma Bank, E7-c, diluted at 1:500) were used for the co-localization assays. Rabbit anti-SPEM1 polyclonal antibody was prepared as described (Zheng et al., 2007) and used at a dilution of 1:1,000. The next day, the slides were moved to RT for at least 30min before being washed 3 times with 1XPBS (10min per wash). The slides were then incubated with the fluorescence-conjugated, species-specific secondary antibodies (Molecular Probes) at RT for 1hr with gently shaking, flowed by washing with 1XPBS 3 times (10min per wash). Finally the slides were counterstained with DAPI for indirect immunofluorescent assay using confocal laser microscope (Olympus, FV1000).
2.9. Western blot analyses
Western blot analyses were performed as described previously (Wang et al., 2006). Briefly, total protein was isolated from mouse testes or cultured COS-7 cells. Protein lysates (100μg/lane) were fractionated on 10% Tris-HCl polyacrylamide gels through electrophoresis and transferred onto nitrocellulose membranes (Bio-Rad). The membrane was then blocked with 5% non-fat milk. Primary antibody incubation was conducted at RT for 1 hour. After three washes with PBST (1XPBS containing 0.1% Tween20), membranes were incubated with a secondary antibody conjugated with horseradish peroxidase (1:1,000, Amersham) for 1 hr, followed by washing. An enhanced chemiluminescence detection system (Amersham) was used for detection of the specific proteins.
3. Results
3.1. Identification of UBQLN1 as a SPEM1-interacting partner
We used SPEM1 as bait and screened an adult mouse testis library using the yeast 2-hybrid system (Fig. 1A). Y2H screening yielded 80 clones, 49 of which contained cDNA sequences for Ubiquilin1 (Ubqln1). Among the remaining clones analyzed, 17 contained cDNAs for Ranbp17 (encoding Ran-binding protein 17) and 12 for Tex12 (encoding testis-expressed protein12). Co-transformation assays were performed on non-selective (SD/-Leu/-Trp) and selective (SD/-Leu/-Trp/-Ade/-His) medium plates by co-transforming pGADT7/Ubqln1 with the empty bait vector pGBKT7, or with pGBKT7/Spem1 (Fig. 1B). The non-selective plates showed yeast cell growth among all the cotransformations, indicating that the yeast cells were viable and the plasmid constructs were non-toxic. The selective plates only showed yeast cell growth in the cotransformations between pGBKT7/Spem1 and pGADT7/Ubqln1 (Fig. 1B). These results confirmed interactions between SPEM1 and UBQLN1 in the yeast cells.
Fig.1.
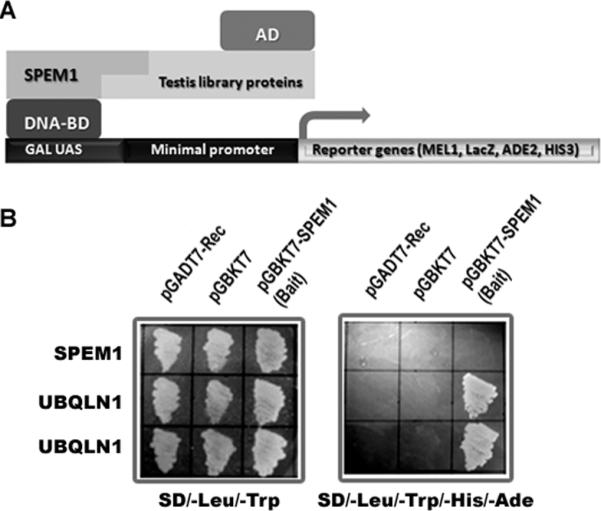
Identification of UBQLN1 as a SPEM1-interacting protein using yeast 2-hybrid (Y2H) assay. (A) In the Y2H assay, SPEM1 was used as bait and an adult mouse testis library was screened. Interactions between SPEM1 and any testicular proteins would activate the expression of reporter genes, leading to yeast cell growth on selective medium. (B) Yeast co-transformation assays to confirm interactions between SPEM1 and two independent clones expressing UBQLN1. On the non-selective plates (SD/-Leu/-Trp) all transformed yeast cells grew, indicating viability of the cells and non-toxicity of the plasmids. Only cells co-transformed with pGBKT7-SPEM1 and pGDT7-UBQLN1 grew on the selective plates (SD/-Leu/-Trp/-His/-Ade), demonstrating true interactions between UBQLN1 and SPEM1 in yeast cells.
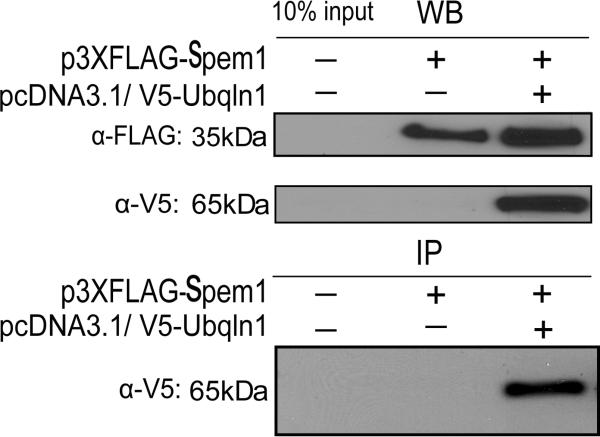
To further examine the interaction between SPEM1 and UBQLN1 in the mammalian system, we transfected plasmids expressing FLAG-tagged SPEM1 and/or V5-tagged UBQLN1into COS-7 cells. Expression of the proteins with correct sizes was verified using Western blot analyses (Fig. 2, upper panels). In the FLAG pull-down products, V5-tagged UBQLN1 was detected (Fig. 2, lower panel), demonstrating that SPEM1 and UBQLN1 interact not only in the yeast cells, but also in mammalian cells.
Fig. 2.
Pull-down assays to verify the UBQLN1-SPEM1 interactions in mammalian cells. FLAG-tagged SPEM1 and V5-tagged UBQLN1 were both expressed in the COS-7 cells, as shown by Western blot analyses (WB). In the immunoprecipitation (IP) assays, V5-tagged UBQLN1 was detected in the anti-FLAG immunoprecipitants, suggesting UBQLN1 indeed interacts with SPEM1 in mammalian cells.
3.2. Expression and localization of Ubqln1 mRNA and protein in the mouse testes
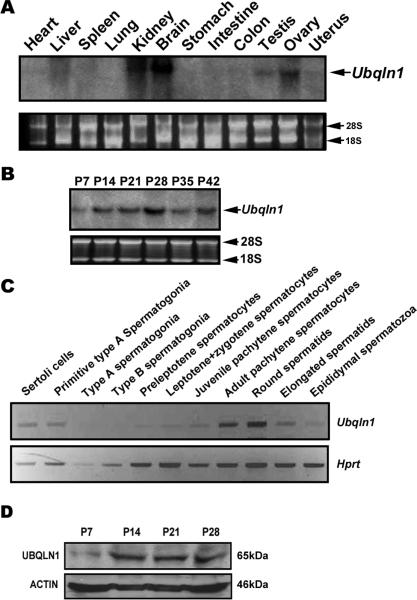
We next examined the expression profiles of Ubqln1 mRNAs and proteins. Northern blot analyses detected Ubqln1 mRNAs in multiple organs including heart, liver, kidney, brain, testis and ovary (Fig. 3A). Ubqln1 mRNA was detectable in developing testes with a noticeable increase at postnatal day 14 (P14). Levels of Ubqln1 mRNAs peaked at P21-P28 in the testes (Fig. 3B), in which spermiogenesis has progressed into elongation stages (Clermont, 1972). Since proportionally haploid male germ cells (round and elongating spermatids) become dominant at P28, the increased expression of Ubqln1 mRNA suggests that haploid cells are the major source of Ubqln1 mRNA in the testis. Using a semi-quantitative PCR method, we also investigated levels of Ubqln1 mRNAs in purified testicular cell populations (Fig. 3C). Ubqln1 mRNA levels were the highest in pachytene spermatocytes and round spermatids, consistent with the expression patterns of Ubqln1 mRNA in developing testes (Fig. 3B). Modest levels were detected in Sertoli cells and early spermatocytes including preleptotene, leptotene and zygotene spermatocytes. Low to undetectable levels were observed in type A and type B spermatogonia. Consistent with Ubqln1 mRNA expression patterns, UBQLN1 levels increased at P14 and remained constantly high thereafter in the testes (Fig. 3D).
Fig. 3.
Expression profiles of Ubqln1 mRNA and protein. (A) Northern blot analyses of Ubqln1 mRNAs in 12 mouse organs. 28S and 18S rRNA bands were shown as loading and mRNA quality controls. (B) Northern blot analyses of Ubqln1 mRNA in developing testes in mice. Testes of postnatal days 7-42 (P7-P42) were analyzed, and 28S and 18S rRNA bands were shown as loading and mRNA quality controls. (C) Levels of Ubqln1 mRNA in purified testicular cell populations determined using a semi-quantitative PCR method. The hypoxanthine-guanine phosphoribosyltransferase (Hprt) gene was used as loaning control. PCR cycle number for Ubqln1 and Hprt were 26 and 20, respectively, which were tested to be within the exponential range. (D) Western blot analyses of UBQLN1 in mouse developing testes. ACTIN was used as a loading control.
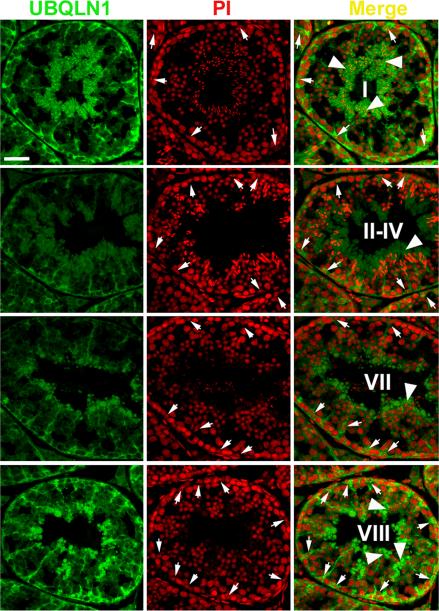
We then performed immunofluorescent microscopy to reveal the localization of UBQLN1 in the mouse testes (Fig. 4). The specificity of the UBQLN1 antibody used was evaluated by incubating the antibody with excessive amount of antigen (100 times more molecules of polypeptides used for immunization than IgG) followed by Western blot and immunofluorescent analyses, results of which showed the disappearance of the specific bands and signals (data not shown). UBQLN1 is mainly localized to the cytoplasm of Sertoli cells and elongating/elongated spermatids. The tree-like patterns of the green fluorescent signals reflects the boundary and the shape of the cytoplasm of Sertoli cells, which extends extensively from the basal membrane to the lumen of the seminiferous tubules (Fig.4). Immunoreactivity of UBQLN1 was also high in the cytoplasm of late spermatids (arrowheads in Fig. 4) and UBQLN1 was also detected in residual bodies in stage VIII tubules, in which spermiation was occurring. A closer look suggested that UBQLN1 may also be localized to the manchette of the elongating spermatids (see below).
Fig. 4.
Immunofluorescent detection of UBQLN1 in the adult mouse testes. Green signals represent the immunoreactivity of UBQLN1 and the cell nuclei were counter-stained with propidium iodide (PI). Stages of the seminiferous epithelial cycle were shown in Roman numerals. Arrows point to the distinct, usually double-dotted nucleoli of Sertoli cells. Arrow heads indicate the signals of UBQLN1 immunoreactivity in the cytoplasm of elongating and elongated spermatids. All panels are in the same magnification and the scale bar represents 20 μm.
3.3. Both SPEM1 and UBQLN1 are localized to the manchette of elongating spermatids
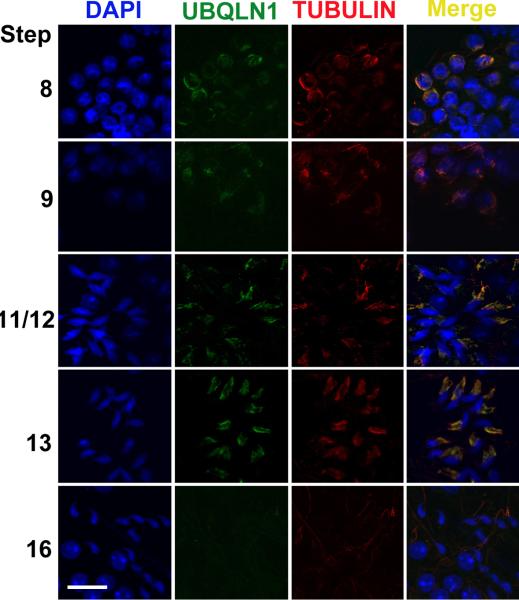
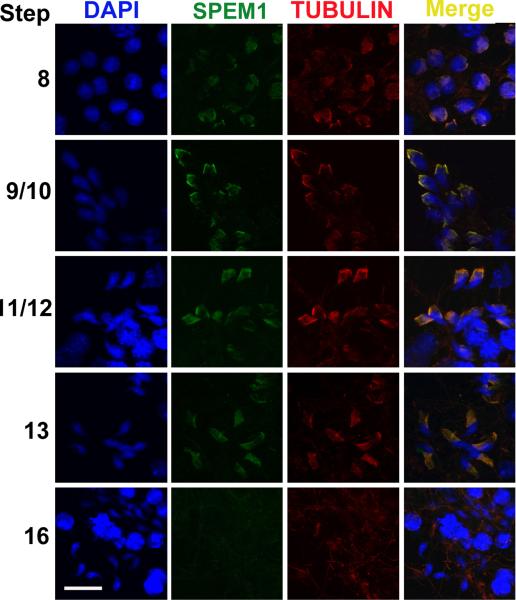
To further examine the subcellular localization of UBQLN1, we dissected seminiferous tubules from defined stages and the stage-defined seminiferous tubule fragments were squashed onto slides to generate a monolayer of spermatogenic cells followed by immunofluorescent staining using antibodies against UBQLN1 and β-Tubulin (a maker for the manchette of elongating spermatids) (Cole et al., 1988; Kierszenbaum, 2002b; Mochida et al., 1999; Rivkin et al., 1997)(Figs. 5-6). The manchette is an apparatus which develops in step 8 spermatids and disappears in step 15 spermatids (Cole et al., 1988; Meistrich et al., 1990; Mochida et al., 1999; Russell et al., 1991). It is mainly composed of F actins and microtubules and its function remains largely unknown (Kierszenbaum, 2002a; Kierszenbaum et al., 2007). UBQLN1 is colocalized with β-Tubulin to the manchette of elongating spermatids (Fig. 5). Our previous data have shown that SPEM1 is also localized to the cytoplasm of elongated spermatids (Zheng et al., 2007). To examine whether SPEM1 is also localized to the manchette, we investigated SPEM1 expression in elongating spermatids of wild-type (Fig. 6). Similar to UBQLN1, SPEM1 is indeed localized to the manchette of elongating spermatids (Fig. 6). In Spem1-null spermatids, SPEM1 was not detected (Supplemental Fig. 1), confirming the specificity of the SPEM1 antibody used. In the Spem1-null elongating spermatids, the manchette is present during the correct developmental window (between steps 8-13) as revealed by β-Tubulin staining (Supplemental Figs 1 and 2). Interestingly, UBQLN1 remained to be detected on the manchette of Spem1-null elongating spermatids (steps 8-13) (Supplemental Fig. 2), suggesting that the lack of SPEM1 expression does not affect the localization of UBQLN1 to the manchette.
Fig. 5.
Immunofluorescent localization of UBQLN1 to the manchette of elongating spermatids. Green fluorescence represents the UBQLN1 immunoreactivity and red fluorescence indicates the immunoreactivity of β-Tubulin, a marker for the manchette. Cell nuclei were counterstained using DAPI. All panels are in the same magnification. Scale bar = 10μm.
Fig. 6.
Immunofluorescent localization of SPEM1 to the manchette of elongating spermatids. Green fluorescence represents the SPEM1 immunoreactivity and red fluorescence reflects the immunoreactivity of β-Tubulin, a marker for the manchette. Cell nuclei were counterstained using DAPI. All panels are in the same magnification. Scale bar = 10μm.
3.4. UBQLN1 is upregulated in Spem1- knockout testes
Since UBQLN1 interacts with SPEM1 and both proteins are colocalized to the manchette of developing spermatids, we further examined levels of UBQLN1 in SPEM1 knockout testes using Western blot analyses (Fig. 7). At light microscopic levels, the adult Spem1-null testes are indistinguishable from that of the adult wild-type or Spem1+/- mice (Zheng et al., 2007). The cellular composition among testes of these three genotypes is therefore comparable and Western blot-based protein quantification should reveal the differences in UBQLN1 levels. In Spem1-null testes, UBQLN1 levels were almost doubled comparing to those in the WT testes. The upregulation of UBQLN1 in the Spem1-null testes may represent a compensatory effect due to the loss of SPEM1, as has been shown in many cases where levels of one protein are upregulated when its interacting partners are absent (Matzuk and Lamb, 2002, 2008; Yan, 2009). This finding is in agreement with the fact that UBQLN1interacts with SPEM1.
Fig. 7.
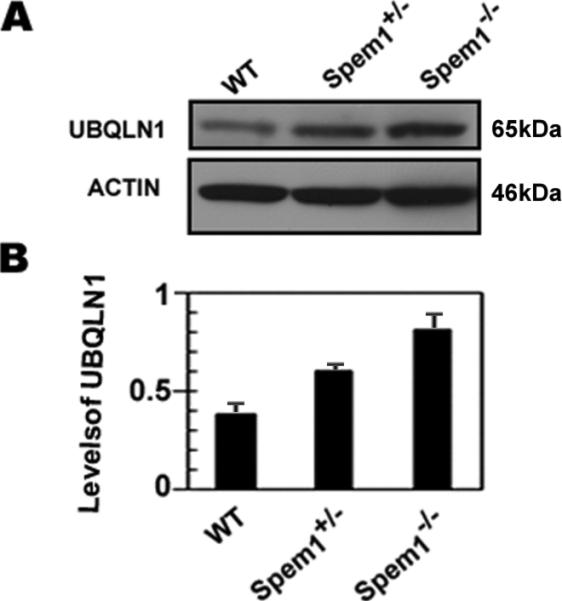
Upregulation of UBQLN1 levels in the Spem1-/- testes. (A) A representative result of Western blot analyses of UBQLN1 levels in adult wild-type (WT), Spem1+/- and Spem1-/- testes. ACTIN was used as a loading control. (B) Quantitative analyses of the Western blot results on UBQLN1 levels in WT, Spem1+/- and Spem1-/- testes. Bars represent mean ± SEM (n=3).
4. Discussion
Spermiogenesis represents a dramatic cellular morphogenesis, through which round-shaped spermatids undergo multiple steps of differentiation and are transformed into tadpole-shaped spermatozoa, the final male gametes. Earlier studies have estimated that up to 1,000 proteins are involved in this process (Schultz et al., 2003; Shima et al., 2004). The orderly appearance and disappearance of the structural proteins and the quality control of these proteins have to be tightly regulated by eliminating unwanted and misfolded proteins via the ubiquitin-proteasome pathway (Su and Lau, 2009; Sutovsky, 2003). Ubiquitin is a small and highly conserved protein that can covalently modify proteins and thereby mark them for destruction by the 26S proteasome (Su and Lau, 2009; Zhang and Saunders, 2009). The process is termed ubiquitination, which has been found to be involved in the regulation of many cellular processes. Ubiquitination is achieved through multiple steps of ubiquitin ligation. Ubiquitin is first activated by an ubiquitin activating enzyme (E1) and the activated ubiquitin is then transferred to an ubiquitin conjugating enzyme (E2). An ubiquitin ligase (E3) usually associates with both the substrate protein and E2, thereby facilitating the covalent attachment of one or more ubiquitin molecules to a surface lysine of a target protein via an isopeptide linkage (Madura, 2002; Su and Lau, 2009). The attachment of ubiquitin can be in a form of single or multiple mono-ubiquitin signals, or in the form of a poly-ubiquitin chain consisting of several ubiquitin molecules. The length of the chain and the lysine residue involved in the isopeptide linkage between two successive ubiquitin molecules determine the outcome of the ubiquitin signals. For example, K48-linked poly-ubiquitin (>4 ubiquitin) attached to a substrate protein marks an efficient signal for proteasome degradation, whereas K63-linked chains affect DNA repair and NFk-B signal transduction (Chai et al., 2004; Wang et al., 2008).
Ubiquilins belong to a protein family that is conserved among all eukaryotes examined to date (Zhang and Saunders, 2009). These proteins contain an ubiquitin-like (UBL) domain at its N-terminus, a central domain containing short repeats, and an ubiquitin-associated domain (UBA) at its C-terminus (Zhang and Saunders, 2009). Ubiquilin proteins have been reported to interact with numerous proteins that do not appear to have any functional relations (Ficklin et al., 2005; Ford and Monteiro, 2006; Ko et al., 2002; Regan-Klapisz et al., 2005; Thomas et al., 2006). It is currently believed that UBQLN1 may function as a shuttle factor acting to deliver poly-ubiquitinated proteins to the proteasome for degradation (Zhang and Saunders, 2009). In this setting, the UBA domain of ubiquilin binds poly-ubiquitin chains of the ubiquitinated protein and the UBL domain binds the S5a subunit of the 26S proteasome (Wilkinson et al., 2001).
Interactions between UBQLN1 and SPEM1 suggest that they are part of the ubiquitin-proteasome (UBP) system active in spermiogenesis. Their co-localization to the manchette is indicative of a potential role in late spermiogenesis. The manchette is a structure developed only in elongating spermatids (steps 8-13 spermatids) (Cole et al., 1988; Meistrich et al., 1990; Mochida et al., 1999; Russell et al., 1991). Given its microtubule-enriched nature, it has been suggested that the manchette may play a role in shaping the head of sperm through the physical forces generated by the microtubules of the manchette and the acroplaxome, which is a cytoskeletal plate linking the acrosome to the spermatid nuclear envelop, in conjunction with the exogenous forces from Sertoli cells (Kierszenbaum, 2002a; Kierszenbaum and Tres, 2004). This notion is partially supported by the fact that several mutant mouse lines with abnormal manchettes also display deformed head shapes. Another potential function of the manchette as a transporting devise has been suggested based upon the fact that many motor proteins (i.e. kinesins and dyneins), or nuclear-cytoplasmic shuttle proteins (Ran-GTP) are localized to the manchette (Kierszenbaum, 2002a). One hypothesis is that the manchette is responsible for shipping structural proteins to the centrosome and developing tail. Another hypothesis is that the manchette represents a more efficient way of transporting large quantity of proteins in and out of the condensing nucleus. The latter hypothesis is plausible because the time window during which the manchette is developed coincides with spermatid nuclear elongation and condensation (Kierszenbaum et al., 2007; Rivkin et al., 2009). Proteins required for these processes have to be transported into the nucleus and unwanted and/or defective proteins have to be shipped out of the nucleus (Kierszenbaum, 2002b). These two hypotheses are not mutually exclusive because in many mutant mice (e.g. Tnp1-/-/Tnp2-/-, Spem1-/-, Azh-/- and Hrb-/-), sperm head deformation and tail abnormalities co-exist (Kierszenbaum, 2002a, b; Kierszenbaum and Tres, 2004; Meistrich et al., 1990; Yan, 2009).
On one hand, the manchette is implicated in the delivery of building blocks for developing spermatids, as discussed above. On the other hand, the newly synthesized proteins have to be folded into the correct conformation. Misfolded proteins have to be eliminated because they are toxic and may have adverse effects on late spermiogenesis. The ubiquitin-proteasome (UBP) system is a very efficient means to eliminate unwanted or misfolded proteins. Studies by Kierszenbaum and colleagues have shown that the UBP system is indeed localized to the manchette (Mochida et al., 2000; Rivkin et al., 1997; Rivkin et al., 2009). An ubiquitin protein ligase (E3) called RNF19A and PSMC3, a component of the 26S proteasome, are recently found to be colocalized to the manchette (Rivkin et al., 2009). The existence of the UPB system on the manchette suggests that it may also has a function through degrading unwanted protein and/or monitoring the quality of the newly synthesized structural proteins for the developing spermatids. The manchette is thus involved in the control of both protein turnover and protein quality. It is therefore not surprising to find UBQLN1in the developing spermatids. Its localization to the manchette suggests that it participates in the UBP pathway by binding to ubiquitinated proteins and leading them to the proteasome for destruction. Its interaction with SPEM1 suggests that SPEM1 is involved in UBQLN1-mediated ubiquitination events. SPEM1is exclusively expressed in the cytoplasm of elongating/elongated spermatids (Zheng et al., 2007). Subcellularly SPEM1 and UBQLN1 directly interact with each other and are colocalized to the manchette. Given that the UBP system does exist within the manchette and UBQLN1 functions as a shuttle factor to deliver poly-ubiquitinated proteins to proteasome for degradation (Su and Lau, 2009; Zhang and Saunders, 2009), the SPEM1 may thus serve as a scaffold protein that can introduce ubiquitinated unwanted/defective proteins to UBQLN1, which then delivers these proteins to the proteasome for degradation. In the absence of SPEM1, many proteins destined to be degraded by the UBP system may remain, causing conflicting signals leading to aberrant development of late spermatids and abnormal cytoplasm removal. Further investigation is required to understand the “head-bent-back” deformation of Spem1-null sperm and to elucidate the physiological roles of the SPEM1-UBQLN1 complex in spermiogenesis.
Supplementary Material
Acknowledgement
This work was supported by a grant from the NIH (HD050281) and by the University of Nevada School of Medicine Startup Fund to W.Y.. Dr. John R. McCarrey is acknowledged for providing us with the purified testicular cells. C.X. was supported by a grant for the Shanghai Leading Academic Discipline Project (Project#: S30201). J.B. was supported by a scholarship from the Chinese Scholarship Council.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen KM, Hofmann K, Hartmann-Petersen R. Ubiquitin-binding proteins: similar, but different. Essays Biochem. 2005;41:49–67. doi: 10.1042/EB0410049. [DOI] [PubMed] [Google Scholar]
- Chai Y, Berke SS, Cohen RE, Paulson HL. Poly-ubiquitin binding by the polyglutamine disease protein ataxin-3 links its normal function to protein surveillance pathways. J Biol Chem. 2004;279:3605–3611. doi: 10.1074/jbc.M310939200. [DOI] [PubMed] [Google Scholar]
- Clermont Y. Kinetics of spermatogenesis in mammals: seminiferous epithelium cycle and spermatogonial renewal. Physiol Rev. 1972;52:198–236. doi: 10.1152/physrev.1972.52.1.198. [DOI] [PubMed] [Google Scholar]
- Cole A, Meistrich ML, Cherry LM, Trostle-Weige PK. Nuclear and manchette development in spermatids of normal and azh/azh mutant mice. Biol Reprod. 1988;38:385–401. doi: 10.1095/biolreprod38.2.385. [DOI] [PubMed] [Google Scholar]
- Conklin D, Holderman S, Whitmore TE, Maurer M, Feldhaus AL. Molecular cloning, chromosome mapping and characterization of UBQLN3 a testis-specific gene that contains an ubiquitin-like domain. Gene. 2000;249:91–98. doi: 10.1016/s0378-1119(00)00122-0. [DOI] [PubMed] [Google Scholar]
- Cooke HJ, Saunders PT. Mouse models of male infertility. Nat Rev Genet. 2002;3:790–801. doi: 10.1038/nrg911. [DOI] [PubMed] [Google Scholar]
- Esponda P. Spermiogenesis and spermatozoa in mammals. Revis Biol Celular. 1985;6:1–99. [PubMed] [Google Scholar]
- Ficklin MB, Zhao S, Feng G. Ubiquilin-1 regulates nicotine-induced up-regulation of neuronal nicotinic acetylcholine receptors. J Biol Chem. 2005;280:34088–34095. doi: 10.1074/jbc.M506781200. [DOI] [PubMed] [Google Scholar]
- Ford DL, Monteiro MJ. Dimerization of ubiquilin is dependent upon the central region of the protein: evidence that the monomer, but not the dimer, is involved in binding presenilins. Biochem J. 2006;399:397–404. doi: 10.1042/BJ20060441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszenbaum AL. Intramanchette transport (IMT): managing the making of the spermatid head, centrosome, and tail. Mol Reprod Dev. 2002a;63:1–4. doi: 10.1002/mrd.10179. [DOI] [PubMed] [Google Scholar]
- Kierszenbaum AL. Sperm axoneme: a tale of tubulin posttranslation diversity. Mol Reprod Dev. 2002b;62:1–3. doi: 10.1002/mrd.10139. [DOI] [PubMed] [Google Scholar]
- Kierszenbaum AL, Rivkin E, Tres LL. Molecular biology of sperm head shaping. Soc Reprod Fertil Suppl. 2007;65:33–43. [PubMed] [Google Scholar]
- Kierszenbaum AL, Tres LL. The acrosome-acroplaxome-manchette complex and the shaping of the spermatid head. Arch Histol Cytol. 2004;67:271–284. doi: 10.1679/aohc.67.271. [DOI] [PubMed] [Google Scholar]
- Ko HS, Uehara T, Nomura Y. Role of ubiquilin associated with protein-disulfide isomerase in the endoplasmic reticulum in stress-induced apoptotic cell death. J Biol Chem. 2002;277:35386–35392. doi: 10.1074/jbc.M203412200. [DOI] [PubMed] [Google Scholar]
- Madura K. The ubiquitin-associated (UBA) domain: on the path from prudence to prurience. Cell Cycle. 2002;1:235–244. [PubMed] [Google Scholar]
- Mah AL, Perry G, Smith MA, Monteiro MJ. Identification of ubiquilin, a novel presenilin interactor that increases presenilin protein accumulation. J Cell Biol. 2000;151:847–862. doi: 10.1083/jcb.151.4.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk MM, Lamb DJ. Genetic dissection of mammalian fertility pathways. Nat Cell Biol. 2002;4(Suppl):s41–49. doi: 10.1038/ncb-nm-fertilityS41. [DOI] [PubMed] [Google Scholar]
- Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med. 2008;14:1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meistrich ML, Trostle-Weige PK, Russell LD. Abnormal manchette development in spermatids of azh/azh mutant mice. Am J Anat. 1990;188:74–86. doi: 10.1002/aja.1001880109. [DOI] [PubMed] [Google Scholar]
- Mochida K, Tres LL, Kierszenbaum AL. Structural and biochemical features of fractionated spermatid manchettes and sperm axonemes of the azh/azh mutant mouse. Mol Reprod Dev. 1999;52:434–444. doi: 10.1002/(SICI)1098-2795(199904)52:4<434::AID-MRD13>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Mochida K, Tres LL, Kierszenbaum AL. Structural features of the 26S proteasome complex isolated from rat testis and sperm tail. Mol Reprod Dev. 2000;57:176–184. doi: 10.1002/1098-2795(200010)57:2<176::AID-MRD9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Regan-Klapisz E, Sorokina I, Voortman J, de Keizer P, Roovers RC, Verheesen P, Urbe S, Fallon L, Fon EA, Verkleij A, et al. Ubiquilin recruits Eps15 into ubiquitin-rich cytoplasmic aggregates via a UIM-UBL interaction. J Cell Sci. 2005;118:4437–4450. doi: 10.1242/jcs.02571. [DOI] [PubMed] [Google Scholar]
- Rivkin E, Cullinan EB, Tres LL, Kierszenbaum AL. A protein associated with the manchette during rat spermiogenesis is encoded by a gene of the TBP-1-like subfamily with highly conserved ATPase and protease domains. Mol Reprod Dev. 1997;48:77–89. doi: 10.1002/(SICI)1098-2795(199709)48:1<77::AID-MRD10>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Rivkin E, Kierszenbaum AL, Gil M, Tres LL. Rnf19a, a ubiquitin protein ligase, and Psmc3, a component of the 26S proteasome, tether to the acrosome membranes and the head-tail coupling apparatus during rat spermatid development. Dev Dyn. 2009;238:1851–1861. doi: 10.1002/dvdy.22004. [DOI] [PubMed] [Google Scholar]
- Russell LD, Russell JA, MacGregor GR, Meistrich ML. Linkage of manchette microtubules to the nuclear envelope and observations of the role of the manchette in nuclear shaping during spermiogenesis in rodents. Am J Anat. 1991;192:97–120. doi: 10.1002/aja.1001920202. [DOI] [PubMed] [Google Scholar]
- Schultz N, Hamra FK, Garbers DL. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc Natl Acad Sci U S A. 2003;100:12201–12206. doi: 10.1073/pnas.1635054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- Song R, Ro S, Michaels JD, Park C, McCarrey JR, Yan W. Many X-linked microRNAs escape meiotic sex chromosome inactivation. Nat Genet. 2009;41:488–493. doi: 10.1038/ng.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su V, Lau AF. Ubiquitin-like and ubiquitin-associated domain proteins: significance in proteasomal degradation. Cell Mol Life Sci. 2009;66:2819–2833. doi: 10.1007/s00018-009-0048-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutovsky P. Ubiquitin-dependent proteolysis in mammalian spermatogenesis, fertilization, and sperm quality control: killing three birds with one stone. Microsc Res Tech. 2003;61:88–102. doi: 10.1002/jemt.10319. [DOI] [PubMed] [Google Scholar]
- Thomas AV, Herl L, Spoelgen R, Hiltunen M, Jones PB, Tanzi RE, Hyman BT, Berezovska O. Interaction between presenilin 1 and ubiquilin 1 as detected by fluorescence lifetime imaging microscopy and a high-throughput fluorescent plate reader. J Biol Chem. 2006;281:26400–26407. doi: 10.1074/jbc.M601085200. [DOI] [PubMed] [Google Scholar]
- Wang H, Matsuzawa A, Brown SA, Zhou J, Guy CS, Tseng PH, Forbes K, Nicholson TP, Sheppard PW, Hacker H, et al. Analysis of nondegradative protein ubiquitylation with a monoclonal antibody specific for lysine-63-linked polyubiquitin. Proc Natl Acad Sci U S A. 2008;105:20197–20202. doi: 10.1073/pnas.0810461105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zheng H, Esaki Y, Kelly F, Yan W. Cullin3 is a KLHL10-interacting protein preferentially expressed during late spermiogenesis. Biol Reprod. 2006;74:102–108. doi: 10.1095/biolreprod.105.045484. [DOI] [PubMed] [Google Scholar]
- Wilkinson CR, Seeger M, Hartmann-Petersen R, Stone M, Wallace M, Semple C, Gordon C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- Wolgemuth DJ, Rhee K, Wu S, Ravnik SE. Genetic control of mitosis, meiosis and cellular differentiation during mammalian spermatogenesis. Reprod Fertil Dev. 1995;7:669–683. doi: 10.1071/rd9950669. [DOI] [PubMed] [Google Scholar]
- Wu S, Mikhailov A, Kallo-Hosein H, Hara K, Yonezawa K, Avruch J. Characterization of ubiquilin 1, an mTOR-interacting protein. Biochim Biophys Acta. 2002;1542:41–56. doi: 10.1016/s0167-4889(01)00164-1. [DOI] [PubMed] [Google Scholar]
- Yan W. Male infertility caused by spermiogenic defects: lessons from gene knockouts. Mol Cell Endocrinol. 2009;306:24–32. doi: 10.1016/j.mce.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Suominen J, Toppari J. Stem cell factor protects germ cells from apoptosis in vitro. J Cell Sci. 2000;113(Pt 1):161–168. doi: 10.1242/jcs.113.1.161. [DOI] [PubMed] [Google Scholar]
- Zhang C, Saunders AJ. An emerging role for Ubiquilin 1 in regulating protein quality control system and in disease pathogenesis. Discov Med. 2009;8:18–22. [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Stratton CJ, Morozumi K, Jin J, Yanagimachi R, Yan W. Lack of Spem1 causes aberrant cytoplasm removal, sperm deformation, and male infertility. Proc Natl Acad Sci U S A. 2007;104:6852–6857. doi: 10.1073/pnas.0701669104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.