Abstract
Phosphorylation of the C-terminal domain of RNA polymerase II controls the co-transcriptional assembly of RNA processing and transcription factors. Recruitment relies on conserved CTD-interacting domains that recognize different CTD phosphoisoforms during the transcription cycle, but the molecular basis for their specificity remains unclear. We show that the CTD-interacting domains of two transcription termination factors, Rtt103 and Pcf11, achieve high affinity and specificity both by specifically recognizing the phosphorylated CTD and by cooperatively binding to neighboring CTD repeats. Single amino acid mutations at the protein-protein interface abolish cooperativity and affect recruitment at the 3′-end processing site in vivo. We suggest that this cooperativity provides a signal-response mechanism to ensure that its action is confined only to proper polyadenylation sites where Serine 2 phosphorylation density is highest.
Introduction
Synthesis of mature mRNA by RNA polymerase II (Pol II) requires the mRNA to be processed in a series of reactions that occur concomitantly with transcription1. These processes are physically linked to the transcribing polymerase by the C-terminal domain (CTD) of the largest subunit of Pol II2,3. The CTD promotes efficient RNA maturation by raising the local concentration of processing complexes near the nascent transcript; by kinetically coupling the rate of assembly of RNAprotein complexes with the rate of transcription4,5; and by allosterically regulating the activity of processing complexes6,7,8.
The CTD contains 26 (in yeast) to 52 (in human) heptad peptide repeats of sequence: Y1-S2-P3-T4-S5-P6-S7 (ref. 9). Residues Ser2, Ser5, and Ser7 are dynamically phosphorylated and dephosphorylated by specific kinases and phosphatases during the transcription cycle10. Changes in the CTD phosphorylation pattern during transcription are both temporally and functionally coupled to the recruitment and activation of RNA processing complexes11. At the promoter, the Pol II CTD is unphosphorylated11. Coincident with early initiation, the CTD becomes phosphorylated at Ser5 (Ser5P), which promotes recruitment and activation of the 5′-capping enzymes6,12,13. As Pol II progresses into the elongation phase, levels of Ser2 phosphorylation (Ser2P) increase while Ser5P levels decrease11. Near the 3′-end of genes, Ser2P predominates leading to increased recruitment of the protein complexes responsible for cleavage and polyadenylation14,15. Phosphorylation of Ser7 has been recently linked to termination events for the small nuclear snRNA genes16-18.
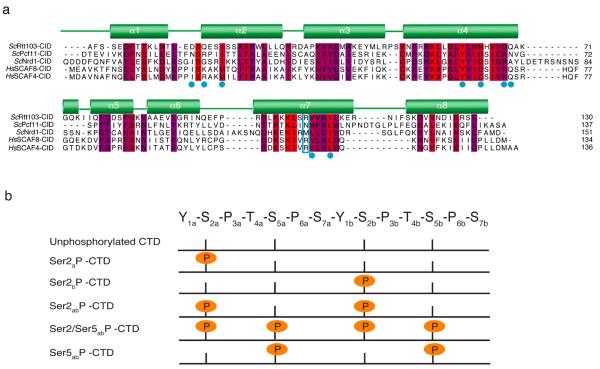
The functional unit of the CTD is thought to consist of two consecutive heptad peptide sequences19 and each CTD-interacting domain (CID) from different transcription factors appears to have a preferred phosphoepitope20-23. Several structures of CIDs bound to diheptad CTD peptides revealed common themes for recognition of the CTD22,24; however, the molecular details for how the domain achieves specificity for a particular phosphoepitope remain elusive because all CIDs bind the CTD with a conserved binding surface (Fig. 1a).
Figure 1.
Sequence alignment of CIDs and diheptad CTD phosphopeptides used in this study. (a) Sequence alignment of yeast Rtt103, Pcf11, Nrd1, and human SCAF8 and SCAF4 CIDs. Red represents identical residues, while purple identifies conserved amino acids. The secondary structure of Rtt103 is shown above the figure. Blue dots identify Rtt103 residues that bind to the CTD in our structure. The cyan box shows the positions of Arg108 and Asn107 in Rtt103 and Pcf11, respectively, as well as the corresponding residues in the Nrd1 and the SCAF CIDs. (b) Schematic diagram of the diheptad CTD peptides used in this study; the sequence is indicated at the top and the sites of serine phosphorylation are shown as orange circles.
To understand how the phosphorylation state of the CTD is recognized specifically by mRNA transcription termination factors, we measured the binding affinities of the CIDs of S. cerevisiae Pcf11 and Rtt103 for an exhaustive set of CTD peptides containing all possible Ser2 and Ser5 phosphorylation states. We were surprised to observe that Pcf11 binds to Ser2 phosphorylated diheptad CTD peptides much more weakly than Rtt103 and with little preference over other phosphoisoforms, because Pcf11 plays a particularly critical function in transcription termination21,25-27 and its concentration is sharply peaked near the 3′-end of genes where the CTD is primarily phosphorylated at Ser2 (ref. 28). However, when Rtt103-CID and Pcf11-CID were presented with longer CTD mimics containing four heptad peptide repeats, we observed stronger binding affinity to the CTD and evidence for cooperativity between adjacent CID molecules on these longer CTD mimics. By mutating the observed cooperative protein-protein interaction sites, we were able to demonstrate that protein-protein interactions between CIDs along the CTD are important for the recruitment of these proteins to the 3′ processing site, particularly for Rtt103-CID. Several lines of evidence suggest cooperative interactions may be important for recruitment of Pcf11 as well, but additional interactions with other components of the cleavage factor Ia (CFIa) complex are also likely to aid Pcf11 recruitment to the 3′ processing site making this mechanism less prominent. Overall, our data indicate that the ability to cooperatively bind to the Ser2 CTD provides an additional effective molecular mechanism to sharply regulate recruitment to the CTD only near 3′-end processing sites of genes, where a critical density of Ser2 CTD phosphorylation sites occur, and to suppress premature termination at cryptic sites.
Results
Rtt103 and Pcf11 recognize CTD phosphoisoforms differently
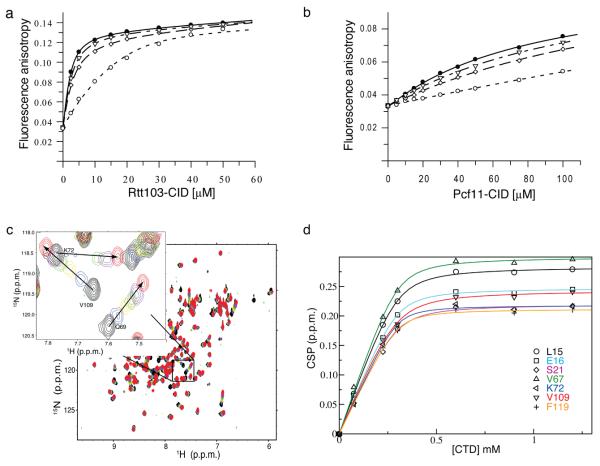
To investigate the molecular basis for Pol II CTD specificity, we compared the affinity of Pcf11-CID and Rtt103-CID for an exhaustive panel of CTD phosphoisoforms. We initially probed the binding affinity of these two proteins against a library of synthetic diheptad CTD peptides (Fig. 1b) using two independent assays, fluorescence anisotropy (FA) and NMR. Representative binding curves generated using a competitive fluorescence anisotropy titration are shown in Figure 2a and b. Changes in NMR chemical shift were plotted against the concentration of added CTD peptide for every resonance exhibiting significant chemical shift changes upon peptide addition (Fig. 2c, d). The average value of these chemical shift changes for each CTD peptide with Pcf11-CID and Rtt103-CID is given in Table 1 next to the data generated by FA experiments (see Supplementary Methods for a detailed description).
Figure 2.
Binding of Rtt103 and Pcf11 to diheptad CTD phosphopeptides monitored by fluorescence anisotropy and NMR. (a) Rtt103 titrated into 2 μM FAM-labeled Ser2P-CTD diheptad repeat peptide (●) which competes against 25 μM Ser2P-CTD diheptad repeat peptides (○) 50 μM Ser5P- CTD diheptad repeat peptides (▽) and 50 μM Ser2P and Ser5P-CTD diheptad repeat peptides (◇) for binding. (b) Titration of Pcf11, as described in (a) except that 300 μM of unlabeled peptides were used. (c) Superposition of 1H-15N HSQC spectra of Rtt103 at several points in the titration with the doubly Ser2 phosphorylated CTD peptide showing changes in chemical shift upon peptide binding. The inset zooms into part of the spectrum to better illustrate that binding occurs in the so-called fast exchange NMR regime, indicative of Kd greater than about 1-10 μM. (d) HSQC perturbations were used to calculate binding affinities on a residue-by-residue basis for all amino acids displaying significant chemical shift changes upon peptide binding by plotting the change in chemical shift vs the peptide concentration and by fitting the data to equation 1 (Supplementary Methods).
Table 1.
Affinities of Rtt103-CID and Pcf11-CID for diheptad repeat CTD peptides measured by NMR and fluorescence anisotropy for each of the diheptad repeat peptides tested (as identified in Figure 1b).
| CTD Peptide | Rtt103 (μM) | Pcf11 (μM) | ||
|---|---|---|---|---|
| NMR | FA | NMR | FA | |
|
| ||||
| Unphosphorylated | 1200 ± 300 | > 1000 | 1500 ± 300 | > 1000 |
| Ser2aP-CTD | 420 ± 70 | N.A. | 1400 ± 650 | N.A. |
| Ser2bP-CTD | 76 ± 16 | N.A. | 240 ± 60 | N.A. |
| Ser2abP-CTD | 15 ± 10 | 2.1 +0.1 | 160 ± 50 | 130 ± 35 |
| Ser2abP/Ser5abP-CTD | 41 ± 19 | 53 ± 10 | 630 ± 170 | 370 (+160/−110) |
| Ser5abP-CTD | 600 ± 300 | 122 (+2/−3) | 1200 ± 500 | > 1000 |
The FA and NMR data consistently demonstrate that Rtt103-CID displays higher affinity for Ser2P CTD peptides compared to the unphosphorylated CTD or to any other phosphoisoform (Table 1). For example, it binds to peptides having Ser2P in both repeats with an approximate increase by a factor of 100 in affinity over unphosphorylated CTD. Rtt103-CID also discriminates Ser2P against Ser5P peptides, showing an increase by a factor of 20-30 in affinity for Ser2P peptides, and an increase by a factor of 5-10 for a peptide with Ser2P, Ser5P in both repeats. Pcf11-CID binds to all peptides much more weakly than Rtt103-CID, and requires Ser2P for sub-millimolar binding (Table 1). It also discriminates less effectively than Rtt103-CID between Ser2P and unphosphorylated CTD peptides, with a difference of only a factor of 10, compared to a factor of 100 for Rtt103-CID.
Rtt103-CID–CTD structure reveals Ser2P CTD specificity
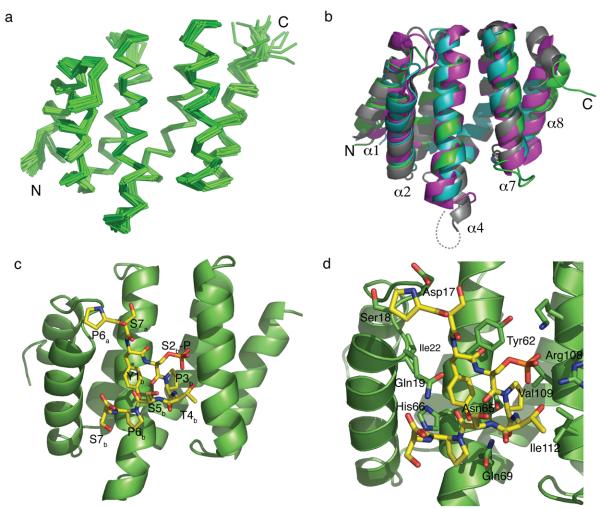
In order to understand why Rtt103 displays much higher affinity and specificity for Ser2P CTD compared to Pcf11, we determined the structure of the CTD interacting domain from yeast Rtt103 (residues 1-131) both free and bound to a Ser2P CTD peptide (Fig. 3 & Supplementary Fig. 1 online). As expected from the sequence conservation, the Rtt103-CID is very similar to the structures of previously solved CIDs with a 1.9 Å Cα RMSD to Pcf11-CID, 2.2 Å Cα RMSD to the SCAF8-CID, and 2.4 Å Cα RMSD to Nrd1-CID over the entire length of the polypeptide chain (Fig. 3b). The primary differences between the Pcf11-CID and the Rtt103-CID are the presence of an additional turn in helix 4 and the much shorter loop connecting helix 7 and 8 in Rtt103-CID. The SCAF8 and Nrd1 CIDs share with Rtt103-CID the extension of helix 4 and the short loop between helices 7 and 8, but they differ in the loop connecting helices 1 and 2 due to the presence an additional 310-helix in the SCAF8-CID and an additional turn of helix 1 in the Nrd1-CID which is not observed in Rtt103-CID or Pcf11-CID (Fig. 3b).
Figure 3.
Structure of Rtt103-CID and recognition of the Ser2-phosphorylated CTD. (a) Ensemble of the 20 lowest energy structures of the free Rtt103-CID as determined by NMR. (b) Superposition of the four existing CID structures: Rtt103 (green, this study), Pcf11 (cyan)24, SCAF8 (pink)22 and Nrd1 (grey)20. All four structures are very similar, but Rtt103, SCAF8 and Nrd1 have an additional turn in helix 4 and shorter loops between helices 7 and 8. SCAF8 and Nrd1 both have extended loops between helices 1 and 2 compared to Pcf11 and Rtt103. (c) Structure of Rtt103 bound to the Ser2 phosphorylated CTD (yellow). The CTD peptide binds in a β-turn conformation along helices 2, 4 and 7. (d) Close-up view of the recognition of the phosphorylated CTD by Rtt103 highlighting residues that directly contact the peptide. This and all subsequent structural figures were generated with PyMOL36.
The structure of Rtt103-CID was also determined bound to a diheptad CTD peptide phosphorylated at both Ser2 positions, by combining experimental NMR restraints with molecular docking routines in HADDOCK (see Methods). The lowest-energy docked structure of Rtt103-CID bound to the Ser2P CTD (Fig. 3c) shows that the peptide binds along helices 2, 4, and 7 of the Rtt103-CID similarly to previously determined CTD–CID structures. As seen in other CID–CTD complexes, the peptide adopts a β-turn conformation formed by Ser2bP, Pro3b, Thr4b, and Ser5b22,24. The structure of the Rtt103-CID complex displays a core set of conserved residues responsible for the recognition of the CTD β-turn, primarily through interactions with Tyr1b and Pro3b in the second CTD repeat (Fig. 1a, 3d). In the Rtt103-CID structure, Tyr1b makes hydrophobic contacts to Ile22, stacks against the aromatic ring of Tyr62, and hydrogen bonds to the side chain carboxyl of Asn65 to the side chain hydroxyl group of Tyr1b (Fig. 3d, Supplementary Fig. 2 online). Asn65 is occupied by a conserved aspartate in all other CIDs that have been characterized structurally, yet this residue makes an equivalent hydrogen bond interaction to the phenolic tyrosine hydroxyl group on the CTD. Pro3b is recognized in a hydrophobic pocket formed by residues Tyr62, Val109, Ile112, and Leu113 (Fig. 3d, Supplementary Fig. 3 online). With the exception of Asn65, these residues are generally conserved across CIDs (Fig. 1a) and make similar contacts in both the SCAF8 and Pcf11-CID structures (Supplementary Fig. 2 online). The CTD β-turn conformation is further stabilized by intra-molecular hydrogen bonding that occurs between the hydroxyl group of Thr4b and a phosphate oxygen in Ser2bP and by two hydrogen bonds between the backbone amides of Thr4b and Ser5b within repeat 2 to Ser2bP (Fig. 4a).
Figure 4.
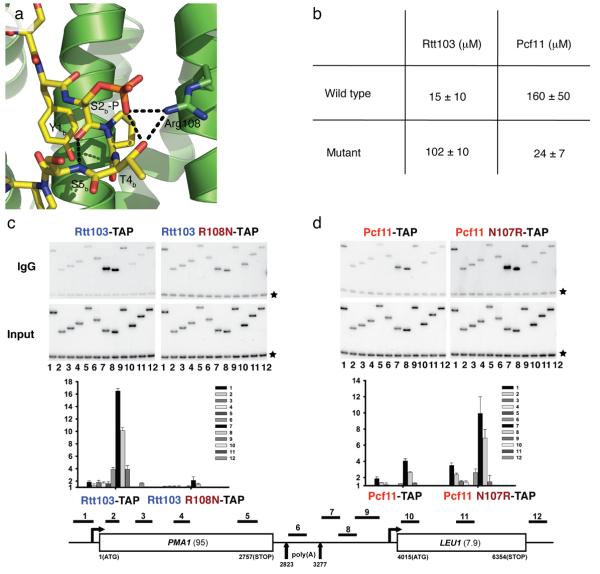
CTD specificity can be altered by a single amino acid change. (a) Arg108 in Rtt103 makes several direct contacts to the CTD Ser2 phosphate; it is within hydrogen bonding distance of both the phosphoserine and the threonine residue in the CTD (dotted black lines). Several other hydrogen bonds (dotted black lines) are characteristic of the β-turn structure. (b) Affinities (± standard deviation) for wild type and mutant Rtt103 and Pcf11 CIDs determined by anisotropy and NMR, respectively, with the Ser2ab phosphorylated diheptade CTD peptide. (c) Recruitment of C-terminally TAP tagged Rtt103 and Rtt103 (R108N) to the highly transcribed PMA1 gene was monitored by ChIP assay. Immunoprecipitated DNA was amplified with PMA1 primers as diagrammed in the bottom panel. The top band is the PMA1-specific band, while the common lower band (marked by an asterisk) is an internal background control from a non-transcribed region on chromosome VI. The middle panel shows quantification of ChIP data, expressed as fold enrichment over the background. (d) Recruitment of wild type Pcf11 and Pcf11 (N107R) to PMA1 gene was also monitored by ChIP assay.
The structure of Rtt103–CTD complex clearly shows that the β-turn in the second CTD heptad peptide repeat provides the majority of intermolecular contacts. However, our binding data (Table 1) suggest that weak interactions with the phosphoserine within repeat 1 increase affinity by a factor of 3 for Rtt103-CID arguing for additional weak contacts outside the second repeat. In contrast, our Pcf11-CID binding affinities are unaffected by the Ser2a phosphorylation state, suggesting this protein does not contact the first phosphoserine at all.
Rtt103-CID forms a key salt-bridge interaction with the Ser2bP phosphate that is absent in the Pcf11–CTD complex. Arg108 in helix 7 of Rtt103-CID (Fig. 4a) consistently shows greater than average chemical shift changes in all of the peptides tested, while the corresponding residue in Pcf11-CID, Asn107, does not. In the structure of Rtt103-CID bound to the Ser2 phosphorylated peptide, Arg108 is within hydrogen bonding distance of the Ser2 phosphate and primed to stabilize the hydrogen bond between the CTD Thr4b hydroxyl and the Ser2bP phosphate oxygen (Fig. 4a). Additionally, the close proximity of the positively charged Arg108 guanidinium group to the negatively charged phosphate leads to a salt bridge not observed in the Pcf11-CID crystal structure.
One mutation enhances Pcf11's affinity for Ser2P CTD
A clear difference in the Rtt103–CTD complex compared to Pcf11–CTD complex is the direct interaction of an arginine side chain (Arg108) with the CTD Ser2bP phosphate. Thus, we mutated Pcf11-CID to have an arginine in place of Asn107 and Rtt103-CID to have an asparagine in place of Arg108, and then determined the binding affinities of each mutant. For Rtt103-CID, the R108N mutation reduced the affinity for the Ser2abP CTD to approximately 100 μM, as determined by FA, which is similar to the affinity of wild type Pcf11-CID for the Ser2abP CTD peptide (Fig. 4b). The Pcf11-CID (N107R) mutant was more difficult to study by FA because the protein aggregates as its concentration is increased. Thus, we measured its affinity by NMR, because this technique allowed the protein concentration to be kept constant, below levels where aggregation occurs, and the peptide was titrated instead. The Pcf11-CID (N107R) mutant had an affinity of 24 μM (Fig. 4b) for the diheptad peptide with both Ser2 residues phosphorylated, which is nearly identical to the wild-type Rtt103-CID affinity for the same peptide. These results strongly suggest that the difference in affinity for Ser2P CTD peptides between Rtt103-CID and Pcf11-CID is due to the presence of Arg108, and that a single amino acid change reverses the binding properties of these two proteins.
To test the effect of these mutants in vivo, we used chromatin immunoprecipitation (ChIP) to compare 3′-end recruitment of wild-type proteins with the Rtt103 (R108N) and Pcf11 (N107R) mutant proteins. Replacement of wild type TAP-tagged Rtt103 with the tagged Rtt103 (R108N) mutant did not cause any notable growth differences or changes in expression level (not shown). This is not surprising given that the RTT103 gene is not essential for viability. However, 3′-end recruitment of the mutant was significantly reduced (by a factor of 8 at the PMA1 gene and by a factor of 5 at ADH1) (Fig. 4c and Supplementary Fig. 4a online). At the same time, 3′-end recruitment for the Pcf11 (N107R) mutant significantly increased (greater than a factor of 2 at the PMA1 and ADH1 genes), compared to the wild type Pcf11-TAP (Fig. 4d and Supplementary Fig. 4b online). An untagged Pcf11 (N107R) strain showed no obvious growth defect (not shown). However, the TAP tagged mutant could not support viability, despite being expressed at levels indistinguishable from the wild-type protein (data not shown), indicating that the Pcf11 (N107R) mutant is partially defective in vivo. Accordingly, the Pcf11-TAP ChIP experiments were done in the presence of an untagged wild-type Pcf11 protein to rule out any indirect effects. When normalizing the results for the Pcf11 and Rtt103 mutants to account for differences in Pol II levels, we observe a similar pattern of recruitment as for the un-normalized data (Supplementary Fig. 5 online). The results of these experiments demonstrate that direct recognition of the Ser2 phosphate by an arginine residue in the CID affects the recruitment of Pcf11 and Rtt103 to 3′-end processing sites. However, we found no evidence for a termination defect for either mutant, since we only observed small effects on read through termination (data not shown).
Multiple CTD repeats enable cooperative CID–CID interactions
We were surprised by the observation that the essential termination factor Pcf11-CID does not share Rtt103-CID's ability to bind strongly to Ser2P CTD peptides and to discriminate between the Ser2P CTD and other phosphoisoforms. We reasoned that the repeated nature of the CTD might be able to influence recruitment of these proteins.
In order to explore the possibility of cooperative binding to the CTD, we repeated our titrations with longer peptides containing four instead of two heptad peptide repeats and performed NMR and fluorescence anisotropy measurements with both Rtt103-CID and Pcf11-CID (Fig. 5a). The fluorescence anisotropy results for both CIDs are shown in Table 2. The data for both proteins with the longer peptides were fit using a 2:1 binding model where Kd1 ≠ Kd2. We observed one binding event tighter than the binding of the original 2-repeat CTD peptides, and a second binding event, Kd2, with affinity similar to that observed with 2-repeat peptides (compare Table 1 and Table 2).
Figure 5.
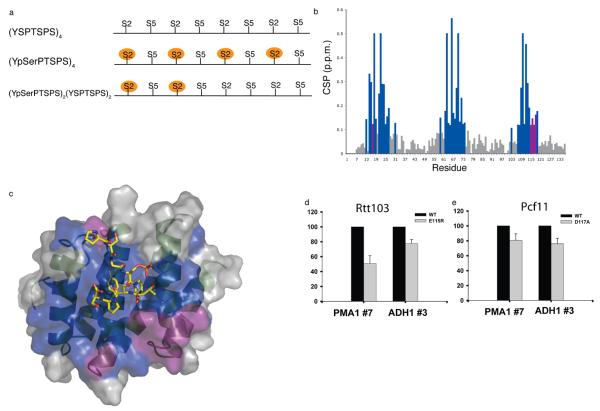
Cooperative binding of the CID to phosphorylated CTD phosphoisoforms. (a) Schematic of the extended CTD phosphopeptides composed of four CTD heptad peptide repeats; phosphorylated serines are shown in orange circles. (b) Chemical shift perturbations (CSP) of Rtt103 with the four heptad repeat peptide phosphorylated at Ser2. Resonances that show increased CSPs with the four repeat peptides are shown in purple, while resonances that show changes with both 2 and 4 repeat peptides are in blue. (c) Residues showing chemical shift perturbations due to protein-protein interactions (purple) are primarily localized near the C-terminus of helix 7; coloring is the same as in (b). ChIP assays were carried out for the HA-tagged protein and Rpb3 from cells expressing (d) HA-tagged Rtt103 (WT) [YSB2537] or Rtt103 (E115R) [YSB2538], and (e) HA-tagged Pcf11 (WT) [YSB2535] or Pcf11 (D117A) [YSB2536]. For quantitation, the fold enrichment of Pcf11 or Rtt103 proteins were normalized to that of Rpb3 protein, and the ratio of wild-type protein level over Rpb3 level was set to 100 %. Results are only shown for the polyA region where Rtt103 or Pcf11 association is strongest (primer set 7 for PMA1 and 3 for ADH1). Error bars show standard deviation from three repeats of the experiment.
Table 2.
FA data with four heptad repeat Ser2P CTD peptide. The data were fit using a 2:1 (protein:peptide) binding model. Kd1 is the dissociation constant for the first binding event and Kd2 is the dissociation constant for the second binding event. All units are μM unless otherwise indicated.
| Protein | Kd1 | Kd2 |
|---|---|---|
|
| ||
| wtRtt103-CID | 3 (+1.1/−0.9) | 20 (+16/−8) |
| Rtt103(E115R)-CID | 1.8 (+1/−0.6) | > 1 mM |
| wtPcf11-CID | 50 (+30/−20) | 140 (+230/−80) |
| Pcf11(D117A)-CID | 40 (+5/−10) | 430 (+1040/−220) |
| Pcf11(D117K)-CID | 30 (+10/−6) | 130 (+130/−60) |
To assess whether these effects are simply due to avidity (the presence of multiple neighboring binding sites) or if instead this 2:1 CID–CTD complex behaves as a single cooperative unit held together by proteinprotein interactions, we used NMR relaxation measurements to estimate the overall rotational correlation times of the various CID–CTD complexes. If the two CIDs were cooperatively associated with the 4-heptad repeat CTD, we would expect an overall tumbling rate consistent with a single complex of approximately 30 kDa. In contrast, the independent, non-cooperative association of two CID monomers with the same 4-repeat peptide would produce tumbling rates comparable to those observed for the 1:1 complex of about 16 kDa formed with a diheptad repeat CTD peptide. For the free Rtt103-CID, we measured the rotational correlation time (τc=10 ns) expected for a 16-kDa protein. This rate increases to τc=12 ns for Rtt103-CID bound to a 2-repeat Ser2abP peptide, consistent with the slight increase in molecular weight. For Rtt103-CID bound to a 4-repeat Ser2P peptide, we observe a much larger increase to τc=16 ns, consistent with a single cooperative complex of two proteins bound to the peptide. For Pcf11-CID we see a 50 % increase in the rotational correlation time with the longer peptides, similar to what is observed for Rtt103-CID, but accurate measurements of the rotational correlation times could not be obtained because the Pcf11-CID partially dimerizes at the concentrations used for NMR, as determined by analytical ultracentrifugation (data not shown). These results suggest that when two CID molecules are bound to the four heptad repeat CTD peptide, this complex tumbles as a single complex held together by proteinprotein interactions, and not as two independent domains tethered to the CTD and tumbling independently of each other.
To identify regions of the Rtt103-CID structure involved in the proteinprotein interactions, we compared the NMR chemical shifts of Rtt103-CID bound to two or four heptad repeat CTD peptides. We identified several residues within helix 7 that show significant chemical shift perturbations in the presence of the four heptad repeat peptide but not with the two-repeat peptide (Fig. 5b). These are residues Glu115, Arg116 and Asn117 near the carboxyl terminus of helix 7 (Fig. 5c) and Asp17, located in the loop connecting helix 1 and helix 2. Based on this observation, we mutated residue Glu115 to arginine within the Rtt103-CID and then tested its ability to bind to the four heptad repeat Ser2P peptide using fluorescence anisotropy. Consistent with the structural analysis, the fluorescence anisotropy data for the Rtt103 E115R mutant could no longer be reliably fit by a 2:1 binding model (Supplementary Fig. 6a online). Instead, it was better fit using a 1:1 binding model. To confirm a 1:1 stoichiometry, we repeated our NMR relaxation experiment with this mutant bound to a four heptad repeat Ser2P CTD peptide. The relaxation data demonstrated that the Rtt103-CID (E115R) mutant behaves similar to the 1:1 Rtt103-CID complex bound to the diheptad repeat CTD peptide, giving a rotational correlation time of 13.3 ns. In the CTD–Rtt103-CID complex, Glu115 points away from the CTD binding site and mutation to arginine has little effect on the affinity of Rtt103-CID for the first binding event (Table 2); this mutation only affects binding of a second Rtt103-CID molecule to the CTD. Thus, this single amino acid change appears to directly affect binding of a second Rtt103-CID molecule to the CTD peptide without substantially altering the affinity of the first binding event or the structure of the Rtt103-CID, as judged by small changes in the 15N-HSQC (data not shown). This result strongly suggests that we are observing a cooperative interaction between neighboring Rtt103-CIDs templated by the CTD.
To assess the generality of this observation, we tested mutants in the corresponding region of the Pcf11-CID. We mutated Asp117 to alanine or lysine in the loop connecting helices 7 and 8 because this region is involved in crystal contacts in the structure of Pcf11-CID, and tested recognition of the four heptad repeat Ser2P CTD using fluorescence anisotropy (Table 2). Mutation of D117A resulted in decreased affinity for the second binding event, but the effect is not as dramatic as observed for the glutamate to arginine mutation in Rtt103-CID and the data can still be reliably fit with a 2:1 binding model. Mutation of D117K results in affinity similar to the wild type Pcf11-CID.
Finally, we tested binding of wild type Pcf11-CID to a four heptad repeat peptide where the two amino terminal heptads have Ser2P and the two carboxyl terminal heptads are unphosphorylated (Fig. 5a). With this peptide, we observe that the Pcf11-CID binds with wild type affinity for the first repeat, but has substantially weaker affinity for the second repeat; its binding is best fit using 1:1 stoichiometry (Supplementary Fig 6b online). Thus, it appears that for the Pcf11-CID, cooperative binding of a second molecule to the longer CTD peptides requires Ser2P on adjacent diheptad peptide units.
In order to establish the physiological significance of these findings, we used ChIP to compare the recruitment of Pcf11 (D117A) and Rtt103 (E115R) with wild type proteins. The ChIP results clearly show a strong (~20-50 %) reduction in recruitment of the Rtt103 (E115R) mutant to the 3′-end of the PMA1 and ADH1 genes (Fig. 5d). Pcf11 (D117A) also shows reduced recruitment at the 3′-end processing site (Fig. 5e) but this effect is less dramatic than the observed reduction for Rtt103 (E115R) (~20 % for the PMA1 and ADH1 genes), consistent with the less dramatic loss of affinity observed for this mutant. Expression levels remain unchanged compared to wild type proteins for both point mutants (not shown).
Discussion
Co-transcriptional RNA processing depends on the precisely-timed recruitment of processing factors to the elongating transcript, which is facilitated by recognition of specific CTD phosphoisoforms through specialized CTD binding domains29. Changes in CTD phosphorylation during transcription generate a ‘CTD code’ that can be read by these protein domains30. This ‘thermodynamic’ model of factor recruitment assumes that CTD-binding domains have sufficient affinity and selectivity to bind to and discriminate between different CTD states. However, when we compared binding of Rtt103-CID and Pcf11-CID to peptides containing diheptad repeats (several studies have suggested that the CTD functional unit is composed of pairs of heptad repeats19) phosphorylated at Ser2 and/or Ser5 in all possible combinations (Fig. 1b), we observed that the Pcf11-CID has only modest preference for CTD peptides phosphorylated at Ser2 (Table 1), consistent with previous results31,32. Given that the Pcf11-CID is critically important in transcription termination for mRNA-coding genes23, and that Pcf11 is sharply concentrated near the 3′-end of such genes, we expected, instead, that Pcf11 would bind specifically and with high affinity to the Ser2P form of the CTD that is predominant at or near poly(A) processing sites14.
Examination of the structures of Rtt103-CID and Pcf11-CID bound to the Ser2P CTD indicated that the differing affinities are likely due to a direct contact with the Ser2 phosphate by the Rtt103-CID mediated by Arg108 (Fig. 4a). By mutating Pcf11-CID Asn107 to arginine and correspondingly mutating Arg108 in Rtt103-CID to asparagine, we essentially reversed the relative affinities of Rtt103-CID and Pcf11-CID for the Ser2P CTD (Fig. 4b). The importance of this direct contact is further corroborated in the SCAF8-CID where the corresponding residue, Arg112, makes direct contacts to the Ser2 phosphate as well, and mutation to a threonine residue shows a reduction by a factor of 4 in affinity22. Supporting the functional relevance of the in vitro data, this single amino acid change in Rtt103 and Pcf11 resulted in an altered recruitment to the 3′-end processing site in vivo (Fig. 4c, d).
These results surprised us; during evolution, this single amino acid could have been readily changed to provide Pcf11 with increased specificity for Ser2P CTD. However, when we tested for termination defects with the Pcf11 (N107R) and Rtt103 (R108N) mutants we could not find conclusive evidence for a termination defect. We reasoned that perhaps these proteins also rely on additional clues (proteinprotein, proteinRNA interactions or the distance from the promoter) to identify a proper site to terminate mRNA transcription. One possibility was the untested suggestion that the CTD may facilitate cooperative proteinprotein interactions on neighboring repeats and thereby increase the specificity of these proteins for particular CTD phosphorylation patterns.
We observed that the four heptad repeat peptides could accommodate two CIDs (Table 2), and that these proteins behaved as a single complex in solution. Several observations suggested that protein-protein interactions occur between CIDs bound to the four heptad repeat CTD involving the loops connecting helices 1 and 2 as well as helix 7. Mutation of a single residue in helix 7 of Rtt103-CID dramatically altered the affinity of the second CID molecule to the four heptad repeat CTD, to the point that cooperativity is lost. For Pcf11-CID, mutation of Asp117 to alanine reduced binding of the second Pcf11-CID molecule in a manner consistent with the observed effect for the Rtt103-CID mutants, yet mutation of Asp117 to lysine behaved similarly to wild type in vitro. Since this loop of Pcf11 is quite flexible31, it is possible that mutation to alanine results in a loop conformation that hinders binding of the second CID while a lysine at the same position results in a conformation where the second CID can still bind. Building from these in vitro results, we tested the Rtt103-CID and Pcf11-CID mutants that severely reduced binding of the second CID molecule in vivo, and observed reduced recruitment to the 3′ processing site of several genes (Fig. 5d, e), confirming that the observed cooperativity in vitro is important for factor recruitment in vivo. For the Rtt103 mutants, we observed a significant decrease in protein localization near the 3′-end of genes, which demonstrates the importance of cooperativity for its recruitment. For the Pcf11 mutant, the decrease is significant but not as marked as for Rtt103. Perhaps recruitment of Pcf11 is less dependent on cooperative interactions with the CTD because, as a stable component of CFIa, it relies on multiple proteinprotein and proteinRNA interactions to provide robustness to recruitment at the 3′-end of genes. However, these additional interactions do not fully compensate for the loss of a cooperative interaction with the CTD, since recruitment levels are still reduced significantly compared to wild type. It should also be noted that CFIa may be a dimer23,33,34, thus more than one Pcf11 molecule may be present at the 3′ processing site which could facilitate cooperative interactions with the CTD. Thus, it appears that cooperativity mediated by direct proteinprotein interactions is an important mechanism for recruitment of Rtt103, and to a lesser extent Pcf11, to the 3′-ends of genes.
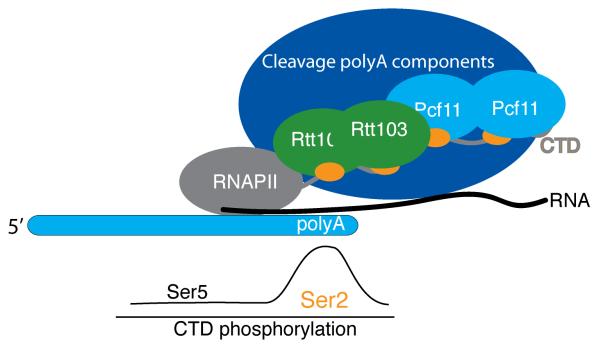
The phosphorylation state of the CTD is a sensor indicating the position of Pol II within a gene14,20,30,35. Two competing transcription termination mechanisms use this positional cue to decide which termination pathway to use. For the Nrd1-dependent pathway of snoRNA-coding genes, high Ser5P density within the CTD predominant at short distances from the transcription start recruits the Nrd1-Nab3-Sen1 complex to act on short RNAs20,35. Near poly(A) sites of mRNA-coding genes, high Ser2P density provides a mechanism to tightly regulate binding of Rtt103 and Pcf11 so that their recruitment occurs only when the polymerase reaches proper mRNA termination sites. Our results suggest that the increasing density of Ser2P heptad peptide repeats facilitate cooperative proteinprotein interactions between neighboring CIDs (Fig. 6). In this model, cooperative binding to neighboring repeats may allow Pcf11 and Rtt103 to respond sigmoidally to changes in the CTD phosphorylation state, providing a potent signal-response mechanism that results in the tight regulation of transcription termination.
Figure 6.
Cooperative model of Pcf11 and Rtt103 recruitment. The density of Ser2-phosphorylated heptad repeats affects the location of transcription termination through cooperative recruitment of Pcf11 and Rtt103. At the polyA site, the density of Ser2-phosphorylation is highest15, allowing efficient recruitment of Pcf11 and Rtt103.
Methods
Protein samples were prepared as described in the Supplementary Methods, and all peptides purchased from AnaSpec (San Jose, CA). NMR experiments on CIDs and their complexes were recorded 15N/13C-labeled samples at ~1 mM protein concentration on various spectrometers at fields of 500-900 Mhz, equipped with cryoprobes, at the University of Washington and at the Environmental Molecular Sciences Laboratory (EMSL) at Pacifc Northwest National Laboratory (PNNL). Backbone and side-chain resonance assignments were obtained from standard heteronuclear NMR experiments37; while NOE data were obtained using 15N- and 13C- edited three-dimensional spectra. Intermolecular NOEs between Rtt103 and the CTD peptide were observed with 2D-filtered-edited NOESY experiments38. Relaxation experiments were collected for the free protein, as well as for Rtt103 and Pcf11 bound to both 2- and 4-repeat Ser2 phosphorylated CTD peptides as described39. NOE assignments and structure calculation for both free Rtt103-CID and for Rtt103-CID bound to the CTD were performed using CYANA2.140. The structure of Rtt103-CID bound to the CTD was determined using multiple samples, as described in the Supplementary Methods. For the Rtt103-CID complex, docking with HADDOCK41 was used starting from structures generated using CYANA2.1 and CNS42. The strength of the interaction between Pcf11-CID and Rtt103-CID with various CTD phosphoisoforms was measured using both NMR titrations, and fluorescence anisotropy assays using N-terminally 5,6-carboxyfluorescein (FAM) labeled CTD peptides, using standard methods as detailed in the Supplementary Methods. ChIP assays of TAP-tagged Rtt103 and Pcf11 were performed using IgG sepharose (GE Healthcare) as previously described28. PMA1 and ADH1 primers used in PCR are as listed in Kim et al43. ChIP assays for 3xHA tagged Pcf11 or Rtt103 were carried out as previously described28. Yeast strains and plasmid construction are provided in the Supplementary Methods.
Supplementary Material
Acknowledgements
We thank Roland Becker for his assistance with the fluorescence anisotropy experiments. We would also like to thank Borries Demeler and Virgil Schirf at the Center for Analytical Ultracentrifugation of Macromolecular Assemblies for assistance with the AUC experiments. This work was supported by grants from the NIH-NIGMS to G.V. (GM64440) and S.B. (GM56663), from the German Research Foundation to A.M. (ME3135/1-1) and by the WCU project (305-20080089) from KMEST and grants from the National Research Foundation of Korea (NRF) (R31-2009-000-10032-0 and 2010-0011750) to M.K. A portion of the research was performed using EMSL, a national scientific user facility sponsored by the Department of Energy's Office of Biological and Environmental Research located at Pacific Northwest National Laboratory.
Footnotes
Accession Codes
Protein Data Bank: the coordinates for Rtt103 structures and Rtt103 bound to the Ser2P CTD have been deposited with accession codes 2KM4 and 2L0I, respectively.
References
- 1.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 4162002:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 2.Corden JL, Patturajan M. A CTD function linking transcription to splicing. Trends Biochem Sci. 221997:413–6. doi: 10.1016/s0968-0004(97)01125-0. [DOI] [PubMed] [Google Scholar]
- 3.Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 1082002:501–12. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 4.de la Mata M, et al. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 122003:525–32. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Howe KJ, Kane CM, Ares M., Jr. Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. RNA. 92003:993–1006. doi: 10.1261/rna.5390803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho CK, Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell. 31999:405–11. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- 7.Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 172005:251–6. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 8.Cho EJ, Rodriguez CR, Takagi T, Buratowski S. Allosteric interactions between capping enzyme subunits and the RNA polymerase II carboxy-terminal domain. Genes Dev. 121998:3482–7. doi: 10.1101/gad.12.22.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corden JL. Tails of RNA polymerase II. Trends Biochem Sci. 151990:383–7. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- 10.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 242008:280–8. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 202006:2922–36. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 12.McCracken S, et al. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 111997:3306–18. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho EJ, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 111997:3319–26. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 142000:2452–60. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell. 132004:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 16.Chapman RD, et al. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 3182007:1780–2. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- 17.Egloff S, et al. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 3182007:1777–9. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akhtar MS, et al. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell. 342009:387–93. doi: 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stiller JW, Cook MS. Functional unit of the RNA polymerase II C-terminal domain lies within heptapeptide pairs. Eukaryot Cell. 32004:735–40. doi: 10.1128/EC.3.3.735-740.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol. 152008:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim M, et al. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 4322004:517–22. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 22.Becker R, Loll B, Meinhart A. Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2832008:22659–69. doi: 10.1074/jbc.M803540200. [DOI] [PubMed] [Google Scholar]
- 23.Sadowski M, Dichtl B, Hubner W, Keller W. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 222003:2167–77. doi: 10.1093/emboj/cdg200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meinhart A, Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature. 4302004:223–6. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- 25.Luo W, Johnson AW, Bentley DL. The role of Rat1 in coupling mRNA 3′-end processing to transcription termination: implications for a unified allosteric-torpedo model. Genes Dev. 202006:954–65. doi: 10.1101/gad.1409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Birse CE, Minvielle-Sebastia L, Lee BA, Keller W, Proudfoot NJ. Coupling termination of transcription to messenger RNA maturation in yeast. Science. 2801998:298–301. doi: 10.1126/science.280.5361.298. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z, Fu J, Gilmour DS. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes Dev. 192005:1572–80. doi: 10.1101/gad.1296305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 232004:354–64. doi: 10.1038/sj.emboj.7600053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P. A structural perspective of CTD function. Genes Dev. 192005:1401–15. doi: 10.1101/gad.1318105. [DOI] [PubMed] [Google Scholar]
- 30.Buratowski S. The CTD code. Nat Struct Biol. 102003:679–80. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- 31.Noble CG, et al. Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nat Struct Mol Biol. 122005:144–51. doi: 10.1038/nsmb887. [DOI] [PubMed] [Google Scholar]
- 32.Ramos A, et al. The structure of the N-terminal domain of the fragile X mental retardation protein: a platform for protein-protein interaction. Structure. 142006:21–31. doi: 10.1016/j.str.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 33.Noble CG, Walker PA, Calder LJ, Taylor IA. Rna14-Rna15 assembly mediates the RNA-binding capability of Saccharomyces cerevisiae cleavage factor IA. Nucleic Acids Res. 322004:3364–75. doi: 10.1093/nar/gkh664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bai Y, et al. Crystal structure of murine CstF-77: dimeric association and implications for polyadenylation of mRNA precursors. Mol Cell. 252007:863–75. doi: 10.1016/j.molcel.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 35.Gudipati RK, Villa T, Boulay J, Libri D. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat Struct Mol Biol. 152008:786–94. doi: 10.1038/nsmb.1460. [DOI] [PubMed] [Google Scholar]
- 36.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific LLC; San Carlos, CA, USA: 2002. [Google Scholar]
- 37.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Mag Res Sp. 341999:93–158. [Google Scholar]
- 38.Zwahlen C, et al. Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: Application to a bacteriophage lambda N-peptide/boxB RNA complex. Journal of the American Chemical Society. 1191997:6711–6721. [Google Scholar]
- 39.Farrow NA, et al. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 331994:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 40.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2782004:353–78. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 41.Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 1252003:1731–7. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 42.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 541998:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 43.Kim M, et al. Distinct pathways for snoRNA and mRNA termination. Mol Cell. 242006:723–34. doi: 10.1016/j.molcel.2006.11.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.