Abstract
Background
Fetal Alcohol Spectrum Disorder (FASD) is a set of developmental defects caused by prenatal alcohol exposure. Clinical manifestations of FASD are highly variable and include mental retardation and developmental defects of the heart, kidney, muscle, skeleton, and craniofacial structures. Specific effects of ethanol on fetal cells include induction of apoptosis as well as inhibition of proliferation, differentiation, and migration. This complex set of responses suggests that a bioinformatics approach could clarify some of the pathways involved in these responses.
Methods
In this study, the responses of fetal stem cells derived from the amniotic fluid (AFSCs) to treatment with ethanol have been examined. Large-scale transcriptome analysis of ethanol-treated AFSCs indicates that genes involved in skeletal development and ossification are up-regulated in these cells. Therefore, the effect of ethanol on osteogenic differentiation of AFSCs was studied.
Results
Exposure to ethanol during the first 48 hours of an osteogenic differentiation protocol increased in vitro calcium deposition by AFSCs and increased alkaline phosphatase activity. In contrast, ethanol treatment later in the differentiation protocol (day 8) had no significant effect on the activity of alkaline phosphatase.
Conclusions
These results suggest that transient exposure of AFSCs to ethanol during early differentiation enhances osteogenic differentiation of the cells.
Introduction
Maternal alcohol (ethanol) consumption during pregnancy may cause abnormal growth and morphogenesis of the conceptus (Astley and Clarren, 2000). The spectrum of defects caused by maternal alcohol consumption is known as Fetal Alcohol Spectrum Disorder (FASD) and occurs in approximately 0.5–2.0 percent of all live births in the United States (Centers for Disease Control and Prevention, 2002). At the severe end of the spectrum of alcohol-induced defects is Fetal Alcohol Syndrome (FAS). Affected individuals typically have mild to moderate mental retardation, growth deficiencies, and craniofacial defects (Habbick, Blakley et al., 1998;Johnson, Swayze, II et al., 1996;Lemoine, Harousseau et al., 2003). Prenatal alcohol exposure can also result in defects in multiple organs and tissues, including the heart, eyes, kidneys, and skeleton (Becker, az-Granados et al., 1996;Herrmann, Pallister et al., 1980;Parnell, Dehart et al., 2006;Randall, Taylor et al., 1977;Sulik, Johnston et al., 1981).
Considering that, during embryonic and fetal life, various types of stem cells are prevalent and critical for normal development (Rice and Barone S Jr, 2000), and that interference with their viability or function may represent a little-explored, yet potentially significant teratogenic pathology, the current study has employed human amniotic fluid-derived stem cells (AFSCs) as a model to study the effects of ethanol on stem cell differentiation. AFSCs are multipotent and have extensive self-renewal potential. They are capable of differentiating into bone, muscle, fat, endothelium, liver, and neuron-like cells in vitro (De Coppi, Bartsch et al., 2007).
Ethanol has many reported teratogenic mechanisms of actions including induction of apoptosis and inhibition of proliferation, differentiation, migration and other cellular functions (Gong and Wezeman, 2004;Li, Lin et al., 2001;Miller, Chiaia et al., 1990;Siegenthaler and Miller, 2004). Additionally, ethanol exposure affects membrane-associated receptor signaling pathways (Resnicoff, Sell et al., 1993) and cell adhesion (Charness, Safran et al., 1994;Vangipuram, Grever et al., 2008), yields free radical-mediated damage (Chen and Sulik, 1996), and alters the binding of transcription factors (Pignataro, Miller et al., 2007). Recognizing this wide range of cellular effects, initial examination of ethanol-mediated insult to AFSCs entailed global gene expression analysis. The results of this work indicate that genes involved in osteogenesis are consistently upregulated in AFSCs exposed to ethanol during early differentiation. An in vitro osteogenic differentiation protocol was subsequently employed to examine the potential effects of ethanol on this process. The data suggest that ethanol exposure during the uncommitted stage lineage restricts AFSCs to osteogenesis.
Materials and Methods
Ethanol Treatment
Human AFSCs were maintained as described previously (De Coppi, Bartsch et al., 2007). They were grown in α-MEM medium (Gibco, Invitrogen, Carlsbad, CA) containing 15% ES-FBS, 1% glutamine, and 1% penicillin/streptomycin (Gibco), supplemented with 18% Chang B and 2% Chang C (Irvine Scientific, Santa Ana, CA). The cells were kept at 37 °C in an atmosphere containing 5% CO2. For growth and viability studies, the AFSCs were treated with 25 mM, 50 mM, 75 mM, or 100 mM ethanol (Sigma-Aldrich, St. Louis, Missouri) and the plates were sealed with Parafilm. Media was replaced every 24 hours. The ethanol concentrations used in these experiments are equivalent to the blood alcohol concentrations achieved by social drinkers to chronic alcoholics (Adachi, Mizoi et al., 1991;Perper, Twerski et al., 1986). They have been utilized in previous in vitro experiments (Chen and Sulik, 1996). The duration of ethanol exposure (48 hr) was determined to induce the maximum effect without causing toxicity. In addition, a study on the disposition of ethanol in the amniotic fluid and maternal blood in early 2nd trimester females showed a delay in the clearance of ethanol from the amniotic fluid (Brien et al., 1983). This suggests that ethanol serves as a reservoir ethanol and that the duration of exposure of ethanol to the fetus may be longer than previously thought. Ethanol concentrations were measured spectrophotometrically using an Ethanol L3K assay (Diagnostic Chemicals Limited, Oxford CT) according to the manufacturer’s instructions.
Cell number and viability
Ethanol was added to the culture media when AFSC reached 3,000 cells/cm2 and the 6 well plates were sealed with Parafilm to prevent evaporation. Untreated cells served as controls. Cell counts were made using a Coulter counter after 48 hours of exposure to the various ethanol concentrations. Cell viability was determined by propidium iodine (PI) exclusion. Briefly, AFSCs were dissociated using a solution of 0.05% trypsin/EDTA (Gibco), centrifuged at 1,500 RPM for 5 minutes, and resuspended in 1 ml of PBS in 15 ml polypropylene tubes. 50 μl of PI, a DNA intercalating agent, was added to each tube followed by incubation for 1 hour at room temperature. Cells were then centrifuged, washed twice with phosphate buffered saline and analyzed by flow cytometry using a FACSCalibur analyzer (BD Biosciences, San Jose, CA) and the FL-1 channel.
RNA Isolation and Real-Time Quantitative PCR
Total RNA was isolated from the AFSCs using the PerfectPure RNA Cultured Cell Kit (5 Prime, Gaithersburg, MD) according to the manufacturer’s protocol. A DNA digestion step was included to eliminate contamination by genomic DNA. The quality of total RNA was assessed by measuring the A260/280 ratio using a spectrophotometer. For the reverse-transcriptase reaction, SuperscriptII reverse transcription reagents (Invitrogen, Carlsbad, CA) were used. Briefly, 1 mg of RNA was converted to cDNA, and PCR amplification of this DNA was performed using the TaqMan Universal Master Mix (Applied Biosystems). Reactions were performed in duplicate and consisted of 1 ml of cDNA, 1.25 ml probe, 12 ml Master Mix, and 10 ml DI water. These reactions were performed in a 96-well optical reaction plate (Applied Biosystems, Foster City, CA). Reactions were amplified and quantified using an ABI 7700 sequence detector and the manufacturer’s software (Applied Biosystems, Foster City, CA). On demand fluorescent probes were produced for the β-actin and osteopontin genes. The threshold cycle (Ct) indicates the fractional cycle number at which the amount of amplified target reaches a defined threshold. ΔCt was obtained by subtracting the Ct values of endogenous controls (β-actin) from the Ct values of the target genes.
Microarray Analysis
Two microarrays were performed on AFSCs cultured for 48 hours with or without 100 mM of ethanol. Fragmented antisense cRNA was used for hybridizing to human U133 A arrays (Affymetrix, Inc. Santa Clara, CA, USA) at the Core Genomic Facility of Wake Forest University School of Medicine. Raw CEL files were provided by the Microarray Core Facility of the Wake Forest University School of Medicine and were analyzed with AffylmGUI (Affymetrix LIMMA, Linear Models for Microarray Data, Graphical User Interfaces) (Wettenhall, Simpson et al., 2006;Wettenhall and Smyth, 2004). Within AffylmGUI, gene expression values were summarized with RMA. RMA adjusts for background noise, performs a quantile normalization, transforms the data into log base 2, and then summarizes the multiple probes into one intensity (Bolstad, Irizarry et al., 2003;Irizarry, Bolstad et al., 2003;Irizarry, Hobbs et al., 2003). Quantification of relative differences in gene expression among the groups of interest was accomplished using AffylmGUI (Wettenhall, Simpson et al., 2006;Wettenhall and Smyth, 2004). AffylmGUI reads the raw Affymetrix CEL files directly and summarizes the gene expression values using RMA. Differentially expressed genes were identified as those with a fold change > 1.8. The data discussed in this publication is deposited in NCBI’s Gene Expression Omnibus (GEO) and accessible through GEO series accession number GSE13569, in accordance with MIAME standards.
DAVID
To uncover enriched processes, data sets were analyzed with DAVID (Database for Annotation, Visualization and Integrated Discovery), a web-based tool that provides statistical methods for identifying over-represented biological themes and pathways within diverse and disparate gene lists (Dennis, Jr., Sherman et al., 2003). DAVID also identifies over-represented biological themes in terms of their Gene Ontology (GO) terms and provides tools to visualize the distribution of genes on BioCarta and KEGG pathway maps. (Ashburner, Ball et al., 2000). GO provides consistent descriptions of genes in terms of biological processes and molecular function. Gene-enrichment analysis computes a modified Fisher exact p-value by comparing the ontological themes identified in our data set to total possible ontological processes present on the U133A chip. Those ontological processes that had a p-value of less than 0.05 were selected.
Osteogenic Induction
Human AFSCs were induced to differentiate into osteogenic cell types as described previously (De Coppi, Bartsch et al., 2007). As described in De Coppi et al., AFSCs develop into osteoblast-like morphology within 1 week of differentiation. By sixteen days, they form bone-like lamellar structures. Furthermore, AFSCs express mRNA and protein for alkaline phosphatase after one week of osteogenic differentiation. Functional assays for calcium deposition show strong histological staining by alizarin red. They also show strong histochemical staining for alkaline phosphatase and secrete this enzyme(De Coppi, Bartsch et al., 2007). Briefly, AFSCs were cultured in low glucose DMEM containing 10% FBS and supplemented with 100nM dexamethasone (Sigma-Aldrich, St. Louise, MO), 10mM b-glycerophosphate (Sigma-Aldrich, St Louise, MO) and 0.05mM ascorbic acid-2-phosphate (Wako Chemicals, Irving, TX). Because osteogenic differentiation requires quiescence and contact inhibition, we plated AFSC at a higher density (Stein, Lian et al., 1989). Cells were grown to a higher density, 6,000 cells/cm2, and then treated with ethanol for the first 48 hours of osteogenic differentiation. Cell number was determined after 24 and 48 hours of ethanol treatment and osteogenic differentiation. Ethanol had no effect on cell number when cells were exposed to osteogenic media (Supplemental Figure 1). Bone differentiation was analyzed by measurement of expression of osteopontin and bone-specific alkaline phosphatase mRNAs, measurement of alkaline phosphatase activity, and with Alizarin Red to quantify extracellular calcium deposition at Day 23 of differentiation.
Alizarin Red Staining
The presence of calcium in the cell cultures was determined by alizarin red (Sigma) staining at day 23 of osteogenic differentiation. Cells were fixed with 70% ethanol for 15 min. Fixed cells were incubated with 0.5% alizarin red solution in water (pH adjusted to 4.0) for 1 minute and then washed three times with deionized water and once with 70% ethanol, then allowed to dry. The alizarin red stain was extracted with 100 mM cetylpyridinium chloride (Sigma-Aldrich, St Louise, MO) at room temperature for three hours. The absorbance of the extracted alizarin red stain was measured at 540nm. The concentration of alizarin red staining in the samples was determined by comparing the absorbance values with those obtained from an alizarin red standard curve.
Alkaline Phosphatase Activity
Alkaline phosphatase activity was measured using a p-nitrophenyl phosphate liquid substrate system (Sigma). Cells grown in 24-well plates were rinsed with PBS and incubated with 0.15% Triton X-100 for 30 mins. Subsequently, 200 ml of p-nitrophenyl phosphate (pNPP) solution (ph = 8.8) were added to the Triton-X 100 solution. The pNPP cleavage rate was demonstrated to be linear in all samples after 30 minutes, 1 hour, and 2 hours of incubation. Cells were incubated in the dark for 1 hour and absorbance was measured at 405 nm using a spectrophotometer.
Data Analysis
Results are expressed as mean ± standard deviation (S.D.) for quantitative data. Analysis of Variance (ANOVA) was used to identify statistically significant differences between groups. Alternatively, two-tailed tests of significance were computed to determine relationships between ethanol-treated and control groups. Statistical significance was defined as p < 0.05.
Results
Effects of ethanol on number and viability of AFSCs
When AFSCs were exposed to ethanol for 48 hours, a dose-dependent reduction in cell number was observed (Figure 1A). The dose range for these studies was chosen to reflect a physiologically relevant range of blood alcohol concentrations (from a “legal” blood alcohol level to levels found in chronic alcoholics) (Adachi, Mizoi et al., 1991;Perper, Twerski et al., 1986). When cells reached a density of 60,000 cells/well, ethanol (25mM–100mM) was added to the growth media. After 48 hr of ethanol exposure, cell counts were made. AFSC that were not exposed to ethanol grew from 60,000 to 151,000 cells while cells exposed to 25mM, 50mM, 75mM and 100mM grew to 118,080, 118,480, 107,560, and 100,560 cells, respectively (Supplement Table 1). This result indicates that AFSC were growing in the presence of ethanol but at a slower rate. Thus, in all ethanol concentrations tested AFS cells continued to proliferate however, their proliferation was slower than cells grown in the absence of ethanol. To determine if the ethanol-induced reduction in cell number was due to cell death, we examined the effect of ethanol exposure on cell viability using propidium iodide exclusion (Figure 1B). Cultures of AFSCs that had not been exposed to ethanol had an average baseline level of 10.3% ± 4.7% non-viable cells. The percentage of non-viable cells in cultures that were exposed to ethanol concentrations ranging from 25mM to 100mM had averages of 8.2% ± 2.8% to 10.1% ± 3.6% non-viable cells. These data indicate that ethanol does not have a significant effect on cell viability and suggests that the observed reduction in the number of AFSCs is a result of a reduced proliferation rate.
Figure 1. Effect of ethanol on cell proliferation and viability.
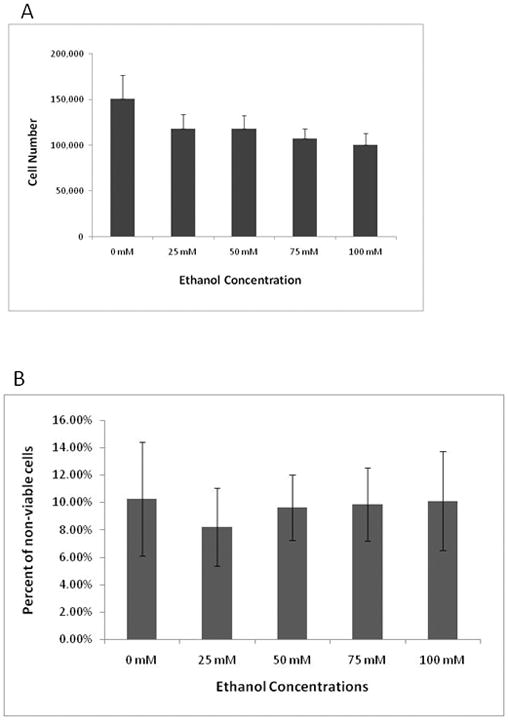
AFSCs were cultured with 0, 25, 50, 75, and 100 mM of ethanol. (A) Cell counts were made after 48 hours of ethanol exposure and compared to counts of cultures that were not treated with ethanol. The results are expressed as percentages relative to cells without ethanol (dark grey bars). The values shown are the mean +/−standard deviation (SD, n=5) of three independent experiments (*, p<0.03 by student T-test). (B) The effect of ethanol on the percentage of non-viable cells was determined by 7-AAD and flow cytometry. Light grey bars indicate the percentage of non-viable cells in the presence of various concentrations of ethanol. The data shown represents the mean number of non-viable cells in 10,000 events from two independent experiments. Error bars represent SDs. Data were not significant. (C) Ethanol concentration in the media was measured by spectrophotometry. Results represent the mean +/− SD from three replicates. (D) Ethanol treatment and osteogenic differentiation began when AFSC reached 60,000 cells. After 24 and 48 hours of 100 mM ethanol, cell counts were performed and showed no significant change in cell number. Ethanol does not have a proliferative effect on AFSC when cultured in osteogenic media.
The effect of ethanol on global gene expression
For large-scale transcriptome analysis, AFSCs were exposed to 100 mM ethanol for 48 hours in growth media. Ethanol concentration present in the culture media did not change over a period of 24 hours (Figure 1C). Standard growth media was used instead of a lineage-specific differentiation medium in order to prevent a bias toward identification of lineage-specific genes. To identify differentially expressed genes, Affymetrix GeneChips were used. The data sets were normalized and subjected to statistical analysis. Using this method, 65 genes that were up-regulated in response to ethanol and 19 genes that were down-regulated in response to ethanol were identified. To uncover enriched processes, data sets were analyzed by DAVID, a web-based tool that identifies over-represented biological themes in a data set based on their Gene Ontology (GO) terms. GO provides consistent descriptions of genes in terms of biological processes and molecular function.
DAVID analysis identified several processes that were up-regulated in response to ethanol, as compared to growth under normal conditions. These included processes involved in skeletal development (5 genes), ossification (5 genes), blood vessel development (4 genes), organ development (12 genes) and developmental processes (17 genes; Table 1). Up-regulated skeletal development genes included osteopontin, osteonectin, ectonucleotide pyrophosphatase, myocyte enhancer factor 2c, and matrix metallopeptidase 14. Osteopontin and osteonectin are secreted phosphoproteins expressed at the early stage of osteogenic differentiation. They have been shown to mediate cell-matrix interactions, cell adhesion, and differentiation (Butler, 1989;Delany, Kalajzic et al., 2003;Strauss, Closs et al., 1990). The increase in osteopontin is evidence of lineage commitment.
Table 1. Enrichment of Biological Themes of Ethanol-responsive Genes.
Genes were selected based on the gene ontologies. Gene ontologies with a modified Fisher Exact P-value < 0.05 were selected. Processes that were identified in genes that were up-regulated in response to 100mM ethanol include skeletal development (A) while genes that were down-regulated in response to ethanol included embryonic development (B).
| Table 1 (A): Enrichment of Biological Themes of Ethanol-responsive Genes | ||
|---|---|---|
| Pathway (p value) | Genes | Fold Change |
| Biomineral and ossification (2.87E-04) | Osteonectin | 18 |
| MMP14 | 18 | |
| Osteopontin | 21 | |
| MEF2C | 19 | |
| ENPP1 | 23 | |
| organmorphogenesis (2.18E-03) | COL3A | 2 |
| FOXF1 | 18 | |
| MMP14 | 18 | |
| NRF3 | 21 | |
| MEF2C | 19 | |
| BAX2 | 28 | |
| IGFR1 | 18 | |
| organ development (2.34E-03) | SPARC | 18 |
| COL3A | 2 | |
| FOXF1 | 18 | |
| MMP14 | 18 | |
| OPN | 21 | |
| NRF2 | 21 | |
| SOX4 | 2 | |
| PIK3R1 | 18 | |
| MEF2C | 19 | |
| ENPP1 | 23 | |
| BAX2 | 27 | |
| IGFR1 | 18 | |
| reproductive developmental process (3.42 E-03) | SRD5A1 | 18 |
| MMP14 | 18 | |
| BAX2 | 27 | |
| IGFR1 | 18 | |
| skeletal development (6.38E-03) | Osteonectin | 1.8 |
| MMP14 | 18 | |
| Osteopontin | 21 | |
| MEF2C | 19 | |
| ENPP1 | 23 | |
| blood vessel development (2.32E-02) | FOXF1 | 18 |
| MMP14 | 18 | |
| NRF2 | 21 | |
| MEF2C | 19 | |
| transmembrane receptor protein tyrosine kinase signaling pathway (2.51E-02) | SPARC | 18 |
| RGPS2 | 24 | |
| PIKRS1 | 18 | |
| IGFR1 | 18 | |
| Insulin-like growth factor receptor signaling pathway (2.89E-02) | PIKRS1 | 18 |
| IGFR1 | 18 | |
| Table 1 (B): Enrichment of Biological Themes of Ethanol-responsive Genes | ||
|---|---|---|
| Pathway (p value) | Genes | Fold Change |
| negative regulation of cellular processes (2.40E-03) | ADAMTS1 | −2.2 |
| NRG1 | −3.3 | |
| DLC1 | −2.2 | |
| FGF2 | −2 | |
| DKK1 | −1.8 | |
| circulatory system process (9.9E-03) | OXTR | −2.2 |
| NRG1 | −3.3 | |
| EDN1 | −1.8 | |
| glucose transport (2.8E-02) | NRG1 | −3.3 |
| EDN1 | −1.8 | |
| embryonic development (3.0E-02) | NRG1 | −3.3 |
| EDN1 | −1.8 | |
| DKK1 | −1.8 | |
| multi-organism process | OXTR | −2.2 |
| PTX3 | −1.8 | |
| EDN1 | −1.8 | |
| anatomical structure development (3.6E-02) | DLC1 | −2.2 |
| FGF2 | −2 | |
| DKK1 | −1.8 | |
| EDN1 | −1.8 | |
Genes that were down-regulated in response to ethanol were also organized by their gene ontology. The predominant pathway identified in this data set was embryonic development, which includes the genes encoding dickkopf homolog 1, neuregulin, and endothelin 1. Basic fibroblast growth factor (bFGF/FGF2) was also down-regulated in response to ethanol. bFGF is a potent mitogen and is an important factor in limb development and neurogenesis (Fallon, Lopez et al., 1994;Raballo, Rhee et al., 2000). The down-regulation of genes associated with these pathways suggests that ethanol restricts the differentiation potential of AFSCs and may interfere with proper embryonic and fetal development.
The effect of ethanol on osteopontin expression
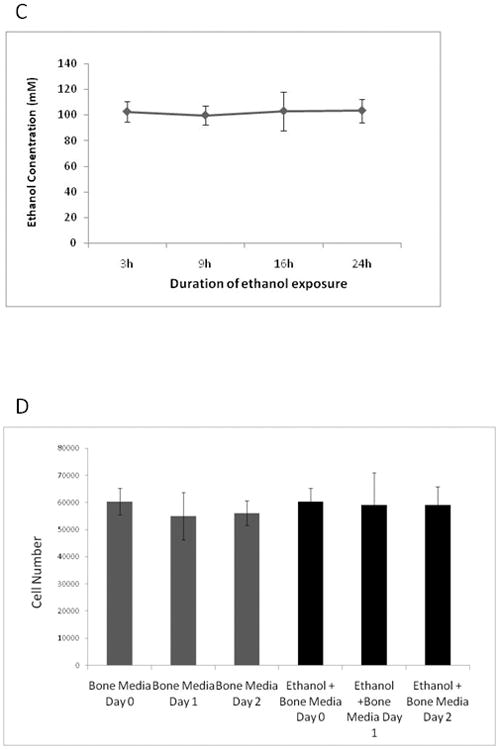
Osteopontin is a phosphorylated glycoprotein that is secreted in the early stages of osteogenic differentiation. It is abundant in mineralized tissue and may be involved in bone formation (Butler, 1989;Kojima, Uede et al., 2004;Strauss, Closs et al., 1990). To characterize the effect of ethanol on osteopontin expression, real time-PCR was performed after 24 and 48 hours of exposure to 100 mM ethanol. Although control AFSCs expressed some osteopontin, ethanol-exposed AFSC showed a significant increase in osteopontin mRNA expression (Figure 2A). After 24 hours of ethanol exposure, osteopontin mRNA levels increased by 2.2-fold (p< 0.02). Exposure to ethanol for 48 hours increased the expression of osteopontin mRNA by 2.8-fold (p< 0.036). These results suggest that the ethanol-induced increase in osteopontin expression may restrict AFSCs to an osteogenic lineage.
Figure 2. Effect of ethanol on OPN expression in AFSCs.
(A) Real-time RT PCR analysis of AFSCs exposed to ethanol for 24 or 48 hours in growth media. CT values were determined from 3 independent experiments from two cell lines and ΔCT values were obtained by subtracting the CT values of β-actin. Mean fold change was determined from four independent experiment-pairs of ethanol-treated AFSCs and non-treated AFSC. Black columns indicate AFSCs without ethanol while grey columns indicate AFSCs exposed to 100 mM ethanol. P-values were determined using a one–tailed paired t test with significance at 24 hours (P < 0.020, n=3) and 48 hours (P < 0.036, n=5). (B) Real-time PCR analysis of AFSCs exposed to ethanol for 24 hours in osteogenic media. CT values were determined from 3 independent experiments using two cell lines and ΔCT values were obtained by subtracting the CT values of β-actin. Mean fold change was determined from 3 independent experiment-pairs of ethanol-exposed AFSCs and non-exposed AFSCs. P-values were determined using a one–tailed paired t test with significance at 24 (P < 0.018).
Next, whether ethanol had similar effects on AFSCs when the cells were cultured in osteogenic differentiation media was examined. To determine whether ethanol interferes with the osteogenic differentiation process, the mRNA expression of osteopontin after a 24 hour exposure to 100 mM ethanol in osteogenic media was analyzed. Ethanol increased the expression of osteopontin by 2- fold in the presence of osteogenic media (Figure 2B). This suggests that ethanol induces osteopontin expression in both non-committed AFSCs and AFSCs that are committed towards an osteogenic phenotype.
The effect of ethanol on alkaline phosphatase activity
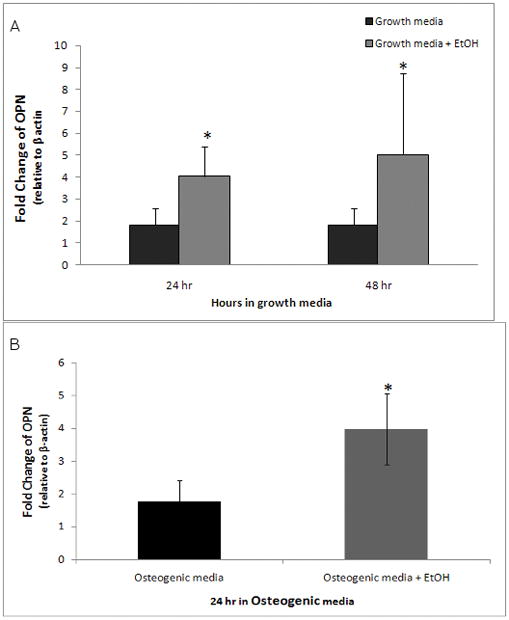
The effect of ethanol on alkaline phosphatase activity, which is an established marker of osteoblasts was also studied (Bellows, Aubin et al., 1991;Fedde, Blair et al., 1999). It had previously been shown that AFSCs begin to express alkaline phosphatase activity after 8 days of osteogenic differentiation. To address whether ethanol exposure had an effect on alkaline phosphatase activity in differentiating AFSCs, they were exposed to 100 mM ethanol for 48 hours in osteogenic media, and then the ethanol was removed. Cells were allowed to differentiate further and were assayed for alkaline phosphatase activity at various time points (Figure 3). AFSCs that were not exposed to osteogenic media expressed a basal level of alkaline phosphatase activity. After 8 days of osteogenic differentiation, alkaline phosphatase activity in untreated cells rose above control levels and continued to increase until day 10 of osteogenic differentiation. However, ethanol exposure during the first 48 hours induced a small, but significant, increase in alkaline phosphatase activity at day 9 and 10. These results suggest that ethanol may have a persistent and enhancing effect on osteogenic differentiation that can be seen several days beyond the ethanol exposure period.
Figure 3. Effect of ethanol on alkaline phosphatase activity in AFSCs.
AFSCs were cultured with or without 100mM ethanol for the first 48 hours of osteogenic differentiation. Ethanol was removed and AFSCs continued to differentiate until days 7–10. At this time, they were assessed for alkaline phosphatase activity. Alkaline phosphatase activity was determined by spectrophotometric measurement of p-nitrophenol conversion. AFSCs exposed to ethanol showed a modest yet significant increase in alkaline phosphatase activity at day 9 and 10 of osteogenic differentiation (*, p < 0.001; ANOVA and two-tail T-test). The values shown are the mean +/− SD from at least ten replicate cultures and similar patterns were observed in a second cell line (data not shown). Differences between treated and non-treated cultures were evaluated by t-tests. Open squares, osteogenic media+ EtOH (48 hours); Black diamonds, osteogenic media; black circles, growth media.
In order to determine whether the effects of ethanol on AFSCs differed depending on the stage of differentiation, AFSCs were cultured in osteogenic media for 8 days without ethanol. Then, beginning on day 8, the cells were treated for 48 hours with 100mM ethanol and the alkaline phosphatase activity was measured. The results showed no statistically significant effect (Figure 4). Collectively, these experiments suggest that ethanol exposure has a significant effect on alkaline phosphatase levels only when it occurs early in the differentiation process.
Figure 4. Alkaline phosphatase activity when AFSCs were exposed to ethanol at the midpoint of differentiation.
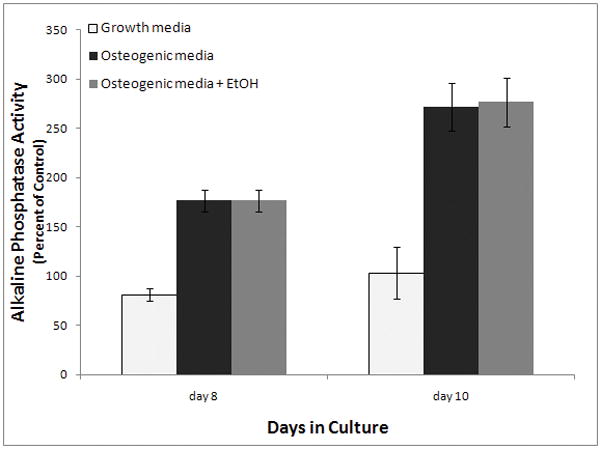
AFSC were treated for 48 hours with 100 mM ethanol beginning at day 8 of osteogenic differentiation. Black columns indicate AFSCs that were not exposed to ethanol while grey bars indicate AFSCs that were exposed to ethanol. Ethanol exposure during the midpoint of differentiation had no significant effect on alkaline phosphatase activity (t test p<0.42). The values shown are the mean +/− SD of twenty cultures.
The effect of ethanol on calcium deposition
Since transient ethanol exposure increased the expression and activation of genes involved in mineralization, the effect of ethanol on calcium deposition was examined. Cells were exposed to osteogenic media with or without 100 mM of ethanol for 48 hours. The ethanol was then removed and the AFSCs were allowed to terminally differentiate. At day 23, calcium deposition in these differentiated cultures was measured by Alizarin Red staining. The effects of ethanol treatment on calcium deposition are shown in Figure 5. AFSCs exposed to ethanol during the first 48 hours of osteogenic differentiation produced 155.06 ± 75.85 mg/mL of calcium, while non-exposed AFSCs produced 77.40 ± 26.85 mg/mL of calcium. Calcium deposition was also detected in non-differentiated AFSCs (52.5 ± 2.223 mg/ml). These data suggest that the effect of transient ethanol exposure on osteogenic genes during early differentiation is directly correlated with the AFSCs’ ability to deposit calcium.
Figure 5. The effect of ethanol on calcium deposition.
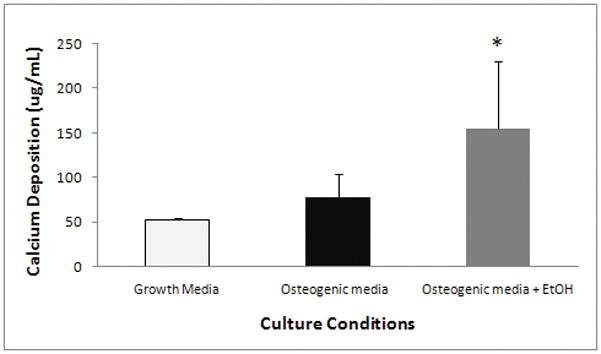
AFSCs were exposed to 100 mM ethanol during the first 48 hours of osteogenic differentiation. At day 23 of differentiation, AFSCs were stained for calcium deposition using alizarin red staining. AFSCs exposed for 48 hours to 100 mM ethanol had significantly increased calcium deposition when compared to non-exposed AFSCs (155.1 ± 75.8 μg/mL vs 77.4 ± 26.9 μg/mL). Calcium deposition was detected in non-differentiated AFSCs at a basal level of 53.3 ± 2.2 μg/ml. The values shown are the mean +/− SD (n=3–6) and similar results were confirmed in another cell line (data not shown). * One-tailed t-test of significance of p<0.006.
Discussion
This work has employed AFSCs as a unique model system for analysis of ethanol’s cellular effects. In this study, exposure of these cells, at their uncommitted stage, to teratogenic concentrations of ethanol has been shown to result in up-regulation of osteogenic genes. Specifically, the sequential increase in the expression of osteopontin, alkaline phosphatase activity, and calcium deposition due to ethanol exposure as cells progress from stem cells into those of an osteogenic lineage has been shown.
The effects of ethanol on a limited variety of stem cells have previously been reported, with some studies showing enhanced and some showing reduced differentiation potential. Neural stem (NSCs) and progenitor cells have been the most commonly studied. Ethanol has been shown to alter the differentiation potential of NSCs by enhancing astrocytic and oligodendrocytic differentiation and decreasing neuronal differentiation (Tateno, Ukai et al., 2005). Adult bone-marrow derived stem cells (BMSCs) have also been employed and, as opposed to the current study, inhibition of osteogenic differentiation has been shown (Gong and Wezeman, 2004). Another study, which used immortalized human fetal osteoblasts to examine the effect of ethanol on skeletal development by analysis of osteogenic gene expression (Maran, Zhang et al., 2001) demonstrated little or no effect of ethanol. As for the BMSCs, the response of these immortalized fetal osteoblasts may not reflect that of prenatal stem cells. Ethanol has been shown to enhance cartilage differentiation in embryonic limb mesenchyme cultures (Kulyk and Hoffman, 1996;Shukla, Velazquez et al., 2008). While these cells seem to be lineage restricted because they spontaneously differentiate into chondrocytes, naive AFSCs are not lineage restricted to osteogenesis. Thus, ethanol may act to lineage restrict AFSCs to osteogenesis by elevating the expression of osteogenesis-specific genes.
We would like to mention that we have previously established that AFSCs express RUNX2 and osteocalcin during osteogenic differentiation (De Coppi, Bartsch et al., 2007). Although these genes that are far more specific as osteogenic markers than ALP and osteopontin, RUNX2 expression, measured by real-time PCR, was not changed after 48 hours of ethanol exposure (Supplement Figure 1). Since chondrocytic differentiation requires a unique protocol (BMP2 and TGF-β), future studies are needed to test whether ethanol affects the commitment into a chondrocytic lineage.
Clearly, gene array analysis is a powerful tool for exploring the pleotrophic effects of agents, or conditions, on gene expression. For the current study, it has allowed characterization of the global effects of ethanol on gene expression, as well as providing the potential to identify unknown processes and pathways.
Although there are many techniques available for large scale evaluation of transcripts, Affymetrix GeneChips were selected because their proven reproducibility and the extensive number of genes included. To identify ethanol exposure-related changes in gene expression, analysis was performed following 48 hours of ethanol exposure. This duration was chosen in order to encompass the entire 36 hr AFSC cell cycle and to allow identification of the late responses of AFSC to ethanol (rather than responses to early oxidative stress). As determined by gene ontology analyses, ethanol exposure was shown to modulate unique pathways that pertain to bone development. This result is consistent with functional analyses that show a predisposition of ethanol-exposed AFSCs to differentiate into an osteogenic lineage.
We would like to mention that the predominant pathway identified in this data set containing genes that were down-regulated after ethanol exposure was embryonic development, which includes the genes encoding dickkopf homolog 1. Since DKK1 is expressed in both embryonic and adult bone tissues, this gene may be important for both embryonic and skeletal development (Li, Sarosi et al., 2006;Monaghan, Kioschis et al., 1999). DKK1 was identified to be an inhibitor of the canonical WNT pathway (Glinka, Wu et al., 1998), thus the down-regulation of DKK1 due to ethanol may be involved in the increase in osteogenesis of AFSC however further studies are needed to determine the role of DKK1 in AFSC.
The conditions necessary for the in vitro induction of osteogenic differentiation of AFSCs have previously been shown to include the presence of ascorbic acid, which is required for collagen synthesis (Murad, Grove et al., 1981); beta-glycerol phosphate, which provides an organic phosphate for the formation of hydroxyapatite (Bellows, Aubin et al., 1986;Bellows, Heersche et al., 1992); and dexamethasone, a glucocorticoid that induces transcription at the promoters of osteogenic genes (Ogata, Yamauchi et al., 1995). Although the mechanism of in vitro osteogenic differentiation is not clearly defined, it has been shown that these media additives can induce gene expression and decrease cell proliferation (Ogata, Yamauchi et al., 1995;Shalhoub, Conlon et al., 1992). These effects are similar to those of ethanol’s and suggest that ethanol acts as a differentiation agent. Premature differentiation of stem cells can deplete the stem cell population, resulting in fewer number of cells which may explain some of the clinical features of FASD such a short stature and craniofacial malformations.
Our results are also consistent with the non-developmental effects of ethanol on bone formation. Bone homeostasis consists of a constant flux between osteogenesis and bone resorption, consisting of osteoblasts and osteoclasts, respectively. Studies have shown that ethanol impairs osteoblast activity associated with normal osteoclast function (Chappard, Plantard et al., 1991). Since osteoclasts are derived from a myeloid lineage, this suggests that ethanol has a lineage-specific/cell type-specific effect. AFSC represents a unique model to examine the effects of ethanol on the differentiation into multiple lineages. Further studies are needed to determine which lineages are affected by ethanol.
Measures of alkaline phosphatase activity, which provided a marker of osteogenic differentiation, were employed to assess the developmental stage-dependency of the AFSC’s osteogenic response to ethanol. It was shown that the stem cells are more susceptible to ethanol prior to and during early differentiation than at later stages. While the reason for this is unknown, it is possible that at early stages of cell differentiation, ethanol affects epigenetic mechanisms, such as DNA methylation and histone methylation and acetylation. Differentiation requires dynamic epigenetic modification, whereas self-renewal requires epigenetic factors that preserve constant gene expression during stem cell division. Although there have been recent studies reporting the effects of ethanol on epigenetic mechanisms (Shukla, Velazquez et al., 2008), the field is relatively new and much work remains to be done. AFSCs may provide a useful model system in this regard.
Identification, in this study, of ethanol-induced osteogenic differentiation in AFSCs provides new information that may be of significance for understanding mechanisms involved in the genesis of FASD. These findings are consistent with the growing view that ethanol exposure alters the differentiation potential of stem cells. Although further studies are needed in order to determine whether ethanol enhances differentiation into other lineages, the decrease in the number of AFSCs and their increased sensitivity to differentiation cues suggest that ethanol acts as a differentiation agent. Importantly, AFSCs are uncommitted fetal-derived stem cells and can be obtained by non-invasive means early in pregnancy. Thus they offer a unique model system with significant promise for pertinent future studies. Acquiring a complete picture of the response of AFSCs to ethanol exposure also offers promise for identification of a biomarker of prenatal insult.
Supplementary Material
Real-time PCR analysis of RUNX2 24 hr after 48 hr after ethanol exposure of AFSCs in osteogenic media. Mean fold change was determined from 4 experiment-pairs of ethanol-exposed AFSCs and non-exposed AFSCs. ΔCT values were obtained by subtracting the CT values of β-actin.
AFSCs were cultured with 0, 25, 50, 75, and 100 mM of ethanol. Cell counts were made after 48 hours of ethanol exposure and compared to counts of cultures that were not treated with ethanol. The results are expressed as number of cells. The values shown are the mean +/− standard deviation (SD, n=5) of three independent experiments.
Acknowledgments
These studies were supported by the NIAAA grant 5F30AA016446-02.
Reference List
- Adachi J, Mizoi Y, Fukunaga T, Ogawa Y, Ueno Y, Imamichi H. Degrees of alcohol intoxication in 117 hospitalized cases. J Stud Alcohol. 1991;52:448–453. doi: 10.15288/jsa.1991.52.448. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astley SJ, Clarren SK. Diagnosing the full spectrum of fetal alcohol-exposed individuals: introducing the 4-digit diagnostic code. Alcohol Alcohol. 2000;35:400–410. doi: 10.1093/alcalc/35.4.400. [DOI] [PubMed] [Google Scholar]
- Becker HC, az-Granados JL, Randall CL. Teratogenic actions of ethanol in the mouse: a minireview. Pharmacol Biochem Behav. 1996;55:501–513. doi: 10.1016/s0091-3057(96)00255-9. [DOI] [PubMed] [Google Scholar]
- Bellows CG, Aubin JE, Heersche JN. Initiation and progression of mineralization of bone nodules formed in vitro: the role of alkaline phosphatase and organic phosphate. Bone Miner. 1991;14:27–40. doi: 10.1016/0169-6009(91)90100-e. [DOI] [PubMed] [Google Scholar]
- Bellows CG, Aubin JE, Heersche JN, Antosz ME. Mineralized bone nodules formed in vitro from enzymatically released rat calvaria cell populations. Calcif Tissue Int. 1986;38:143–154. doi: 10.1007/BF02556874. [DOI] [PubMed] [Google Scholar]
- Bellows CG, Heersche JN, Aubin JE. Inorganic phosphate added exogenously or released from beta-glycerophosphate initiates mineralization of osteoid nodules in vitro. Bone Miner. 1992;17:15–29. doi: 10.1016/0169-6009(92)90707-k. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Brien JF, Loomis CW, Tranmer J, McGrath M. Disposition of ethanol in human maternal venous blood and amniotic fluid. Am J Obstet Gynecol. 1983;146:181–186. doi: 10.1016/0002-9378(83)91050-5. [DOI] [PubMed] [Google Scholar]
- Butler WT. The nature and significance of osteopontin. Connect Tissue Res. 1989;23:123–136. doi: 10.3109/03008208909002412. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Fetal alcohol syndrome. Alaska, Arizona, Colorado, and New York: 2002. pp. 433–435. [PubMed] [Google Scholar]
- Chappard D, Plantard B, Petitjean M, Alexandre C, Riffat G. Alcoholic cirrhosis and osteoporosis in men: a light and scanning electron microscopy study. J Stud Alcohol. 1991;52:269–274. doi: 10.15288/jsa.1991.52.269. [DOI] [PubMed] [Google Scholar]
- Charness ME, Safran RM, Perides G. Ethanol inhibits neural cell-cell adhesion. J Biol Chem. 1994;269:9304–9309. [PubMed] [Google Scholar]
- Chen SY, Sulik KK. Free radicals and ethanol-induced cytotoxicity in neural crest cells. Alcohol Clin Exp Res. 1996;20:1071–1076. doi: 10.1111/j.1530-0277.1996.tb01948.x. [DOI] [PubMed] [Google Scholar]
- De Coppi P, Bartsch G, Siddiqui MM, Xu T, Santos CC, Perin L, Mostoslavsky G, Serre AC, Snyder EY, Yoo JJ, et al. Isolation of amniotic stem cell lines with potential for therapy. Nat Biotechnol. 2007;25:100–106. doi: 10.1038/nbt1274. [DOI] [PubMed] [Google Scholar]
- Delany AM, Kalajzic I, Bradshaw AD, Sage EH, Canalis E. Osteonectin-null mutation compromises osteoblast formation, maturation, and survival. Endocrinology. 2003;144:2588–2596. doi: 10.1210/en.2002-221044. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- Fallon JF, Lopez A, Ros MA, Savage MP, Olwin BB, Simandl BK. FGF-2: apical ectodermal ridge growth signal for chick limb development. Science. 1994;264:104–107. doi: 10.1126/science.7908145. [DOI] [PubMed] [Google Scholar]
- Fedde KN, Blair L, Silverstein J, Coburn SP, Ryan LM, Weinstein RS, Waymire K, Narisawa S, Millan JL, MacGregor GR, et al. Alkaline phosphatase knock-out mice recapitulate the metabolic and skeletal defects of infantile hypophosphatasia. J Bone Miner Res. 1999;14:2015–2026. doi: 10.1359/jbmr.1999.14.12.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391:357–362. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- Gong Z, Wezeman FH. Inhibitory effect of alcohol on osteogenic differentiation in human bone marrow-derived mesenchymal stem cells. Alcohol Clin Exp Res. 2004;28:468–479. doi: 10.1097/01.alc.0000118315.58404.c1. [DOI] [PubMed] [Google Scholar]
- Habbick BF, Blakley PM, Houston CS, Snyder RE, Senthilselvan A, Nanson JL. Bone age and growth in fetal alcohol syndrome. Alcohol Clin Exp Res. 1998;22:1312–1316. [PubMed] [Google Scholar]
- Herrmann J, Pallister PD, Opitz JM. Tetraectrodactyly and other skeletal manifestations in the fetal alcohol syndrome. Eur J Pediatr. 1980;133:221–226. doi: 10.1007/BF00496080. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Johnson VP, Swayze VW, II, Sato Y, Andreasen NC. Fetal alcohol syndrome: craniofacial and central nervous system manifestations. Am J Med Genet. 1996;61:329–339. doi: 10.1002/(SICI)1096-8628(19960202)61:4<329::AID-AJMG6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Kojima H, Uede T, Uemura T. In vitro and in vivo effects of the overexpression of osteopontin on osteoblast differentiation using a recombinant adenoviral vector. J Biochem. 2004;136:377–386. doi: 10.1093/jb/mvh136. [DOI] [PubMed] [Google Scholar]
- Kulyk WM, Hoffman LM. Ethanol exposure stimulates cartilage differentiation by embryonic limb mesenchyme cells. Exp Cell Res. 1996;223:290–300. doi: 10.1006/excr.1996.0084. [DOI] [PubMed] [Google Scholar]
- Lemoine P, Harousseau H, Borteyru JP, Menuet JC. Children of alcoholic parents--observed anomalies: discussion of 127 cases. Ther Drug Monit. 2003;25:132–136. doi: 10.1097/00007691-200304000-00002. [DOI] [PubMed] [Google Scholar]
- Li J, Sarosi I, Cattley RC, Pretorius J, Asuncion F, Grisanti M, Morony S, Adamu S, Geng Z, Qiu W, et al. Dkk1-mediated inhibition of Wnt signaling in bone results in osteopenia. Bone. 2006;39:754–766. doi: 10.1016/j.bone.2006.03.017. [DOI] [PubMed] [Google Scholar]
- Li Z, Lin H, Zhu Y, Wang M, Luo J. Disruption of cell cycle kinetics and cyclin-dependent kinase system by ethanol in cultured cerebellar granule progenitors. Brain Res Dev Brain Res. 2001;132:47–58. doi: 10.1016/s0165-3806(01)00294-2. [DOI] [PubMed] [Google Scholar]
- Maran A, Zhang M, Spelsberg TC, Turner RT. The dose-response effects of ethanol on the human fetal osteoblastic cell line. J Bone Miner Res. 2001;16:270–276. doi: 10.1359/jbmr.2001.16.2.270. [DOI] [PubMed] [Google Scholar]
- Miller MW, Chiaia NL, Rhoades RW. Intracellular recording and injection study of corticospinal neurons in the rat somatosensory cortex: effect of prenatal exposure to ethanol. J Comp Neurol. 1990;297:91–105. doi: 10.1002/cne.902970107. [DOI] [PubMed] [Google Scholar]
- Monaghan AP, Kioschis P, Wu W, Zuniga A, Bock D, Poustka A, Delius H, Niehrs C. Dickkopf genes are co-ordinately expressed in mesodermal lineages. Mech Dev. 1999;87:45–56. doi: 10.1016/s0925-4773(99)00138-0. [DOI] [PubMed] [Google Scholar]
- Murad S, Grove D, Lindberg KA, Reynolds G, Sivarajah A, Pinnell SR. Regulation of collagen synthesis by ascorbic acid. Proc Natl Acad Sci U S A. 1981;78:2879–2882. doi: 10.1073/pnas.78.5.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata Y, Yamauchi M, Kim RH, Li JJ, Freedman LP, Sodek J. Glucocorticoid regulation of bone sialoprotein (BSP) gene expression. Identification of a glucocorticoid response element in the bone sialoprotein gene promoter. Eur J Biochem. 1995;230:183–192. doi: 10.1111/j.1432-1033.1995.0183i.x. [DOI] [PubMed] [Google Scholar]
- Parnell SE, Dehart DB, Wills TA, Chen SY, Hodge CW, Besheer J, Waage-Baudet HG, Charness ME, Sulik KK. Maternal oral intake mouse model for fetal alcohol spectrum disorders: ocular defects as a measure of effect. Alcohol Clin Exp Res. 2006;30:1791–1798. doi: 10.1111/j.1530-0277.2006.00212.x. [DOI] [PubMed] [Google Scholar]
- Perper JA, Twerski A, Wienand JW. Tolerance at high blood alcohol concentrations: a study of 110 cases and review of the literature. J Forensic Sci. 1986;31:212–221. [PubMed] [Google Scholar]
- Pignataro L, Miller AN, Ma L, Midha S, Protiva P, Herrera DG, Harrison NL. Alcohol regulates gene expression in neurons via activation of heat shock factor 1. J Neurosci. 2007;27:12957–12966. doi: 10.1523/JNEUROSCI.4142-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raballo R, Rhee J, Lyn-Cook R, Leckman JF, Schwartz ML, Vaccarino FM. Basic fibroblast growth factor (Fgf2) is necessary for cell proliferation and neurogenesis in the developing cerebral cortex. J Neurosci. 2000;20:5012–5023. doi: 10.1523/JNEUROSCI.20-13-05012.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall CL, Taylor J, Walker DW. Ethanol-induced malformations in mice. Alcohol Clin Exp Res. 1977;1:219–224. doi: 10.1111/j.1530-0277.1977.tb05876.x. [DOI] [PubMed] [Google Scholar]
- Resnicoff M, Sell C, Ambrose D, Baserga R, Rubin R. Ethanol inhibits the autophosphorylation of the insulin-like growth factor 1 (IGF-1) receptor and IGF-1-mediated proliferation of 3T3 cells. J Biol Chem. 1993;268:21777–21782. [PubMed] [Google Scholar]
- Rice D, Barone S., Jr Critical periods of vulnerability for the developing nervous system: evidence from humans and animal models. Environ Health Perspect. 2000;108(Suppl 3):511–533. doi: 10.1289/ehp.00108s3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalhoub V, Conlon D, Tassinari M, Quinn C, Partridge N, Stein GS, Lian JB. Glucocorticoids promote development of the osteoblast phenotype by selectively modulating expression of cell growth and differentiation associated genes. J Cell Biochem. 1992;50:425–440. doi: 10.1002/jcb.240500411. [DOI] [PubMed] [Google Scholar]
- Shukla SD, Velazquez J, French SW, Lu SC, Ticku MK, Zakhari S. Emerging role of epigenetics in the actions of alcohol. Alcohol Clin Exp Res. 2008;32:1525–1534. doi: 10.1111/j.1530-0277.2008.00729.x. [DOI] [PubMed] [Google Scholar]
- Siegenthaler JA, Miller MW. Transforming growth factor beta1 modulates cell migration in rat cortex: effects of ethanol. Cereb Cortex. 2004;14:791–802. doi: 10.1093/cercor/bhh039. [DOI] [PubMed] [Google Scholar]
- Stein GS, Lian JB, Gerstenfeld LG, Shalhoub V, Aronow M, Owen T, Markose E. The onset and progression of osteoblast differentiation is functionally related to cellular proliferation. Connect Tissue Res. 1989;20:3–13. doi: 10.3109/03008208909023869. [DOI] [PubMed] [Google Scholar]
- Strauss PG, Closs EI, Schmidt J, Erfle V. Gene expression during osteogenic differentiation in mandibular condyles in vitro. J Cell Biol. 1990;110:1369–1378. doi: 10.1083/jcb.110.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulik KK, Johnston MC, Webb MA. Fetal alcohol syndrome: embryogenesis in a mouse model. Science. 1981;214:936–938. doi: 10.1126/science.6795717. [DOI] [PubMed] [Google Scholar]
- Tateno M, Ukai W, Yamamoto M, Hashimoto E, Ikeda H, Saito T. The effect of ethanol on cell fate determination of neural stem cells. Alcohol Clin Exp Res. 2005;29:225S–229S. doi: 10.1097/01.alc.0000190658.56149.d4. [DOI] [PubMed] [Google Scholar]
- Vangipuram SD, Grever WE, Parker GC, Lyman WD. Ethanol increases fetal human neurosphere size and alters adhesion molecule gene expression. Alcohol Clin Exp Res. 2008;32:339–347. doi: 10.1111/j.1530-0277.2007.00568.x. [DOI] [PubMed] [Google Scholar]
- Wettenhall JM, Simpson KM, Satterley K, Smyth GK. affylmGUI: a graphical user interface for linear modeling of single channel microarray data. Bioinformatics. 2006;22:897–899. doi: 10.1093/bioinformatics/btl025. [DOI] [PubMed] [Google Scholar]
- Wettenhall JM, Smyth GK. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Real-time PCR analysis of RUNX2 24 hr after 48 hr after ethanol exposure of AFSCs in osteogenic media. Mean fold change was determined from 4 experiment-pairs of ethanol-exposed AFSCs and non-exposed AFSCs. ΔCT values were obtained by subtracting the CT values of β-actin.
AFSCs were cultured with 0, 25, 50, 75, and 100 mM of ethanol. Cell counts were made after 48 hours of ethanol exposure and compared to counts of cultures that were not treated with ethanol. The results are expressed as number of cells. The values shown are the mean +/− standard deviation (SD, n=5) of three independent experiments.


