Abstract
Escherichia coli RecBCD is a DNA helicase with two ATPase motors (RecB, a 3′ to 5′ translocase, and RecD, a 5′ to 3′ translocase) that functions in repair of double-stranded DNA breaks. The RecBC heterodimer, with only the RecB motor, remains a processive helicase. Here we examined RecBC translocation along single stranded (ss) DNA. Surprisingly, we find that RecBC displays two translocase activities: the primary translocase moves 3′ to 5′, while the secondary translocase moves RecBC along the opposite strand of a forked DNA at a similar rate. The secondary translocase is insensitive to the ssDNA backbone polarity, and we propose that its function may be to fuel RecBCD translocation along double stranded DNA ahead of the unwinding fork, and to ensure that the unwound single strands move through RecBCD at the same rate after interaction with a Chi sequence.
Keywords: allostery, translocation, DNA, fluorescence, recombination
Escherichia coli RecBCD is a molecular motor possessing ATPase, DNA helicase, and nuclease activities. This hetero-trimeric enzyme initiates repair of double strand (ds) DNA breaks via homologous recombination and degrades foreign DNA 1. RecBCD possesses two motor subunits, RecB (134 kDa), a 3′ to 5′ DNA helicase and nuclease, and RecD (67 kDa), a 5′ to 3′ DNA helicase 2–4. RecC (129 kDa) is a processivity and regulatory factor that interacts with both RecB and RecD and is structurally homologous to RecB but with non-functional helicase and nuclease domains 5–6. Although RecB and RecD translocate with opposite directionalities along single stranded (ss) DNA, they function within RecBCD to unwind DNA in the same net direction by translocating along complementary DNA strands of the duplex 6, as depicted in Figure 1. To initiate recombinational DNA repair, RecBCD first binds to a dsDNA break and unwinds the DNA using its bipolar helicase activity. At first, RecD is the faster motor 4,7 and the RecB nuclease activity preferentially degrades the 3′ ssDNA end until the complex recognizes a Chi (crossover hotspot instigator) sequence (5′–GCTGGTGG) within the unwound 3′-ssDNA. At this point, the enzyme pauses then continues to unwind at a reduced rate with RecB as the faster motor. In addition, the nuclease selectivity switches to act exclusively on the 5′-ssDNA end, producing a 3′-ssDNA end onto which RecA enzyme is loaded. The resulting RecA filament then initiates homologous recombination to repair of the dsDNA break 1. RecB and RecD are both superfamily 1 (SF1) DNA helicases, each with a functional ATPase motor2. In the absence of RecD, a stable heterodimeric RecBC complex retains highly processive and rapid helicase activity4,8–10.
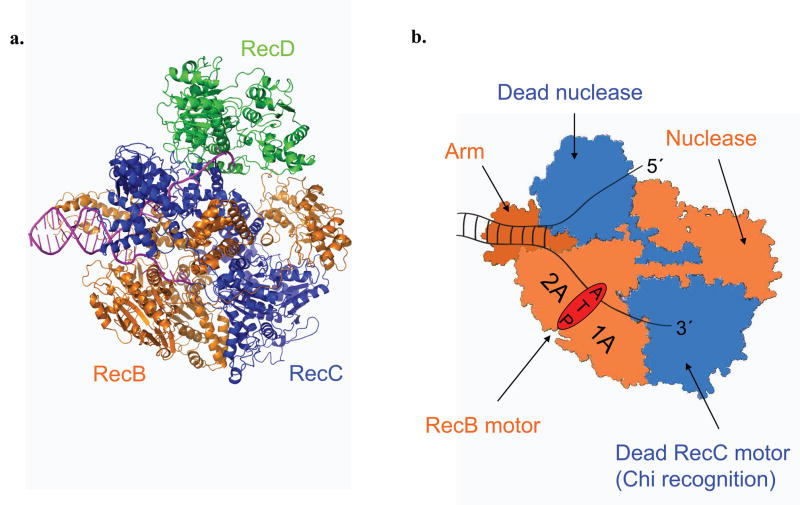
Figure 1. RecBCD and RecBC structures.
RecB (orange), RecC (blue), and RecD (green) subunits are indicated. (a). Ribbon diagram of a RecBCD–DNA complex 6,21. (b). Cartoon depiction of a RecBC–DNA complex. RecB motor, nuclease, and arm domains are indicated along with the catalytically dead RecC motor and nuclease domains. The paths of the 3′- and 5′-terminated unwound ssDNA are shown.
The DNA binding 11–14 and helicase properties of RecBCD and RecBC 3–4,8,10,15–20 have been studied, and structures of RecBCD bound to a duplex DNA end have been determined 6,21 (Fig. 1). Upon binding to a blunt ended DNA duplex, both RecBCD and RecBC 6,11,13,22 melt out 5–6 base pairs (bp) in a Mg2+-dependent, but ATP-independent reaction. However, RecBCD unwinds DNA more rapidly than RecBC 4; (774 ± 16 bp s−1 vs. 348 ± 5 bp s−1) under the same conditions 10,18–19.
To unwind DNA processively, a helicase must also translocate along DNA. From DNA unwinding experiments, Bianco and Kowalczykowski 8 inferred that RecBC translocates along ssDNA with 3′ to 5′ directionality, consistent with the directionality of the RecB motor 23–24. However, the ssDNA translocation properties of RecB, RecBC and RecBCD have not been examined directly, and thus, the relationship between ssDNA translocation and DNA unwinding is not established. Here, we examined ssDNA translocation of RecB and RecBC and show that RecBC not only possesses its expected primary 3′ to 5′ ssDNA translocase activity, but also a second previously undetected translocation activity, both of which are controlled by the RecB motor. This discovery has important implications for the mechanism and regulation of DNA unwinding activities by RecBC and RecBCD.
Results
RecB monomer translocation along ssDNA
A stopped-flow fluorescence approach 25 (see Methods) was used to monitor RecB translocation along a series of ss oligodeoxythymidylates, (dT)L, L nucleotides long with a fluorophore (Cy3 or Oregon Green (OG)) attached on the 5′- or the 3′-end. When RecB reaches the ssDNA end labeled with Cy3 or OG, Cy3 fluorescence is enhanced, whereas OG fluorescence is quenched; hence, one can monitor the kinetics of RecB arrival at the ssDNA ends. Use of a two-fold molar excess of ssDNA over RecB ensures that no more than one RecB monomer is bound to each ssDNA. Translocation was initiated by mixing pre-formed RecB–(dT)L complexes with ATP and heparin, the latter serving as a trap to prevent rebinding to the DNA of any RecB that dissociates during translocation or that was initially free. Hence, a single round of translocation is monitored, although multiple rounds of ATP hydrolysis occur.
The translocation time courses for different DNA lengths (Fig. 2a and 2b) indicate that RecB monomers bind randomly to (dT)L and translocate with 3′ to 5′ directionality 25. Identical experiments performed with a (dT)54 substrate with Cy3 on the 3′ end of the ssDNA, show an exponential decrease in Cy3 fluorescence (Fig. 2c), the rate of which is independent of ssDNA length (data not shown) indicating RecB translocates away from the 3′-end. These data indicate that RecB translocates along ssDNA with biased 3′ to 5′ directionality.
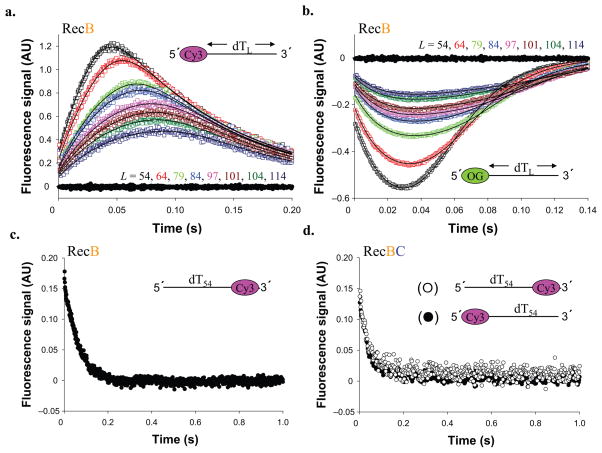
Figure 2. RecB translocates with 3′ to 5′ directionality along ssDNA.
(a) RecB monomer translocation kinetics for a series of 5′–Cy3–(dT)L DNA (Supplementary Table 1, DNA I–VIII). Cy3 fluorescence from DNA alone (DNA I) is shown in filled black circles. (b). RecB monomer translocation kinetics for a series of 5′–OG–(dT)L substrates. OG (Oregon Green) fluorescence from DNA alone (DNA I) is shown in filled black circles. Smooth black curves in panels A and B are simulated time courses using Equation S1 and the best fit kinetic parameters (Table 1). (c). Time course obtained with RecB and (dT)54 –Cy3–3′ (Supplementary Table 1, DNA IX). (d). Time course obtained with RecBC and 5′–Cy3–(dT)54 (filled circles) or (dT)54–Cy3–3′ (opened circles).
The time courses in Figure 2a and 2b were analyzed using the n-step sequential translocation model in Scheme 1 (Eq. S1), to obtain fluorophore-independent estimates of the translocation kinetic parameters (Table 1). A dissociation rate constant, kd = 7.5 ± 0.3 s−1, determined from independent RecB–poly(dT) dissociation experiments (see Supplementary Fig. 1), was constrained in the analysis of the translocation time courses as described 26. This analysis indicates that RecB translocates along ssDNA in the 3′ to 5′ direction with a macroscopic rate of mtkt = 803 ± 13 nucleotides per second (nt s−1). The smooth curves in Figure 2 show that the n-step sequential model provides a good description of the translocation kinetics.
Table 1.
RecB and RecBC translocation kinetics summary
| NLLS fit to Eq S1 (Scheme 1) | mtkt (nt s−1) | kt (s−1) | mt (nt) | kd (s−1) | kend (s−1) | d (nt) | r |
|---|---|---|---|---|---|---|---|
| RecB | 803 ± 13 | 168 ± 25 | 4.8 ± 0.3 | 7.5 ± 0.3 (constrained) | 11 ± 3 | 18 ± 2 | 0.8 ± 0.1 |
| NLLS fit to Eq S2 (Scheme 2) | mtkt (nt s−1) | kt (s−1) | mt (nt) | mUkU (bp s−1) | kU (s−1) | mU (bp) | kend (s−1) |
| RecBC (3′ to 5′) primary along ssDNA | 920 ± 33 | 260 ± 41 | 3.5 ± 0.2 | 348 ± 5 (constrained) | 79 ± 11 (constrained) | 4.4 ± 0.1 (constrained) | 2.7 ± 0.1 |
| “lag time” analysis | mtkt (Cy3) (nt s−1) | mtkt (F) (nt s−1) | |||||
| RecBC (3′ to 5′) primary along ssDNA | 909 ± 51 | 1,030 ± 53 | |||||
| RecBC (5′ to 3′) secondary along ssDNA | 990 ± 49 | 1,187 ± 61 | |||||
| RecBC along both ssDNA strands (5′ Cy3) | 671 ± 47 | ||||||
| RecBC along both ssDNA strands (3′ Cy3) | 621 ± 43 | ||||||
| Fit to Eq S4 | Vmax (nt s−1) | KM (μM) | |||||
| RecB | 860 ± 53 | 125 ± 38 | |||||
| RecBC (3′ to 5′) Primary | 946 ± 64 | 203 ± 32 | |||||
| RecBC (5′ to 3′) Secondary | 1,055 ± 75 | 123 ± 28 | |||||
| NLLS fit to Eq S3 (Scheme 3) | mtkt (nt s−1) | kt (s−1) | mt (nt) | mUkU (bp s−1) | kU (s−1) | mU (bp) | |
| RecBC gap unwinding 3′ to 5′ ssDNA gap | 928 ± 38 | 242 ± 60 | 3.8 ± 0.4 | 396 ± 15(constrained) | 90 ± 25 (constrained) | 4.4 ± 1.7 (constrained) | |
| RecBC gap unwinding 5′ to 3′ ssDNA gap | 919 ± 42 | 262 ± 57 | 3.5 ± 0.2 | 396 ± 15 (constrained) | 90 ± 25 (constrained) | 4.4 ± 1.7 (constrained) | |
The best fit parameters for RecB and RecBC translocation are summarized. The equations and kinetic schemes used to analyze the data are given in Supplementary Data. Errors denote s.d.
ssDNA translocation by RecBC
We next examined ssDNA translocation of RecBC using the same 5′–Cy3–(dT)L substrates; however, no translocation activity was detected. In fact, Figure 2d shows exponential decreases in Cy3 fluorescence when pre-bound RecBC–DNA complexes (5′–Cy3–(dT)54 or (dT)54–Cy3–3′) were mixed with ATP and heparin, suggesting that RecBC is unable to initiate translocation on ssDNA alone and instead dissociates from the ssDNA. Based on the structures of RecBCD DNA complexes 6,21 (Fig. 1), we hypothesize that it is difficult for ssDNA to be threaded into the RecBC complex to properly engage the RecB motor and designed a different DNA substrate to monitor RecBC translocation.
RecBC binds weakly to ssDNA and blunt duplex DNA ends, but binds with high affinity to a DNA end possessing twin 5′-dT6 and 3′-dT6 tails 13 and RecBC can rapidly initiate DNA unwinding from such a site 10. Therefore, we hypothesized that once RecBC initiates DNA unwinding from this loading site, it might continue to translocate in the 3′ to 5′ direction along a (dT)L ssDNA extension. The modified DNA substrates used to test for RecBC translocation along ssDNA are shown schematically in Figure 3a and 3b. These substrates contain a 24 bp duplex with a high affinity RecBC loading site at one end. On the other end of the duplex is a ssDNA ((dT)L) extension of length, L, labeled on its 5′-end with either Cy3 or Fluorescein (F); these substrates should detect 3′ to 5′ translocation of RecBC along the ss-(dT)L extensions. To differentiate between the two DNA strands in these molecules, we refer to them as the 3′-terminated strand and the 5′-terminated strand, where the 3′ or 5′ denotes the duplex end containing the RecBC loading site. Hence, in Figure 3a, the strand with the ssDNA extension is the 3′-terminated strand. To ensure that RecBC initiates at the high affinity loading site, a twofold molar excess of DNA (200 nM) over RecBC (100 nM) was used. If all RecBC enzymes initiate DNA unwinding from this unique site, we expect a measurable lag time before RecBC arrives at the fluorophore-labeled 5′-ssDNA end 27.
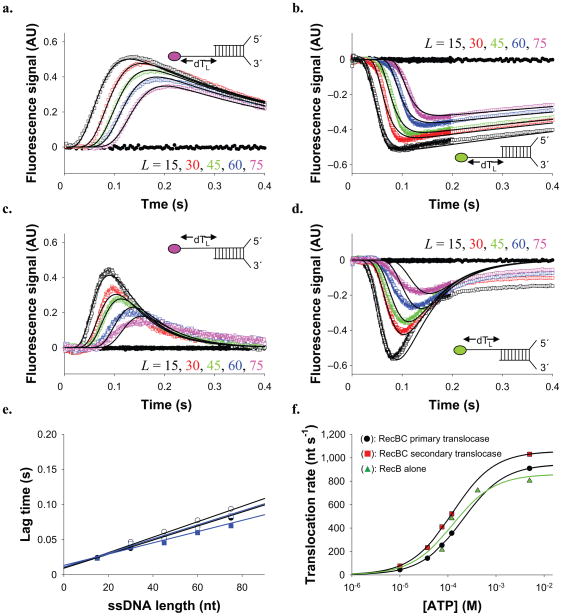
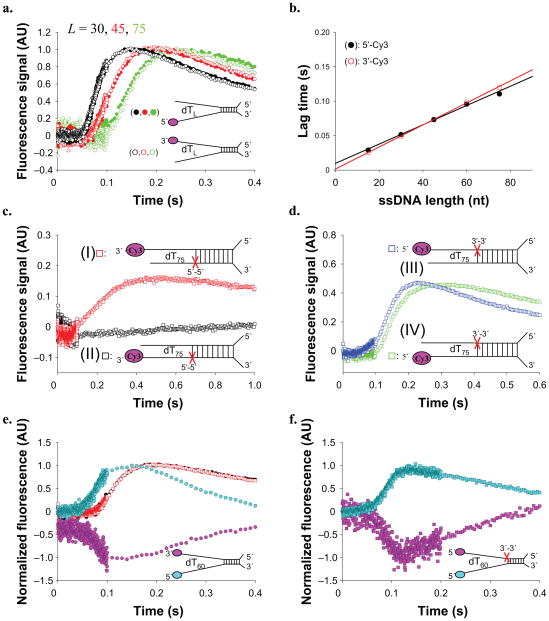
Figure 3. RecBC displays both a primary (3′ to 5′) and secondary (5′ to 3′) translocase activity.
RecBC translocation time courses obtained using the DNA substrates depicted which possesses a 24 bp duplex with a high affinity (twin-dT6) RecBC loading site on one end and either 5′– or 3′–(dT)L ssDNA extensions on the other end. (a). DNA substrates labeled with Cy3 on the 5′ end of the (dT)L extension. (b). DNA substrates labeled with fluorescein (F) on the 5′ end of the (dT)L extension. (c). DNA substrates labeled with Cy3 on the 3′ end of the (dT)L extension. (d). DNA substrates labeled with fluorescein (F) on the 3′ end of the (dT)L extension. Smooth black curves in panels a d are simulated time courses using Equation S2 and the kinetic parameters in Table 1. (e). Dependence of the lag time on ssDNA extension length, L. Cy3 data from panel a (opened circles) (Lag time = 0.00110 L + 0.0095) (909 ± 51 nt s−1). Fluorescein data from panel b (filled circles) (Lag time = 0.000971 L + 0.0110) (1,030 ± 53 nt s−1). Cy3 data from panel c (opened squares) (Lag time = 0.00101 L + 0.0093) (990 ± 49 nt s−1). Fluorescein data from panel d (filled squares) (Lag time = 0.000843 L + 0.0134) (1,187 ± 61 nt s−1). (f). [ATP] dependence of RecBC translocation rates (from lag time analyses) for the (circles)-primary (3′ to 5′) (Vmax = 946 ± 64 nt s−1, KM = 203 ± 32 μM); and (squares)-secondary (5′ to 3′) (Vmax = 1,055 ± 75 nt s−1, KM = 123 ± 28 μM) translocases. (triangles)-Effects of [ATP] on RecB monomer translocation (Vmax = 860 ± 53 nt s−1, KM = 125 ± 38 μM). Smooth curves represent fits to the Michaelis-Menton equation (Eq. S4) and the best fit parameters summarized in Table 1.
The results of experiments with RecBC and these DNA substrates show a lag phase (Fig. 3a–3b), the duration of which increases with ssDNA extension length, L, consistent with RecBC translocating along the extension in the 3′ to 5′ direction after unwinding the 24 bp duplex. For the 5′–Cy3–(dT)L extensions (Fig. 3a), Cy3 fluorescence increases after the lag, reflecting arrival of RecBC at the 5′-end, after which Cy3 fluorescence decreases to its starting value after ~ 2 seconds, reflecting RecBC dissociation. The same trend, but with a transient quenching of fluorescein fluorescence is observed for DNA with 5′–F–(dT)L extensions (Fig. 3b).
We analyzed these time courses using two approaches: an analysis of the “lag time” (defined in Supplementary Fig. 2) and an analysis of the complete time course (described in Supplementary Data). The “lag time” for each time course reflects the average time for RecBC to reach the 5′ end of the ssDNA and is linearly dependent on the (dT)L extension length, L (see Fig. 3e), consistent with directional translocation. The reciprocal slope of these plots yields an estimate of the 3′ to 5′ translocation rate, which is the same for both substrates (909 ± 51 nt s−1 (Cy3); 1,030 ± 53 nt s−1 (F)). Since all DNA molecules contain the same 24 bp duplex, these rates only reflect ssDNA translocation.
We also analyzed the full time courses in Figure 3a and 3b using Scheme 2 (Eq. S2), which combines two tandem n-step sequential schemes, reflecting the unwinding of the 24 bp duplex by RecBC, followed by translocation of RecBC along the (dT)L extensions. In this analysis, the DNA unwinding kinetic parameters (kU and mU) were constrained to the values determined from a previous study of DNA unwinding by RecBC performed under the same solution conditions 10, allowing us to float only the kinetic parameters for RecBC translocation. The resulting ssDNA translocation rate of 920 ± 33 nt s−1 is the same within error as the rates determined from the “lag time” analyses. Both Cy3 and fluorescein time courses are well described by this mechanism and the parameters in Table 1, as shown by the simulated curves in Figure 3a and 3b.
RecBC has a distinct secondary translocase activity
We next performed control experiments with RecBC using a similar set of partial duplex substrates but with the fluorophore-labeled (dT)L ssDNA extending from the 5′-terminated strand. If RecBC translocates along ssDNA with strict 3′ to 5′ directionality, then no length dependent translocation signal should be observed on these substrates since the strand along which RecB translocates stops at the end of the duplex; therefore, RecBC would be expected to either dissociate or become stuck after unwinding the 24 bp duplex. To our great surprise, we observed the characteristic signature of ssDNA translocation (length-dependent lag, followed by a peak (trough) in fluorescence) (Fig. 3c–3d) indicating movement of RecBC in the unexpected 5′ to 3′ direction.
“Lag time” analyses (Fig. 3e) show linear dependences on ssDNA length, consistent with RecBC translocation in the 5′ to 3′ direction with rates of 990 ± 49 nt s−1 (Cy3) and 1187 ± 61 nt s−1 (F), which are the same within error as the rates determined for RecBC translocation in the 3′ to 5′ direction. Although the time courses observed for the two types of DNA substrates ((dT)L extensions on the 3′-terminated strand (Fig. 3a–b) versus on the 5′-terminated strand (Fig. 3c–3d)) show the same qualitative characteristics, they differ in detail and are not superimposable (see Supplementary Fig. 3). The amplitudes of the fluorescence changes (both Cy3 and F) are larger and the dissociation rates of RecBC are slower for the DNA substrates that monitor 3′ to 5′ translocation. Furthermore, the complete time courses for the DNA substrates that monitor 5′ to 3′ translocation are not described as well by the simple tandem n-step sequential model of Scheme 2, suggesting that additional steps (e.g., pausing or conformational rearrangement or multistep dissociation from the ssDNA end) may occur. From here on, we will refer to the 3′ to 5′ translocation activity as the “primary” RecBC translocase, and the apparent 5′ to 3′ translocation activity as the “secondary” RecBC translocase.
We note that when RecB alone is examined using the DNA substrates in Figure 3a and 3c, we observe translocation profiles similar to those shown in Figure 2a and 2c (data not shown), indicating that RecB preferentially loads onto the ssDNA extensions at random sites and moves with 3′ to 5′ directionality. When RecC alone is examined with any of the DNA substrates described above, no translocation activity is observed. We also note that the RecBC used here was purified from E. coli strains that do not express RecD and thus the secondary translocase activity is not due to RecD contamination.
ATP-dependence of the primary and secondary translocase activities
We next examined the effect of [ATP] on the primary and secondary translocase activities of RecBC. Cy3 time courses were obtained for all five (dT)L extension lengths at each [ATP]. Both primary and secondary translocation rates, determined from “lag time” analyses (see Supplementary Fig. 4), display hyperbolic dependences on [ATP] (Fig. 3f) and were fit to the Michaelis-Menton model (Eq. S4). The primary translocase has Vmax = 946 ± 64 nt s−1 and KM = 203 ± 32 μM while the secondary translocase has Vmax = 1,055 ± 75 nt s−1 and KM = 123 ± 28 μM; hence, both KM values are similar. Translocation by the RecB monomer alone has Vmax = 860 ± 53 nt s−1 and KM = 125 ± 38 μM (Fig. 3f).
The primary 3′ to 5′ translocation activity of RecBC was anticipated, but our discovery of a secondary 5′ to 3′ translocase activity was surprising. Both translocase rates are similar over a range of [ATP] and [NaCl] (Supplementary Fig. 6), suggesting that they are both driven by the same motor (RecB). Since RecB ATPase activity is strongly stimulated by binding ssDNA 23,28, it would seem necessary for the primary ssDNA binding site of RecB to remain bound to ssDNA during the course of both translocase activities. This suggests that in order for RecBC to translocate in the 5′ to 3′ direction after unwinding the 24 bp duplex (Fig. 3c), the primary ssDNA binding site within RecB must remain bound to the short unwound 3′-terminated ssDNA strand. If instead, RecBC translocated off the end of the unwound ssDNA, then RecB ATPase activity would decrease dramatically and would not support the secondary translocation activity.
We designed two DNA substrates (see Fig. 4a and 4b) to test whether the primary RecBC translocase stops and remains bound to the end of the short ssDNA after unwinding the 24 bp duplex. The ends of the (dT)60 extensions are labeled with Cy3, while the ends of the shorter strand that forms the 24 bp duplex are labeled with Cy5. The Cy3 (donor) and Cy5 (acceptor) fluorophores can undergo fluorescence resonance energy transfer (FRET) that can be monitored as a change in the Cy5 acceptor fluorescence when the Cy3 donor is excited. Thus, if one of either translocase gets stuck at the end of the 24 bp duplex, the Cy3 will be brought closer to the Cy5 resulting in a transient increase in Cy5 fluorescence. However, if RecBC translocates past the Cy5 donor and releases the short Cy5 labeled strand after unwinding, then a decrease in Cy5 fluorescence should be observed.
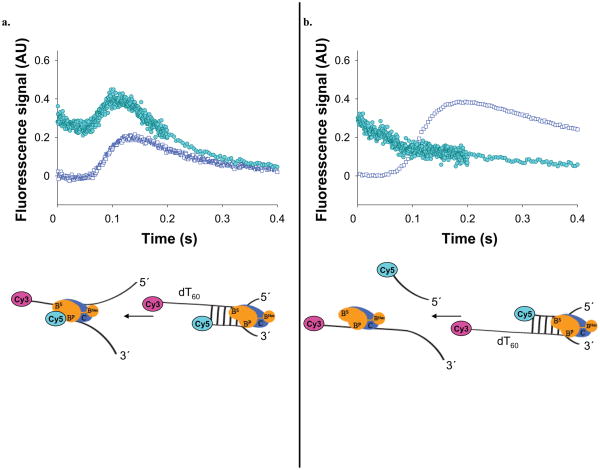
Figure 4. The primary RecBC translocase site remains bound to ssDNA upon reaching a 5′-end, while its secondary translocase continues.
(a) RecBC translocation along a partial duplex substrate with a 5′ to 3′ dT60 ssDNA extension, doubly labeled with Cy3 and Cy5 as shown. Time course monitoring FRET as Cy5 fluorescence (filled circles) (due to exciting Cy3 donor) shows that the 3′-terminated ssDNA remains bound to RecBC while the secondary translocase continues; × Translocation time course for the same DNA, but without the Cy5 label (opened squares). (b). RecBC translocation along a partial duplex substrate with a 3′ to 5′ dT60 ssDNA extension doubly labeled with Cy3 and Cy5 as shown. Time course monitoring FRET as Cy5 fluorescence (filled circles) (due to exciting Cy3 donor) shows dissociation of the 5′-terminated ssDNA; Translocation time course for the same DNA, but without the Cy5 label (opened squares).
The results of the two FRET experiments are shown in Figure 4a and 4b. The substrate in Figure 4a shows a lag followed by a transient increase in Cy5 fluorescence and then a decrease, consistent with the primary translocase remaining bound to the end of the unwound duplex while the secondary translocase continues to move along the other strand. Superimposed on the Cy5 time course is the time course of arrival of RecBC at the 3′–Cy3 labeled end (determined using a DNA containing only the Cy3 fluorophore). The peak in Cy5 fluorescence occurs slightly before the peak in the Cy3 fluorescence, as expected. In contrast, the substrate in Figure 4b shows only a decay in Cy5 fluorescence, suggesting dissociation of the Cy5 labeled strand after the 24 bp duplex is unwound since the primary RecBC translocase can translocate uninterrupted on this DNA. These results indicate that when RecBC reaches an end or gap in the 3′-terminated DNA strand, the primary translocase stops and remains bound to the ssDNA while continuing to hydrolyze ATP which drives the secondary translocase. Hence, we conclude that the RecB ATPase motor drives both the primary and secondary translocase activities.
Simultaneous translocation of RecBC along both strands of ssDNA
Although both translocase activities are fueled by the RecB motor, we anticipate that RecBC uses two distinct DNA binding sites for the two translocation activities. Hence, we designed DNA substrates, shown schematically in Figure 5a, to examine whether RecBC can translocate along the two single strands simultaneously. These substrates possess the same 24 bp duplex with a RecBC loading site on one end, but two non-complementary ssDNA ((dT)L) extensions of equal length on the other. The (dT)L extensions are labeled either on the 5′-end or the 3′-end with Cy3 to independently monitor the primary or secondary translocase, respectively. RecBC shows identical time courses (when normalized to the peak fluorescence) for translocation along either strand, as shown in Figure 5a. In fact, only a slightly (~15%) larger Cy3 enhancement is observed when Cy3 is on the 5′-end of the DNA extension (Supplementary Fig. 5). The fact that the normalized translocation time courses are identical (for a given L), regardless of which strand is monitored, indicates that RecBC translocates along both DNA strands simultaneously and with identical rates.
Figure 5. primary and secondary RecBC translocases operate simultaneously along two ssDNA extensions.
Translocation of RecBC along DNA substrates containing a 24 bp duplex region with high affinity RecBC loading site on one end and two (dT)L extensions of equal length (L = 15, 30, 45, 50, 75 nucleotides) and labeled with Cy3 on one of the two ends as depicted. (a). Normalized time courses for DNA substrates 5′–Cy3 labeled DNA (filled circles) and 3′–Cy3 labeled DNA (opened circles) for L = 30, 45, and 75 nucleotides. (b). Lag time analyses of time courses; 5′–Cy3 labeled DNA (filled circles) (Lag time = 0.00149 L + 0.0098) (671 ± 47 nt s−1). 3′–Cy3 labeled DNA (opened circles) (Lag time = 0.00161 L + 0.0016) (621 ± 43 nt s−1). (c). Backbone polarity of the ssDNA extension along which the primary (3′ to 5′) translocase operates is reversed using a 5′–5′ linkage at the position indicated (red X). DNA I - top strand end-labeled with Cy3; DNA II - bottom strand end-labeled with Cy3. (d). Backbone polarity of the ssDNA extension along which the secondary translocase operates is reversed using a 5′–5′ linkage at the position indicated (red X); DNA III - top strand end-labeled with Cy3; DNA IV - bottom strand end-labeled with Cy3. (e). FRET experiment monitoring Cy3 (purple circles) and Cy5 fluorescence (blue circles) performed using the DNA substrate double labeled with Cy3 and Cy5 as depicted. Red and black squares show the time courses for a DNA possessing both ssDNA extensions, but containing only a Cy3 fluorophore (as in panel (b), with L = 60)). (f). FRET experiment monitoring Cy3 (purple squares) and Cy5 fluorescence (blue squares) performed as in panel (e), but with a DNA substrate in which the backbone polarity of the top ssDNA extension was reversed using a 3′–3′ linkage at the position indicated (red X).
Consistent with directional translocation, we observe lag kinetics for all time courses and the lag times increase with increasing L (Fig. 5b). Interestingly, when RecBC translocates along both strands simultaneously, the rate of translocation is substantially slower (671 ± 47 nt s−1 (5′–Cy3), 621 ± 43 nt s−1 (3′–Cy3)) than when only one strand is present (~900–1,100 nt s−1). This indicates that both strands participate in RecBC translocation. Furthermore, the rates for both the primary and secondary ssDNA translocases are identical, and slightly faster than the DNA unwinding rate of RecBC measured under the same solution conditions (348 ± 5 bp s−1) 10.
RecBC translocation on DNA with reversed polarity backbone linkages
We next examined whether either translocase is affected by reversing the polarity of the phosphodiester backbone in the ssDNA extension. Since the primary ssDNA binding site of the RecB motor (1A and 2A sub-domains) binds ssDNA with a distinct polarity 6,11,21 (see Fig. 1), the primary (3′ to 5′) translocase should stop if the motor encounters a 5′–5′ phosphodiester linkage within the ssDNA extension. We introduced a 5′–5′ linkage in the lower (3′-terminated) strand just after the 24 bp duplex region (Fig. 5c), which reverses the backbone polarity of the (dT)75 extension in that strand. The polarity of the 5′-terminated strand was not changed. We generated two DNA substrates (indicated I and II in Fig. 5c), differing only by which ssDNA extension was labeled with Cy3, so that translocation along each strand could be monitored independently. The time courses in Figure 5c indicate that reversing the backbone polarity of the bottom (3′-terminated) strand (DNA II) blocks the primary RecBC translocase, as expected. However, the secondary translocase still moves along the top (5′-terminated) strand (DNA I).
We then introduced a 3′–3′ linkage in the top (5′-terminated) strand (Fig. 5d, DNA III and IV). Since the backbone polarity of the bottom ssDNA extension is not interrupted there was no effect on the primary translocation activity. However, the secondary translocase was also functional, even though the backbone polarity of the top (5′-terminated) strand was reversed. This indicates that the secondary translocase of RecBC is insensitive to the ssDNA backbone polarity. This surprising conclusion is further supported by comparisons of other variants of reversed polarity substrates (see Supplementary Fig. 5). In all cases, the primary translocase is blocked by a polarity reversal in the 3′-terminated strand, whereas the secondary translocase is unaffected by a polarity reversal in the 5′-terminated strand.
As a further test of whether RecBC translocates along both ssDNA extensions, we examined a DNA substrate with two ssDNA extensions that are both end labeled, one with Cy3 and one with Cy5 (Fig. 5e). With this DNA, changes in FRET between Cy3 and Cy5 should occur if the ends of the ssDNA extensions are brought closer together as RecBC translocates. Figure 5e shows the anti-correlated Cy3 and Cy5 time courses consistent with FRET resulting from the simultaneous translocation of the DNA strands through the RecBC enzyme. Reversing the backbone polarity within the top (5′-terminated) ssDNA extension does not affect the secondary translocase activity (Figure 5f).
Re-initiation of DNA unwinding after RecBC crosses a ssDNA gap
Previous studies inferred a 3′ to 5′ directionality for RecBC ssDNA translocation by examining its ability to unwind two DNA duplexes separated by ssDNA gaps 8. We therefore performed experiments using similar gapped DNA substrates (Fig. 6), which contain a RecBC loading site on one end of a 24 bp (proximal) duplex, followed by a ssDNA ((dT)L) extension and then a 40 bp (distal) duplex. Two types of DNA substrates were made such that a gap occurs in either strand of the duplex DNA, with gap lengths of L = 2, 21 or 41 nucleotides. The 5′-ends of one strand of both DNA duplexes were radiolabeled with 32P so that unwinding of both duplexes could be monitored in the same experiment10,18. RecBC initiates only from the high affinity loading site since it binds weakly and initiates unwinding poorly from blunt ends13,10. Because DNA was in excess over RecBC and single-round conditions were used, the maximum amount of DNA unwinding is limited by the RecBC concentration (2 nM).
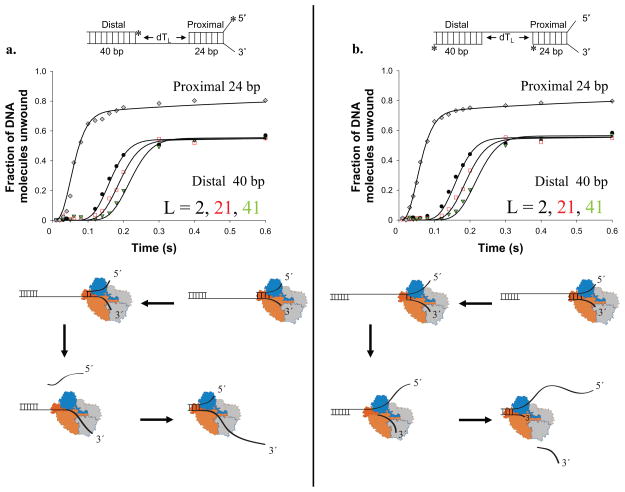
Figure 6. RecBC re-initiation of DNA unwinding after a ssDNA gap.
DNA substrates contain a proximal 24 bp duplex with a RecBC loading site (twin dT6 fork), a ssDNA gap of length, L = 2, 21 or 41 nucleotides, followed by a distal 40 bp duplex DNA. The ssDNA connecting the proximal and distal duplexes runs either 3′ to 5 (panel a) or 5′ to 3′ (panel b) relative to the RecBC loading site. (a). Single round time course for RecBC unwinding the proximal 24 bp duplex (diamonds) and the distal 40 bp duplex connected by a 3′ to 5′ ssDNA with L = 2 nt (circles), 21 nt (squares), or 41 nt (triangles). Smooth curves indicate fits to Scheme 3 (Eq. S3) using the parameters (mtkt = 928 ± 38 nt s−1; mUkU = 396 ± 15 bp s−1 (constrained)). (b). Single round time course for RecBC unwinding the proximal 24 bp duplex (diamonds) and the distal 40 bp duplex connected by a 5′ to 3′ ssDNA with L = 2 nt (circles), 21 nt (squares), or 41 nt (triangles). Smooth curves indicate fits to Scheme 3 (Eq. S3) using the parameters (mtkt = 919 ± 42 nt s−1; mUkU = 396 ± 15 bp s−1 (constrained)). Models for re-initiation of DNA unwinding by RecBC after traversing a ssDNA gap in either strand are shown below each panel.
In the substrates used in Figure 6a, the ssDNA connecting the proximal and distal duplexes is the strand along which the primary RecBC translocates (3′ to 5′). For the substrates in Figure 6b, the ssDNA in the gap is the strand along which the secondary RecBC translocates. For both substrates, RecBC unwinds ~85% of the 24 bp proximal duplex with time courses consistent with previous RecBC studies 10. Furthermore, RecBC can re-initiate unwinding of the distal 40 bp DNA duplex equally well for ssDNA gaps of different lengths, regardless of which strand spans the gap. The extent of the lag phase increases with increasing gap length, L. Since the proximal (24 bp) and the distal (40 bp) duplexes are identical in all substrates, the shifts in lag times reflect only the additional time required for RecBC to traverse the progressively longer ssDNA gaps.
We analyzed these time courses using Scheme 3 (Eq. S3), which assumes three stages, with RecBC starting from the high affinity loading site: (1)-unwinding of the proximal duplex, (2)- translocation along the ssDNA region, and (3)- unwinding of the distal duplex. The RecBC DNA unwinding rates were constrained to those determined previously (396 ± 15 bp s−1) 10, while the ssDNA translocation kinetic parameters were allowed to float. This analysis indicates that RecBC translocates along ssDNA in the 3′ to 5′ direction using its primary translocase with a rate of 928 ± 38 nt s−1, and along the other strand in the 5′ to 3′ direction using its secondary translocase with the same rate (919 ± 42 nt s−1). The observation that the primary and secondary translocases move with the same rates supports our earlier conclusions obtained using the fluorescently labeled DNA (Fig. 3a–d).
Discussion
RecBC has two translocase activities, both controlled by the RecB motor
To date, SF1 translocases containing only one ATPase motor have been observed to translocate uni-directionally along ssDNA29–33. It was therefore unexpected to find that RecBC, possessing only a single canonical motor (RecB), displays two distinct translocase activities. The primary translocase activity is sensitive to the phosphodiester backbone polarity and moves only in a 3′ to 5′ direction, consistent with the translocation properties of the isolated RecB motor. However, the secondary translocase, which moves at the same rate as the primary, is insensitive to the ssDNA backbone polarity. Our studies with the RecB subunit alone show no evidence of the secondary translocase activity. Hence it appears that the interaction of RecB with RecC is needed to form the secondary ssDNA translocase site or to support its interactions with DNA.
The regions of RecBC that form the channel for the unwound 5′-terminated ssDNA, and thus likely involved in the secondary translocase activity, are well removed from the ATP binding site of RecB and the channel for the unwound 3′-terminated ssDNA, along which the primary translocase moves (see Fig. 1)6. Thus, the DNA binding site associated with the secondary translocase must be distinct from the ssDNA binding site responsible for the primary translocase, hence the concerted control of both translocase activities by the RecB motor must occur allosterically. Both RecBC translocases display the same rates of translocation and are ATP-dependent with KM values similar to that measured for RecB translocation, consistent with both activities being driven by the single RecB motor.
Interestingly, when RecBC translocates along only one strand of DNA the rate is ~1,000 nt s−1, whereas the rate is reduced to ~ 650 nt (or “bp”) s−1 when it translocates along both single strands simultaneously. However, this translocation rate is still faster than the DNA unwinding rate of RecBC under the same conditions (~350 bp s−1), although slower than the RecBCD unwinding rate (~750 bp s−1) 10. Importantly, the primary and secondary translocation rates of RecBC are identical when both ssDNA extensions are present. Thus both DNA strands move through the enzyme at the same rates preventing any loop formation during unwinding.
Implications for RecBC translocation and DNA unwinding mechanisms
Bianco & Kowalczykowski8 previously examined the ability of RecBC to initiate unwinding of a short DNA duplex (proximal), followed by unwinding of a second (distal) DNA duplex separated by a ssDNA gap. Using DNA substrates similar to those shown in Figure 6, they observed that the efficiency with which RecBC could traverse the ssDNA gap was lower when the gap was in the strand along which the primary RecBC translocase operates and concluded that RecBC translocation along ssDNA occurred strictly in the 3′ to 5′ direction. This differs from our conclusion that RecBC has two translocase activities. They8 also observed that RecBC could traverse ssDNA gaps in the 3′-terminated strand as large as 23 ± 2 nucleotides and proposed a “quantum inch-worm” unwinding model. However, RecBC was still observed to unwind ~16% of the distal DNA duplexes, even with a gap of 30 nucleotides in that strand. Our finding that RecBC can bypass ssDNA gaps in either strand and then re-initiate DNA unwinding is qualitatively consistent with this observation8. However, we do not observe a difference in unwinding efficiency regardless of which strand contains the gap. These differences may be due to the absence of E. coli single stranded binding (SSB) protein in our experiments.
Interestingly, Bianco & Kowalczykowski8 also observed that the ability to traverse a gap was dependent upon the length of the proximal duplex DNA that preceded the gap, and concluded that RecBC stepping was not only quantized with a periodicity of 23 ± 2 nucleotides, but also that the placement of the next step was determined by where the enzyme initiated. Although our experiments do not address this result, it seems likely that the secondary translocase activity of RecBC that we report is related to this “quantum inch-worm” behavior8.
Where in RecBC is the secondary translocase activity and what is its function?
The general view of the mechanism of ssDNA translocation by an SF1 helicase is that ATP-hydrolysis drives coupled motions between the two RecA-like sub-domains (1A and 2A) whose interface forms the ATP binding site (Fig. 1b). One proposal is that these two sub-domains each contain a sub-site for binding ssDNA and these sub-sites cycle between high and low ssDNA affinity based on the nucleotide binding state (i.e., ATP, ADP–Pi, ADP, etc.), resulting in an inch-worm mechanism that moves the motor uni-directionally along the ssDNA backbone2. This type of motor activity is likely responsible for the primary translocase of RecBC. However, the secondary RecBC translocase must involve a distinctly different region of the enzyme, but one that is controlled allosterically by the same ATPase motor within RecB.
We suggest that the secondary translocase activity resides either in the arm region of RecB and (or) the dead nuclease domain of RecC (see Fig. 1b) 6. These regions contact either the duplex DNA ahead of the fork or the 5′-terminated ssDNA. Although we detect the secondary translocase activity of RecBC as an ability to translocate along the 5′-terminated ssDNA, its actual function may be to interact with and translocate along the duplex DNA ahead of the fork. This would explain the lack of sensitivity of the secondary translocase activity to the backbone polarity of ssDNA. Hence, the RecB “arm” is a likely candidate since it contacts the dsDNA ahead of the fork.
The function of this secondary translocase may be to load the other (5′-terminated) single strand of DNA into the RecD motor. When RecBCD binds to a blunt ended DNA duplex, 5–6 bp are melted in a Mg2+-dependent but ATP-independent process to form an initiation complex6,12–13,22. In this initiation complex, the 3′-terminated ssDNA is bound in a channel containing the RecB motor and thus is ready to be translocated upon ATP binding and hydrolysis. However, the initial length of the 5′-terminated ssDNA (5–6 nt) is not long enough to reach the motor region of RecD, although RecD can be crosslinked to this strand 12. In fact, a 5′-terminated ssDNA of at least 10 nucleotides is needed to functionally engage the RecD motor 10,13,20. As such, the secondary RecBC translocase activity may function to load the 5′-terminated ssDNA into the RecD motor.
The secondary translocase activity may also function after RecBCD recognizes the recombination hotspot, Chi. Following RecBCD initiation of unwinding at a blunt DNA end, the RecD motor moves faster than the RecB motor generating a ssDNA loop ahead of the fork, in the 3′-terminated ssDNA along which the primary RecB translocase operates 4. During this time, the RecB nuclease preferentially digests the 3′-terminated strand. However, once a Chi site is recognized by RecC, the relative speeds of the RecB and RecD motors switch 7 and the nuclease selectively degrades the 5′-terminated strand. After Chi, the now faster RecB motor will eventually catch up to the RecD motor. At this point, if RecB remains faster than RecD, a different loop would form in the 5′-terminated strand ahead of the enzyme; however, this has not been observed 4. The existence of the secondary RecBC translocase may ensure that the two unwound single strands move through RecBCD at the same rates and thus prevent loop formation after Chi recognition. The existence of concerted translocase activities would also prevent dissociation of RecBCD if a ssDNA gap is encountered on either strand during DNA unwinding after Chi.
In addition to RecBCD, two other classes of double strand DNA break resecting enzymes have been described that are hetero-dimeric and thus more similar to RecBC in that they do not possess a second RecD-like motor. AddAB from Bacillus subtilis has one motor subunit, AddA, but two RecB-like nuclease domains, one each on AddA and AddB 34. AdnAB from Mycobacterium smegmatis has two active motors and two active nuclease domains 35. It will be interesting to determine whether these hetero-dimeric enzymes also possess a secondary translocase activity similar to E. coli RecBC.
Methods
Buffers and Reagents
Buffers were prepared with distilled, de-ionized water and filtered through 0.2 micron filters. Buffer M is 20 mM Mops-KOH (pH 7.0 at 25°C), 30 mM NaCl, 10 mM MgCl2, 1 mM 2-mercaptoethanol (2-ME), and 5% (v/v) glycerol. RecB and RecC storage buffer is Buffer C: 20 mM K phosphate (pH 6.8 at 25), 0.1 mM 2-mercaptoethanol, 0.1 mM ethylenediaminetetraacetic acid (EDTA), and 10% (v/v) glycerol. Heparin stock solutions were prepared in Buffer M as described 32. ATP stock solutions were prepared and concentrations determined as described 10.
Proteins
E. coli RecB was overexpressed in E. coli strain V186 (which contains a deletion of the chromosomal recB, recC and recD genes 23 carrying pPB700 (recB+) and pNM52 (lacIq). E. coli RecC was overexpressed E. coli strain V186 carrying pBP500 (recC+) and pNM52. In this way we avoided any possible contamination of the RecBC prep with RecD. RecB and RecC proteins were expressed and purified separately, and stored in Buffer C as described 36. RecB was dialyzed versus Buffer M at 4°C before use and its concentration determined using an extinction coefficient of ε280 = 1.9 × 105 M−1 cm−1 36. RecBC enzyme was reconstituted and dialyzed against Buffer M at 4°C before use, and its concentration determined using ε280 = 3.9 × 105 M−1 cm−1 36.
DNA
Oligodeoxynucleotides, either unlabeled or labeled covalently with fluorescein, Cy3 or Cy5, or containing reversed polarity phosphodiester backbones were synthesized and purified and their concentrations determined as described 10; stock solutions were dialyzed versus Buffer M and stored at −20°C. DNA labeled with Oregon Green was purchased (Integrated DNA Technologies, Coralville, IA). The sequences of the DNA substrates used are given in Supplementary Tables 1–3.
Stopped-flow fluorescence experiments
Translocation kinetics experiments were performed in Buffer M at 25°C using an SX.18MV stopped-flow fluorescence instrument (Applied Photophysics Ltd., Leatherhead, UK). RecB (50 nM) or RecBC (100 nM) were pre-mixed with DNA present in excess (100 nM or 200 nM) in one syringe of the stopped-flow apparatus, and translocation was initiated by mixing with 10 mM ATP and 8 mg mL−1 heparin in the other syringe. Heparin “trap tests” were performed by including the DNA substrate with the ATP and heparin solution. When mixed with RecBC no translocation signal was observed indicating that heparin trapped all of the free RecBC. Cy3 fluorescence was excited at 515 nm, and its emission was monitored at wavelengths > 570 nm using a long pass filter. Oregon Green fluorescence was excited at 508 nm, and its emission was monitored at wavelengths > 520 nm using a long pass filter. For FRET experiments, Cy3 fluorescence was excited at 515 nm and its emission was monitored at 570 nm using an interference filter while Cy5 fluorescence was monitored at wavelengths > 665 nm using a long pass filer. Analysis of translocation time courses is described in Supplementary Data.
DNA Unwinding
DNA unwinding kinetics was performed in Buffer M at 25°C using a KinTek RQF-3 rapid quenched-flow instrument (University Park, PA). DNA substrates were composed of three DNA strands as depicted in Figure 6 (sequences in Supplementary Table 3, Supplementary Data) which when annealed together, form a proximal (24 bp) and distal (40 bp) duplex DNA regions that are separated by a ssDNA (dT)L region of length L nucleotides with either 3′ to 5′ or 5′ to 3′ polarity 8. Reporter strands from the proximal and distal duplex were radiolabeled on the 5′ end with 32P as described 10. An excess of annealed DNA unwinding substrate (20 nM) was pre-incubated with RecBC (2 nM) in one syringe and reactions were initiated by rapid mixing with an equal volume of 10 mM ATP and 8 mg mL−1 heparin, followed by mixing with an equal volume of 0.4 M EDTA and 10% (v/v) glycerol to quench the reaction after a predefined time interval (Δt). Unwound DNA products were resolved from the native substrates using non-denaturing polyacrylamide gel electrophoresis (PAGE) and quantified as described 10,18 Analysis of unwinding time courses is described in Supplementary Data.
Supplementary Material
Acknowledgments
We thank Drs. R. Galletto, C. Fischer, A. Lucius, K. Maluf, E. Galburt, N. Baker, P. Burgers, G. Smith, T. Ellenberger, S. Kowalczykowski, and E. Antony for valuable discussions and comments on the manuscript, Drs. G. Smith (Fred Hutchinson Cancer Research Center, Seattle, WA), A. Taylor (Fred Hutchinson Cancer Research Center, Seattle, WA) and D. Julin (University of Maryland, College Park, MD) for plasmids and cell lines and T. Ho (Washington University School of Medicine, St. Louis, MO) for synthesis and purification of DNA. This work was supported in part by NIH GM045948 (to TML).
Footnotes
Author contributions
C.W. and T.M.L. designed the experiments. C.W. purified the protein, pe6rformed the stopped-flow and DNA unwinding experiments, and analyzed the data; C.B. and C.W. performed the FRET and RecBC translocation experiments as a function of [NaCl]. T.M.L. supervised the study and C.W. and T.M.L. wrote the manuscript.
References
- 1.Dillingham MS, Kowalczykowski SC. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol Mol Biol Rev. 2008;72:642–671. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singleton MR, Dillingham MS, Wigley DB. Structure and Mechanism of Helicases and Nucleic Acid Translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 3.Dillingham MS, Spies M, Kowalczykowski SC. RecBCD enzyme is a bipolar DNA helicase. Nature. 2003;423:893–897. doi: 10.1038/nature01673. [DOI] [PubMed] [Google Scholar]
- 4.Taylor AF, Smith GR. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature. 2003;423:889–893. doi: 10.1038/nature01674. [DOI] [PubMed] [Google Scholar]
- 5.Rigden DJ. An inactivated nuclease-like domain in RecC with novel function: implications for evolution. BMC Struct Biol. 2005;5:9. doi: 10.1186/1472-6807-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 7.Spies M, Amitani I, Baskin RJ, Kowalczykowski SC. RecBCD Enzyme Switches Lead Motor Subunits in Response to chi Recognition. Cell. 2007;131:694–705. doi: 10.1016/j.cell.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bianco PR, Kowalczykowski SC. Translocation step size and mechanism of the RecBC DNA helicase. Nature. 2000;405:368–372. doi: 10.1038/35012652. [DOI] [PubMed] [Google Scholar]
- 9.Korangy F, Julin DA. Efficiency of ATP Hydrolysis and DNA Unwinding by the RecBC Enzyme from Escherichia coli. Biochemistry. 1994;33:9552–9560. doi: 10.1021/bi00198a022. [DOI] [PubMed] [Google Scholar]
- 10.Wu CG, Lohman TM. Influence of DNA end structure on the mechanism of initiation of DNA unwinding by the Escherichia coli RecBCD and RecBC helicases. J Mol Biol. 2008;382:312–326. doi: 10.1016/j.jmb.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganesan S, Smith GR. Strand-specific binding to duplex DNA ends by the subunits of the Escherichia coli RecBCD enzyme. J Mol Biol. 1993;229:67–78. doi: 10.1006/jmbi.1993.1008. [DOI] [PubMed] [Google Scholar]
- 12.Farah JA, Smith GR. The RecBCD Enzyme Initiation Complex for DNA Unwinding: Enzyme Positioning and DNA Opening. J Mol Biol. 1997;272:699–715. doi: 10.1006/jmbi.1997.1259. [DOI] [PubMed] [Google Scholar]
- 13.Wong CJ, Lucius AL, Lohman TM. Energetics of DNA end binding by E. coli RecBC and RecBCD helicases indicate loop formation in the 3′-single-stranded DNA tail. J Mol Biol. 2005;352:765–782. doi: 10.1016/j.jmb.2005.07.056. [DOI] [PubMed] [Google Scholar]
- 14.Wong CJ, Rice RL, Baker NA, Ju T, Lohman TM. Probing 3′-ssDNA Loop Formation in E. coli RecBCD/RecBC-DNA Complexes Using Non-natural DNA: A Model for “Chi” Recognition Complexes. J Mol Biol. 2006;362:26–43. doi: 10.1016/j.jmb.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 15.Taylor A, Smith GR. Unwinding and Rewinding of DNA by the RecBC Enzyme. Cell. 1980;22:447–457. doi: 10.1016/0092-8674(80)90355-4. [DOI] [PubMed] [Google Scholar]
- 16.Bianco PR, et al. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature. 2001;409:374–378. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- 17.Spies M, et al. A molecular throttle: the recombination hotspot chi controls DNA translocation by the RecBCD helicase. Cell. 2003;114:647–654. doi: 10.1016/s0092-8674(03)00681-0. [DOI] [PubMed] [Google Scholar]
- 18.Lucius AL, et al. DNA unwinding step-size of E. coli RecBCD helicase determined from single turnover chemical quenched-flow kinetic studies. J Mol Biol. 2002;324:409–428. doi: 10.1016/s0022-2836(02)01067-7. [DOI] [PubMed] [Google Scholar]
- 19.Lucius AL, Lohman TM. Effects of temperature and ATP on the kinetic mechanism and kinetic step-size for E. coli RecBCD helicase-catalyzed DNA unwinding. J Mol Biol. 2004;339:751–771. doi: 10.1016/j.jmb.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 20.Dillingham MS, Webb MR, Kowalczykowski SC. Bipolar DNA translocation contributes to highly processive DNA unwinding by RecBCD enzyme. J Biol Chem. 2005;280:37069–37077. doi: 10.1074/jbc.M505520200. [DOI] [PubMed] [Google Scholar]
- 21.Saikrishnan K, Griffiths SP, Cook N, Court R, Wigley DB. DNA binding to RecD: role of the 1B domain in SF1B helicase activity. EMBO J. 2008;27:2222–2229. doi: 10.1038/emboj.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong CJ, Lohman TM. Kinetic control of Mg2+-dependent melting of duplex DNA ends by Escherichia coli RecBC. J Mol Biol. 2008;378:759–775. doi: 10.1016/j.jmb.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boehmer PE, Emmerson PT. The RecB subunit of the Escherichia coli RecBCD Enzyme Couples ATP Hydrolysis to DNA Unwinding. J Biol Chem. 1992;267:4981–4987. [PubMed] [Google Scholar]
- 24.Phillips RJ, Hickleton DC, Boehmer PE, Emmerson PT. The RecB protein of Escherichia coli translocates along single-stranded DNA in the 3′ to 5′ direction: a proposed ratchet mechanism. Mol Gen Genet. 1997;254:319–329. doi: 10.1007/pl00008605. [DOI] [PubMed] [Google Scholar]
- 25.Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent translocation of E. coli UvrD monomers along single-stranded DNA. J Mol Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 26.Fischer CJ, Lohman TM. ATP-dependent translocation of proteins along single-stranded DNA: models and methods of analysis of pre-steady state kinetics. J Mol Biol. 2004;344:1265–1286. doi: 10.1016/j.jmb.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Lucius AL, Maluf NK, Fischer CJ, Lohman TM. General methods for analysis of sequential “n-step” kinetic mechanisms: application to single turnover kinetics of helicase-catalyzed DNA unwinding. Biophys J. 2003;85:2224–2239. doi: 10.1016/s0006-3495(03)74648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hickson ID, Robson CN, Atkinson KE, Hutton L, Emmerson PT. Reconstitution of RecBC DNase activity from purified Escherichia coli RecB and RecC proteins. J Biol Chem. 1985;260:1224–1229. [PubMed] [Google Scholar]
- 29.Brendza KM, et al. Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc Natl Acad Sci U S A. 2005;102:10076–10081. doi: 10.1073/pnas.0502886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dillingham MS, Wigley DB, Webb MR. Direct measurement of single-stranded DNA translocation by PcrA helicase using the fluorescent base analogue 2-aminopurine. Biochemistry. 2002;41:643–651. doi: 10.1021/bi011137k. [DOI] [PubMed] [Google Scholar]
- 31.Niedziela-Majka A, Chesnik MA, Tomko EJ, Lohman TM. Bacillus stearothermophilus PcrA Monomer Is a Single-stranded DNA Translocase but Not a Processive Helicase in Vitro. J Biol Chem. 2007;282:27076–27085. doi: 10.1074/jbc.M704399200. [DOI] [PubMed] [Google Scholar]
- 32.Tomko EJ, Fischer CJ, Niedziela-Majka A, Lohman TM. A Nonuniform Stepping Mechanism for E. coli UvrD Monomer Translocation along Single-Stranded DNA. Molecular Cell. 2007;26:335–347. doi: 10.1016/j.molcel.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Antony E, et al. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol Cell. 2009;35:105–115. doi: 10.1016/j.molcel.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeeles JT, Dillingham MS. A Dual-nuclease Mechanism for DNA Break Processing by AddAB-type Helicase-nucleases. J Mol Biol. 2007;371:66–78. doi: 10.1016/j.jmb.2007.05.053. [DOI] [PubMed] [Google Scholar]
- 35.Unciuleac MC, Shuman S. Characterization of the mycobacterial AdnAB DNA motor provides insights into the evolution of bacterial motor-nuclease machines. J Biol Chem. 2010;285:2632–2641. doi: 10.1074/jbc.M109.076133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lucius AL, Jason Wong C, Lohman TM. Fluorescence stopped-flow studies of single turnover kinetics of E. coli RecBCD helicase-catalyzed DNA unwinding. J Mol Biol. 2004;339:731–750. doi: 10.1016/j.jmb.2004.04.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.