Abstract
Chromosomal inversions have been of long-standing interest to geneticists because they are capable of suppressing recombination and facilitating the formation of adaptive gene complexes. An exceptional inversion polymorphism (ZAL2m) in the white-throated sparrow (Zonotrichia albicollis) is linked to variation in plumage, social behavior and mate choice, and is maintained in the population by negative assortative mating. The ZAL2m polymorphism is a complex inversion spanning >100 Mb and has been proposed to be a strong suppressor of recombination, as well as a potential model for studying neo-sex chromosome evolution. To quantify and evaluate these features of the ZAL2m polymorphism, we generated sequence from 8 ZAL2m and 16 ZAL2 chromosomes at 58 loci inside and 4 loci outside the inversion. Inside the inversion we found that recombination was completely suppressed between ZAL2 and ZAL2m, resulting in uniformly high levels of genetic differentiation (FST=0.94), the formation of two distinct haplotype groups representing the alternate chromosome arrangements and extensive linkage disequilibrium spanning ∼104 Mb within the inversion, whereas gene flow was not suppressed outside the inversion. Finally, although ZAL2m homozygotes are exceedingly rare in the population, occurring at a frequency of <1%, we detected evidence of historical recombination between ZAL2m chromosomes inside the inversion, refuting its potential status as a non-recombining autosome.
Keywords: chromosomal inversion, recombination suppression, linkage disequilibrium
Introduction
Chromosomal inversions are known to occur in a wide variety of organisms, in which they are associated with instances of adaptive evolution, speciation, selfish genes, sex chromosomes, human disease and disease susceptibility (Hartl, 1975; Lahn and Page, 1999; Noor et al., 2001; Hoffmann et al., 2004; Stefansson et al., 2005; Dyer et al., 2007). Although inversions have been studied for nearly a century, only recently have researchers begun to elucidate their effects on patterns of molecular evolution. Their influence on sequence evolution is derived from their unique ability to suppress recombination within the inverted interval in individuals heterozygous for the inversion (Sturtevant, 1921; Sturtevant and Beadle, 1936). Dobzhansky (1937, 1950) proposed that natural selection would favor inversions if they captured a set of positively interacting alleles, which he referred to as a ‘supergene' complex (Dobzhansky and Sturtevant, 1938). Alternatively, Kirkpatrick and Barton (2006) showed that an inversion can enhance the fitness of its carrier, in the absence of positive epistasis, if it simply captures two or more locally adapted alleles. Because inversions suppress recombination, linkage disequilibrium (LD) between the beneficial alleles within the inversion would be preserved even when the alleles are not in close proximity to each other. Thus, the influence of inversions on the rate and pattern of recombination is fundamental to their adaptive significance and evolution.
The suppression of recombination between the alternative chromosomal arrangements along the inverted segment will eventually lead to the formation of two distinct haplotype groups: the standard and the inverted. However, population genetic studies in Drosophila, in which inversions have been most intensely studied, have found that genetic differentiation is non-uniform across an inversion and that gene flow usually does occur between the standard and inverted chromosomes inside the inversion (Novitski and Braver, 1954; Hasson and Eanes, 1996; Schaeffer et al., 2003; Schaeffer and Anderson, 2005). In these examples, genetic differentiation is typically highest near the inversion breakpoints. Double recombination events or gene conversion mediated by the formation of an inversion loop between the inverted and standard chromosomes will, over time, lead to significant gene flow within the inversion (Navarro et al., 1997; Kovacevic and Schaeffer, 2000; Andolfatto et al., 2001; Schaeffer et al., 2003; Kennington et al., 2006). The patterns of gene flux associated with an inversion can be particularly useful in identifying the targets of selection, which are expected to be in LD with each other as well as the inversion (see White et al., 2007). Thus, the extent and pattern of gene flow associated with simple inversion polymorphisms will be influenced by the strength of selection to maintain LD, the size of the inversion, the rate of recombination and the age of the inversion.
Chromosomal polymorphisms involving more than one inversion have been found in natural populations and can be associated with patterns of gene flow that are distinct from the pattern described above for simple inversions. For example, the mouse t-complex comprises four non-overlapping inversions on mouse chromosome 17 and has been studied for decades with respect to its effect on recombination and association with meiotic drive (Lyon, 2003). Although gene conversion and some rare recombinants have been reported between wild-type and t chromosomes (Erhart et al., 2002; Wallace and Erhart, 2008), strong suppression of recombination extends over the length of the t-complex. As a result, genetic differentiation between the t and wild-type chromosomes is uniformly high across the entire ∼30–40 Mb t-complex (Lyon, 2003). Similarly, the XD chromosome in Drosophila recens is composed of a complex set of inversions and is associated with meiotic drive (Dyer et al., 2007). The XD completely suppresses recombination between the XD and its non-distorting homolog, XST, resulting in dramatic chromosome-wide LD spanning ∼130 c (Dyer et al., 2007). Unlike the classic model of gene flow for simple inversions in Drosophila, chromosomal polymorphisms involving more than one inversion can suppress recombination over the entire length of the rearrangement for prolonged periods of time and lead to genetically distinct haplotypes associated with exceptionally large blocks of LD.
Although chromosomal polymorphisms comprising complex inversions can drastically reduce the frequency of recombination within the rearranged regions in individuals heterozygous for the polymorphism, normal recombination levels are expected within the alternative chromosomal rearrangements in individuals homozygous for the standard or inverted chromosomes. However, if homozygosity for one of the alternative arrangements, for example, the inverted arrangement, causes sterility or early lethality, the region within the inversion will not have the opportunity to recombine during meiosis. As a non-recombining segment of the genome, the inverted chromosome will then become subject to a series of population genetic forces that will result in the accumulation of deleterious mutations and genetic degeneration (Rice, 1994; Charlesworth and Charlesworth, 2000). A dramatic example of the long-term consequence of suppressed recombination is the mammalian Y, which has undergone massive genetic degeneration and lost almost all of its original genes except in the small pseudoautosomal region(s) in which recombination still occurs with the X chromosome (Graves, 2006). Newly arisen neo-Y chromosomes also typically show distinct signatures of genetic degeneration (Filatov et al., 2000; Peichel et al., 2004; Kondo et al., 2006; Marais, 2007; Zhou et al., 2008), as do other rare examples of non-recombining regions of the genome (Slawson et al., 2006). These examples illustrate that under extreme circumstances, complex inversion polymorphisms can eventually lead to regions of the genome in which recombination is rare or never occurs.
Recently, we described the first modern genetic and genomic characterization of a chromosomal polymorphism in the white-throated sparrow (Zonotrichia albicollis) that is extraordinary in respect to its phenotypic effects and genetic properties (Thomas et al., 2008). In particular, the two alternative arrangements of the second chromosome, which will be referred to in this study as ZAL2 and ZAL2m, are linked to a plumage polymorphism such that individuals homozygous for ZAL2 are invariably associated with the tan-stripe (TS) morph, whereas individuals of the white-stripe (WS) morph are either heterozygous for the polymorphism (ZAL2/ZAL2m) or very rarely ZAL2m homozygotes (Thorneycroft, 1966, 1975). In addition to the genetic association with plumage, the chromosomal polymorphism is linked to variation in social behavior such that WS individuals are, on average, more aggressive and less parental than their same-sex TS counterparts (Tuttle, 2003). WS and TS individuals occur at similar frequencies in both sexes and show an exceptionally strong negative assortative mating pattern in which >96% of all breeding pairs comprise individuals of opposite morphs (Falls and Kopachena, 1994). As a consequence of this breeding pattern, and perhaps because of reduced viability (Thorneycroft, 1975), ZAL2m/ZAL2m birds are rare in the population with only a single ZAL2m homozygote having been detected in studies that combined karyotyped >600 birds (Thorneycroft, 1975; Romanov et al., 2009). Another consequence of this mating pattern is that ZAL2m is in a near constant state of heterozygosity maintained at the population level by balancing selection. At the molecular level, ZAL2m differs from ZAL2 by at least two nested inversions that, together, are predicted to span ∼100 Mb and encompass ∼1000 genes (Thomas et al., 2008). In addition, limited population-based re-sequencing indicated that recombination was suppressed between ZAL2 and ZAL2m within the inverted interval and that as a result of this suppression of recombination between the chromosome types, as well as a paucity of ZAL2m homozygotes, ZAL2m could be a non-recombining segment of the genome and a model for the early stages of sex chromosome evolution (Thomas et al., 2008). However, these conclusions were based on only a few loci (n=10), and thus it remained unclear whether suppression of recombination between the chromosome types extended across the entire inverted interval, and whether or not ZAL2m truly contained a non-recombining segment of the genome. We report the results of our study in which we have sampled sequenced diversity at 62 loci distributed at ∼1.7 Mb intervals across ZAL2 and ZAL2m to definitely establish the patterns of genetic differentiation and recombination associated with this chromosomal polymorphism.
Materials and methods
Source of DNA
White-throated sparrows were collected on the campus of Emory University in Atlanta, Georgia, during November and December of 2005–2007. A small blood sample was taken from a wing vein for DNA extraction and the morph of each bird was determined according to the criteria described in Watt (1986) and Piper and Wiley (1989) and confirmed by PCR using the method described by Michopoulos et al. (2007. DNA from a single dark-eyed junco (Junco hyemalis) was collected from a locally captured bird. All procedures involving animals were approved by the Institutional Animal Care and Use Committee of the Emory University.
DNA sequencing
PCR primers were designed based on publicly available zebra finch genomic sequence (The Genome Center at Washington University, http://genome.wustl.edu) in regions conserved with chicken as described by Thomas et al. (2008). Primer sequences, orthologous position in zebra finch and specific annealing temperatures are listed in Supplementary Table 1. Each 25 μl PCR contained final concentrations of 1 × PCR buffer, 1.5 m MgCl2, 20 pmol of each primer, 0.2 m of each deoxynucleotide triphosphate, 1.5 units of Taq or Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA) and approximately 12.5–25 ng of genomic DNA. PCR cycling started with an initial denaturation at 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 55 °C or 58 °C for 30 s, 72 °C for 1 min and ended with a final extension at 72 °C for 7 min. PCR products were treated with shrimp alkaline phosphatase and exonuclease I (USB) before direct sequencing using the PCR primers or internal primers. All sequences were deposited in GenBank (accession numbers GU449138–GU449818 and GU450220–GU450272).
Single-nucleotide polymorphism (SNP) discovery and sequence annotation
Polymorphisms were automatically called using SNPdetector (Zhang et al., 2005) and all variants were manually confirmed before further analyses. Insertion and deletion polymorphisms were scored but not included in the subsequent analyses. The sequence at each locus was annotated for gene features based on annotation of the orthologous chicken genomic segments (ICGSC, 2004), as well as the presence of evolutionarily conserved regions as predicted by the PhastCons track on the genome browser of the University of California, Santa Cruz (Siepel et al., 2005). Individual loci were ordered and spaced relative to one another based on a previously established white-throated sparrow–chicken comparative map (Thomas et al., 2008) and the assembled zebra finch genome (taeGut1).
DNA sequence analysis
Haplotypes were reconstructed from raw genotype data using PHASE v.2.1.1 (Stephens et al., 2001) under the default parameters. Each locus was phased individually and SNPs with <0.55 phasing confidence (primarily singletons) were assigned randomly to alternative haplotypes that were used to generate population genetic statistics for each locus in Supplementary Table 2. We also phased the concatenated data set to generate a complete haplotype, which was used to build the haplotype network and estimate LD and recombination. Although incorrectly phased haplotypes do not affect measures of diversity, they can subtly influence estimates of LD and recombination. In TS birds, all haplotypes were identified as ZAL2 chromosomes. In WS birds, within the inversion we used fixed differences to distinguish between the ZAL2 and ZAL2m chromosomes.
Population genetic statistics were calculated in DnaSP v.4.50.3 unless otherwise noted (Rozas et al., 2003). We defined neutral sites as non-coding and synonymous sites located outside of segments orthologous to regions detected as being evolutionarily conserved in the chicken genome by PhastCons (Siepel et al., 2005). Summary statistics for the intervals inside and outside the inversion were calculated using the phased haplotype generated from the concatenated sequences except π and θ, which were calculated as a weighted average of the individual loci. The neighbor-joining haplotype network was generated in Splitstree4 (Huson and Bryant, 2006). Pairwise comparisons of non-random associations were performed in Haploview (Barrett et al., 2005) and the r2 color scheme was used to illustrate LD between pairs of sites. Estimates and standard deviations for the population recombination rate parameter ρ=4Ner were assessed by Monte Carlo coalescent simulations in the interval algorithm in LDhat (Auton and McVean, 2007) conditioned on θW from DnaSP sampling every 2000 of 106 iterations after an initial burn-in of 100 000. Finally, we tested for gene conversion using the algorithm developed by Betrán et al. (1997) implemented in DnaSP (Rozas et al., 2003).
Results
Data set
Previously, we reported an initial survey of the genetic differentiation between ZAL2 and ZAL2m based on sequencing a small number (n=10) of loci (Thomas et al., 2008). To carry out a more detailed study of the genetic differentiation, nucleotide diversity and recombination associated with this chromosomal polymorphism, we expanded our study to include a total of 62 loci that were spaced on average every 1.7 Mb along the ZAL2/2m chromosome in 4 TS and 8 WS birds, corresponding to a sample size of 16 ZAL2 and 8 ZAL2m chromosomes. Of the sequenced loci, 58 mapped within the inversion and totaled ∼35 kb, whereas 4 loci mapped outside the inversion and totaled ∼1.7 kb (Table 1). On the basis of annotation of the loci and our criteria for defining neutral sequence (see Materials and methods for details) the data set included ∼14 and ∼0.8 kb of neutral sites inside and outside the inversion, respectively (Table 1). Overall, we identified 297 SNPs, of which 279 mapped within the inversion and 18 mapped outside the inversion (Table 1). After excluding three tri-allelic SNPs, the final data set available for analyses comprised 277 SNPs within and 17 SNPs outside the inversion.
Table 1. Diversity, divergence and summary statistics for the standard ZAL2 and inverted ZAL2m chromosomes in the white-throated sparrow.
| Region | Loci | Length (bp) | Segregating sites, S | π (All) | π (ZAL2) | π (ZAL2m) | FSTa | dxyb | Tajima's Dc | Tajima's D (ZAL2) | Tajima's D (ZAL2m) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Inside inversion | 58 | 34 823 (13 789.3) | 277 (167) | 0.00293 (0.00449) | 0.00044 (0.00075) | 0.00030 (0.00042) | 0.94 | 0.00583 (0.00871) | 0.65 | −0.74 | −0.84 |
| Outside inversion | 4 | 1655 (777) | 17 (14) | 0.00191 (0.00296) | 0.00160 (0.00311) | 0.00238 (0.00455) | 0.21 | 0.00190 (0.00296) | −0.82 | −0.90 | −0.22 |
Numbers in parentheses are calculations for neutral sites, noncoding, synonymous sites outside most conserved elements.
FST is calculated by comparing all ZAL2 chromosomes with all ZAL2m chromosomes.
Divergence (dxy) between the ZAL2 and ZAL2m chromosomes is calculated as the average number of nucleotide substitutions per site.
Tajima's D values were calculated using four tan-stripe (TS) and four white-stripe (WS) birds, thus representing the natural population frequency of the ZAL2 and ZAL2m chromosomes.
Genetic differentiation between ZAL2 and ZAL2m
In our earlier study we found high levels of genetic differentiation between ZAL2 and ZAL2m within the inversion and evidence of gene flow outside the inversion (Thomas et al., 2008). Consistent with these findings we found that the majority (185/277) of SNPs within the inversion were fixed differences between the chromosomal arrangements (that is, one allele was invariably linked to ZAL2 and the alternative allele to ZAL2m), and only one SNP was a shared polymorphism (that is, both alleles were present in both chromosomal arrangements). In contrast, outside the inversion we identified 17 SNPs, none of which were classified as fixed differences between the chromosomal arrangements and two were shared based on the complete haplotype phase estimate and not the individual locus phasing.
To formally quantify the level of genetic differentiation between the chromosomal arrangements inside and outside the inversion, we calculated the population genetic statistics FST and dxy treating ZAL2 and ZAL2m as two separate populations (Table 1). As expected, based on the prevalence of fixed differences between ZAL2 and ZAL2m, FST inside the inversion was near the theoretical maximum of 1 (FST=0.94), consistent with a high degree of genetic differentiation and extremely limited gene flow between the two chromosomal arrangements within the inversion. Indeed, the average pairwise divergence in neutral sequence between ZAL2 and ZAL2m chromosome inside the inversion was elevated relative to outside the inversion (dxy=0.0087 versus 0.0029). Outside the inversion, the FST value between the arrangements was 0.21. However, because we had no method to reliably assign ZAL2 and ZAL2m identity outside the inversion, we tested for significant population structure between TS and WS groups. Inside the inversion, population differentiation between WS and TS was significant (FST=0.41, P=0.009), whereas outside the inversion, there was no significant structure (FST=0.06, P=0.09; Excoffier et al., 2005). Thus, these results show that genetic differentiation between ZAL2 and ZAL2m is uniformly high across the entire ∼104 Mb inverted interval and low within the region outside the inversion, consistent with gene flow between the chromosomal arrangements being restricted to the small segment of the chromosome outside the inversion.
Patterns of haplotype and nucleotide diversity within and between ZAL2 and ZAL2m
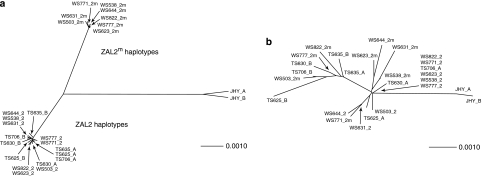
The striking genetic differentiation between the chromosomal rearrangements inside compared with outside the inversion is consistent with the presence of two highly divergent haplotype groups each associated with the inverted segments of the ZAL2 and ZAL2m chromosomes. To visualize the relationships among the haplotypes, we constructed separate haplotype networks based on the phased concatenated haplotype inside and outside the inversion (Figure 1). As expected, the ZAL2 and ZAL2m haplotypes clustered into two distinct groups that were clearly differentiated from one another, whereas outside the inversion no clustering based on chromosomal arrangement was observed (Figure 1). Moreover, the short branches of the ZAL2 and ZAL2m groups illustrate the relatively low diversity observed inside the inversion within both chromosomal rearrangements compared with outside the inversion (Table 1 and Figure 1a).
Figure 1.
ZAL2 and ZAL2m haplotype networks. Haplotype networks were constructed using the concatenated phased haplotypes from all loci within (a) and outside (b) the inversion. For the white-striped (WS) birds the ZAL2 and ZAL2m haplotypes are labeled as 2 or 2m. For the tan-striped (TS) birds and Junco (JHY), the haplotypes for each individual were arbitrarily labeled A and B.
The haplotype networks clearly indicate distinct differences in the genetic diversities of ZAL2m, ZAL2 and the region outside the inversion. To assess this difference we considered that natural populations of white-throated sparrows comprise ∼50% TS (ZAL2/ZAL2) individuals and ∼50% WS (ZAL2/ZAL2m) individuals (Lowther, 1961). Hence, at the population level, the inverted ZAL2m region, the homologous standard ZAL2 region and the region outside the inversion exist in a 1:3:4 ratio. Under the neutral model, in which we assume the absence of selection and comparable rates of recombination, nucleotide diversity is directly proportional to the effective population size (θ=4Neμ). In this context, we predicted that the ratio of neutral nucleotide diversity of ZAL2m, ZAL2 and the region outside the inversion would also be 1:3:4. The observed ratio of neutral nucleotide diversity between ZAL2m and ZAL2 was ∼1:2, similar to our expected ratio. However, the ratio of diversity of ZAL2m, ZAL2 and the region outside the inversion was 1:2:10 (Table 1), which was lower than predicted by the model. This result could be caused by lower than expected diversity for ZAL2m and ZAL2, higher than expected diversity outside the inversion, or both.
Linkage disequilibrium
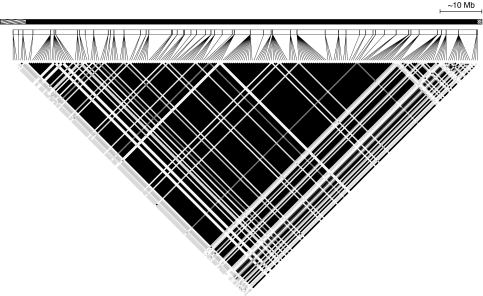
The suppression of recombination between the chromosomal arrangements across the length of the inverted interval is expected to result in a strong signature of LD extending over ∼100 Mb inversion. To quantify the extent of LD associated with this chromosomal polymorphism, we measured LD between pairs of informative sites along the entire length of the chromosome (Figure 2). A total of 240 SNPs, 232 inside the inversion and 8 outside the inversion, were informative and, as expected, under a scenario of long-term suppressed recombination between the arrangements, a large fraction of the pairwise comparisons inside the inversion showed significant nonrandom association, whereas outside the inversion we did not detect significant patterns of LD (Figure 2). To summarize the level of LD inside and outside the inversion, we calculated ZnS between informative sites (Kelly, 1997), which was high inside the inversion (ZnS=0.69) and much lower outside (ZnS=0.10) the inversion. Thus, these results establish that the suppression of recombination between ZAL2 and ZAL2m has resulted in widespread LD that, based on our most proximal and distal markers, spans a minimum of ∼104 Mb inside the inversion.
Figure 2.
Linkage disequilibrium between informative sites across the ZAL2/ZAL2m chromosomes. The concatenated phased haplotypes were used to generate an LD plot across the ZAL2/2m chromosomes. Each of the 239 informative SNPs are plotted to scale with respect to their predicted position along the ZAL2 chromosome. Black squares indicate perfect LD between pairs of SNPs (r2=1), gray squares indicate pairs of SNPs with r2 between 0 and 1 and white squares show no LD. The striped segments of the line above the LD plot indicate regions outside the inversion and the solid (black) segment of the line indicates the region inside the inversion.
Rates of recombination within the ZAL2 and ZAL2m haplotype groups
Our data strongly support a scenario in which recombination in WS (ZAL2/ZAL2m) birds is restricted to the interval outside the inversion. Because ZAL2m/ZAL2m individuals are extremely rare (<1% of the population; Thorneycroft, 1975; Falls and Kopachena, 1994; Romanov et al., 2009), we had previously predicted that the inverted segment of the ZAL2m chromosome might be a non-recombining autosome (Thomas et al., 2008). To test this hypothesis we applied the four-gamete test (Hudson and Kaplan, 1985) to analyse evidence of recombination within ZAL2m haplotypes, as well as ZAL2 haplotypes and the haplotypes outside the inversion on both chromosomes. As expected, we detected recombination within the ZAL2 haplotype (⩾12 events), and within haplotypes outside the inversion (⩾1 event). In contrast to our prediction, however, the four-gamete test indicated that recombination had also occurred within the ZAL2m haplotypes (⩾2 events).
Given the evidence for recombination within both ZAL2 and ZAL2m haplotypes, we next attempted to quantify the difference in population rate of recombination, ρ, between ZAL2 and ZAL2m. In particular, because of the large difference in the population frequency of ZAL2m/ZAL2m birds (<1%) to ZAL2/ZAL2 birds (∼50%) (Thorneycroft, 1975; Falls and Kopachena, 1994; Romanov et al., 2009), we expected a lower rate of recombination inside the inversion within the ZAL2m haplotypes versus within the ZAL2 haplotypes. Estimates of the population recombination rate among all ZAL2m chromosomes was similar to that of the ZAL2 chromosomes (ρ±s.d.=8.48 × 10−9±8.59 × 10−9 and 4.40 × 10−8±5.05 × 10−8 per base, respectively), although it should be noted that diversity levels within the inversion on ZAL2 and ZAL2m were low and our small sample size did not support accurate measurements of ρ (that is, note standard errors). Finally, we also predicted that outside the inversion, the rate of recombination would be greater than within the inversion because the presumed obligatory recombination event required for proper chromosome segregation in ∼50% of the population that are ZAL2/ZAL2m heterozygotes (WS) (Lowther, 1961; Thorneycroft, 1975) must occur in this small interval. Consistent with our prediction, ρ for this segment was estimated to be higher than observed inside the inversion (1.55 × 10−7±2.18 × 10−7 per base); again, note the large confidence intervals.
Relative rate of evolution on the ZAL2 versus ZAL2m chromosomes
The pattern of extensive recombination suppression between ZAL2 and ZAL2m within the inverted region suggests that Hill–Robertson interference among the linked loci could work to reduce the efficacy of selection and subsequently lead to the genetic degeneration of the ZAL2m chromosome (Hill and Robertson, 1966; Rice, 1994; Gordo and Charlesworth, 2001). Hence, we were interested in whether ZAL2m showed any signs of degeneration. Specifically, we tested for differences in the rates of evolution in the ZAL2 versus ZAL2m lineages, applying Tajima's relative rate method using the χ2-test (Tajima, 1993) to fixed differences that mapped inside the inversion and could be unambiguously polarized using sequence from a closely related outgroup (J. hyemalis, Table 2). Although a greater number of mutations were inferred to have occurred in the ZAL2m versus ZAL2 lineage in all sequence classes examined, including nonsynonomous and other presumably functional sites, the observed differences were not statistically significant (Table 2). Thus, although the ZAL2m lineage may be associated with a slightly higher overall mutation rate than the ZAL2 lineage, we did not observe any overt signs of genetic degeneration expected on a chromosome with reduced levels of recombination.
Table 2. Mutations occurring along the ZAL2 and ZAL2m lineages.
| All | Nonsynon | Synon | Conserved | Neutral | |
|---|---|---|---|---|---|
| ZAL2 | 78 | 1 | 6 | 31 | 47 |
| ZAL2m | 98 | 5 | 8 | 36 | 62 |
| P-valuea | 0.13 | 0.102 | 0.593 | 0.54 | 0.15 |
This test includes the subset of fixed differences in which ancestral state could be unambiguously inferred using junco sequence. Conserved category includes all single-nucleotide polymorphisms (SNPs) within most conserved elements and neutral category includes all SNPs outside most conserved elements.
P-values were calculated using Tajima's relative rate test (Tajima, 1993).
Discussion
Gene flow, genetic differentiation and population structure between ZAL2 and ZAL2m
Unlike most simple inversion polymorphisms in which, over time, gene flow occurs between the standard and inverted arrangements except near the inversion breakpoints, complex inversion polymorphisms can be effective suppressors of recombination (Lyon, 2003; Munté et al., 2005; Dyer et al., 2007). For example, the O3+4 inversion polymorphism in Drosophila subobscura differs from the standard OST arrangement by two overlapping inversions and suppresses gene flow between the two chromosome types, producing uniformly high genetic differentiation across the ∼4 Mb region (Munté et al., 2005). Although evidence for gene conversion between the alternative arrangements is prevalent in this system, the limited gene flow in the absence of double recombination events is clearly shown by the high proportion of fixed differences (35%) and the level of divergence (dxy=1.5%) between the alternate arrangements (Munté et al., 2005). Similarly, in the case of an even larger complex inversion polymorphism, the XD chromosome in D. recens, over half of the polymorphisms identified on the XD chromosome were fixed differences between the distorting and wild-type chromosomes (Dyer et al., 2007). We also observed a lack of gene flow between ZAL2 and ZAL2m as indicated by a high frequency of fixed differences (67%) and divergence between the chromosomes approaching 1%. Moreover, the frequency of shared polymorphisms within the inversion was very low (<1%), and unlike the O3+4 and OST system, we failed to detect any gene conversion tracts between the alternative arrangements or between ZAL2m chromosomes within the inversion interval. In these examples from Drosophila and the ZAL2m polymorphism, there is no evidence for genetic exchange toward the center of the inversion polymorphisms, as predicted by models developed by Navarro et al. (1997) for single inversions, suggesting that multiple overlapping inversions are likely to completely suppress double crossovers and can prevent gene flow across the entire inversion interval. Moreover, the very high FST value (0.94) between ZAL2 and ZAL2m inside the inversion is also quite uncommon. Indeed, similar values of FST in excess of >0.8 have been associated with special circumstances, such as homologous loci in non-recombining segments of sex chromosomes (Ironside and Filatov, 2005) or between cryptic species (Terry et al., 2000). Thus, the degree of genetic differentiation observed between ZAL2 and ZAL2m chromosomes is very high even for complex inversions, and is comparable to values observed in the most extreme cases of suppressed gene flow.
Dating and origin of ZAL2m
Using the level of neutral divergence between ZAL2 and ZAL2m chromosomes, we estimated the age of the current ZAL2m haplotypes (that is, time since recombination stopped) assuming a genomic mutation rate equivalent to that described for zebra finch, 2.95 × 10−9 substitutions per site per year (Balakrishnan and Edwards, 2009). Our estimate of ∼2.95±0.3 MY is similar to our previous estimate based on phylogenetic comparisons of noncoding and synonymous sequence between white-throated sparrows and the J. hyemalis outgroup (Thomas et al., 2008). The time of ZAL2m origin predates the estimated time of divergence of the white-throated sparrow from other birds in the genus and is at apparent odds with the hypothesis that the inversion occurred in the white-throated sparrow lineage (Thorneycroft, 1975). Although the polymorphism may be ancient and was simply lost in the other Zonotrichia sparrow lineages, an alternative explanation for its restricted presence in the white-throated sparrow is through introgression as the result of hybridization. Hybridization in birds is well established (Grant and Grant, 1992; Mallet, 2005), and fertile offspring are common in cases in which the genetic divergence between the species is on the order observed between ZAL2 and ZAL2m, that is, <2% (Price and Bouvier, 2002). Although there is precedent for hybridization between the white-throated sparrow and the Junco (Dickerman, 1961), our haplotype networks do not support a recent introgression of either chromosome arrangement from that species. However, relatively recent introgression resulting from the hybridization with another species might explain the high level of genetic divergence between ZAL2 and ZAL2m, as well as the lack of recombination within the inversion. Studies of hybrid genomes have shown that inversions can produce patterns of heterogeneous recombination and high genetic divergence (Rieseberg et al., 1999; Feder et al., 2003; Panithanarak et al., 2004; Stump et al., 2005; Noor et al., 2007). In addition, because collinear regions are expected to homogenize more quickly than non-collinear regions (Rieseberg et al., 1999), time since hybridization is another factor to take into consideration. However, note that the accurate molecular dating of the inversion may be confounded by the unique genetic attributes of the polymorphism, and a direct comparison to the speciation events within this genus were not carried out. Thus, we acknowledge that the above scenarios are not the only possible histories for the inversion. Future studies focused on genome-wide patterns of nucleotide diversity and haplotype structures should help clarify whether or not the ZAL2/ZAL2m polymorphism could have resulted from a recent hybridization event.
Extreme LD associated with the ZAL2/ZAL2m system
Theoretically, the adaptive significance of inversions lies in their capability to suppress recombination, thereby maintaining LD between multiple alleles favored by natural selection (Dobzhansky, 1937; Kirkpatrick and Barton, 2006), and the pattern of LD is dependent on the rate of recombination as well as the strength of selection to maintain linkage between the alleles. In the case of ZAL2/ZAL2m system, we detected two dominant haplotypes representing each arrangement that spanned ∼100 Mb inside the inversion, linking alleles from ∼1000 genes into a super-gene complex. The extended LD we observed in this system is more extreme than that observed in the mouse t-complex, which spans ∼30–40 Mb (Lyon, 2003), and comparable to that observed in the distorting (XD) in D. recens (Dyer et al., 2007). In that case, LD was found to essentially extend across the entire X chromosome (∼130 cM Dyer et al., 2007). As far as we are aware, the pattern of LD associated with ZAL2/ZAL2m is the most extreme example of long-range LD yet to be reported in vertebrates.
Comparison of the ZAL2/ZAL2m chromosome pair to sex chromosomes
Previously, we proposed that the ZAL2/ZAL2m chromosomes were mimicking the early stages of sex chromosome evolution and that ZAL2m shared a number of features with the Y (W) chromosome (Thomas et al., 2008). In particular, these features included suppression of recombination between ZAL2 and ZAL2m over most of their length due to inversions, a negative assortative mating system in which >96% of breeding pairs consist of heterogametic (ZAL2/ZAL2m) × homogametic (ZAL2/ZAL2) individuals and the maintenance of ZAL2m in a near constant state of heterozygosity (Thomas et al., 2008). Although our results from this study do not require us to reconsider these shared features, the detection of recombination within ZAL2m haplotypes refutes the additional prediction that the region inside the inversion on ZAL2m may represent a non-recombining autosome. Thus, although ZAL2m/ZAL2m individuals in the population are rare, the detection of historical recombination events associated with this haplotype suggest that at least some of these individuals are fertile. Indeed, a male ZAL2m homozygote has been described that was able to reproduce (Falls and Kopachena, 1994).
Given the presence of recombination, however rare it may be on ZAL2m, it was therefore not surprising that we did not detect a significant difference in the accumulation of potentially deleterious mutations on ZAL2m compared with ZAL2. The complete cessation of recombination, as with neo-Y chromosomes, such as those in Drosophila miranda (Bachtrog, 2004), stickleback and medaka fish (Peichel et al., 2004; Kondo et al., 2006), the black muntjac (Zhou et al., 2008) and Silene latifolia (Filatov and Charlesworth, 2002; Marais, 2007), leads to reduced efficacy of selection and accumulation of mutations (Charlesworth and Charlesworth, 2000). However, even very low levels of recombination can effectively prevent the process of genetic degeneration observed in non-recombining chromosomes (Haddrill et al., 2007). In this context, and in light of the results from the loci sampled in this study, we can conclude that ZAL2m is unlikely to be mimicking the initial phases of genetic deterioration observed on neo-Y chromosomes.
It should be noted that our inference of recombination within ZAL2m chromosomes relies on the accurate reconstruction of haplotypes from genotype information. Because the number of predicted recombination events within ZAL2m chromosomes is small and the frequency of the minor alleles of these sites is low, incorrect phase estimation could lead to false positives. In addition, the four-gamete test relies on the assumption of infinite sites, which excludes the possibility of independent recurrent mutations. Considering that the minor allele frequency of the sites involved is generally low, independent mutations within the ZAL2 and ZAL2m lineages could produce patterns of variation that resemble those resulting from recombination. However, given that we did not detect the genetic degeneration that is characteristic of non-recombining sequences, we believe that these are true signatures of recombination generated by rare ZAL2m homozygotes.
Although we no longer consider ZAL2m a non-recombining chromosome, we did note potentially low levels of diversity associated with the ZAL2/ZAL2m system that are typical of sex chromosomes. For example, the mammalian and plant Y and the avian W chromosomes, as well as other regions of low recombination, can show 20- to 30-fold reductions in diversity even after correcting for differences in Ne (Begun and Aquadro, 1992; Filatov et al., 2000; Jensen et al., 2002; Berlin and Ellegren, 2004; Hellborg and Ellegren, 2004; Betancourt et al., 2009). Similarly, the W chromosome has been associated with lower than expected levels of nucleotide diversity in other birds (Montell et al., 2001; Berlin and Ellegren, 2004). In the case of ZAL2/ZAL2m polymorphism we observed that diversity within the inversion on ZAL2 and ZAL2m was 5 and 10 times lower, respectively, than outside the inversion. It has been argued that intense sexual selection could be responsible for reductions in diversity on sex chromosomes by further reducing Ne of the Y chromosome (Caballero, 1995; Nagylaki, 1995; Charlesworth, 1996); however, such a mechanism cannot explain the reduced diversity observed on ZAL2m. Hence, natural selection is the only factor that can explain the reduced variability of sex chromosomes and regions of low recombination alike (Hellborg and Ellegren, 2004). Further efforts to quantify patterns of recombination and diversity in the white-throated sparrow genome will provide the necessary context to understand how rates of recombination have influenced the regions inside and outside the inversion.
Implications for identifying specific genes underlying the phenotypes associated with the ZAL2m polymorphism
One of the compelling reasons to study ZAL2m is the opportunity to identify the genetic basis of the phenotypic variation associated with the inversion. Previous studies have localized candidate regions within inversions by examining patterns of recombination between the wild-type and inverted arrangements and identifying region in LD as targets of selection. For example, in a study of the 2La inversion in Anopheles gambiae, White et al. (2007) examined patterns of divergence and LD to localize candidate regions involved in aridity tolerance. In the case of ZAL2/ZAL2m polymorphism, LD across the entire inversion will preclude further localization of candidate regions or genes by standard recombination-based mapping. In addition, if we consider that divergence between ZAL2 and ZAL2m within the inversion is on the order of 1%, conservatively estimate that 50% of the differences are fixed or near-fixed differences between the arrangements, and that the inversion is ∼100 Mb, then we can predict that there are at least 500 000 single-nucleotide differences that distinguish ZAL2 from ZAL2m. Thus, it is possible that the TS and WS birds differ from each other in many traits.
Conclusions
Our population genetic analysis reveals that the ZAL2m arrangement suppresses recombination in the heterokaryotype, resulting in reduced gene flow, high levels of genetic differentiation, extensive population structure and LD between the alternate arrangements. Although we no longer consider ZAL2m a non-recombining chromosome, we believe that it will be valuable as a model for understanding how selection acts to reduce diversity in genomic regions with low recombination rates. Future studies of recombination and diversity at unlinked autosomal and sex-linked loci will likely shed light on the evolution of the ZAL2/ZAL2m system.
Acknowledgments
We thank RT Sullivan, GK Tharp and J Griffin for their computational support, and M Cáceres, D Cutler, K Dyer, T Schlenke and Y Tao for advice on population genetic analyses and comments on the paper. LYH was supported by a Ford Foundation Pre-doctoral Fellowship and a grant from the Center of Behavioral Neuroscience awarded to DLM. DLM and JWT were supported by a grant from the National Institutes of Health (1R21MH082046).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Andolfatto P, DePaulis F, Navarro A. Inversion polymorphisms and nucleotide variability in Drosophila. Genet Res Camb. 2001;77:1–8. doi: 10.1017/s0016672301004955. [DOI] [PubMed] [Google Scholar]
- Auton A, McVean G. Recombination rate estimation in the presence of hotspots. Genome Res. 2007;17:1219–1227. doi: 10.1101/gr.6386707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. Evidence that positive selection drives Y-chromosome degeneration in Drosophila miranda. Nat Genet. 2004;36:518–522. doi: 10.1038/ng1347. [DOI] [PubMed] [Google Scholar]
- Balakrishnan CN, Edwards SV. Nucleotide variation, linkage disequilibrium and founder-facilitated speciation in wild-populations of the zebra finch (Taeniopygia guttata) Genetics. 2009;181:645–660. doi: 10.1534/genetics.108.094250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Aquadro CF. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature. 1992;356:519–520. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- Berlin S, Ellegren H. Chicken W: a genetically uniform chromosome in a highly variable genome. Proc Natl Acad Sci USA. 2004;101:15967–15969. doi: 10.1073/pnas.0405126101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt AJ, Welch JJ, Charlesworth B. Reduced effectiveness of selection caused by a lack of recombination. Curr Biol. 2009;19:655–660. doi: 10.1016/j.cub.2009.02.039. [DOI] [PubMed] [Google Scholar]
- Betrán E, Rozas J, Navarro A, Barbadilla A. The estimation of the number and length distribution of gene conversion tracts from population DNA sequence data. Genetics. 1997;146:89–99. doi: 10.1093/genetics/146.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero A. On the effective size of populations with separate sexes, with particular reference to sex-linked genes. Genetics. 1995;139:1007–1011. doi: 10.1093/genetics/139.2.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. Background selection and patterns of genetic diversity in Drosophila melanogaster. Genet Res. 1996;68:131–149. doi: 10.1017/s0016672300034029. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Phil Trans R Soc B. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerman RW. Hybrids among the fringillid genera Junco-Zonotrichia and Melospiza. Auk. 1961;78:627–632. [Google Scholar]
- Dobzhansky T. Genetics and the Origin of Species. Columbia University Press: New York; 1937. [Google Scholar]
- Dobzhansky T. Genetics of natural populations. XIX. Origin of heterosis through natural selection in populations of Drosophila pseudoobscura. Genetics. 1950;35:288–302. doi: 10.1093/genetics/35.3.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T, Sturtevant AH. Inversions in the chromosomes of Drosophila pseudoobscura. Genetics. 1938;23:28–64. doi: 10.1093/genetics/23.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer K, Charlesworth B, Jaenike J. Chromosome-wide linkage disequilibrium as a consequence of meiotic drive. Proc Natl Acad Sci USA. 2007;104:1587–1592. doi: 10.1073/pnas.0605578104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhart MA, Lekgothoane S, Grenier J, Nadeau JH. Pattern of segmental recombination in the distal inversion of the mouse t-haplotypes. Mamm Genome. 2002;13:438–444. doi: 10.1007/s00335-002-3012-7. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetic data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Falls JB, Kopachena JG.1994White-throated sparrow (Zonotrichia albicollis)In: Poole A, Gill F (eds).The Birds of North America The Academy of Natural Sciences: Philadelphia [Google Scholar]
- Feder JL, Roethele JB, Filchak KE, Niedbalski J, Romero-Severson J. Evidence for inversions related to sympatric host race formation in the apple maggot fly, Rhagoletis pomonella (Diptera: Tephritidae) Genetics. 2003;163:939–953. doi: 10.1093/genetics/163.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov DA, Charlesworth D. Substitution rates in the X- and Y-linked genes of the plants, Silene latifolia and S. dioica. Mol Biol Evol. 2002;19:898–907. doi: 10.1093/oxfordjournals.molbev.a004147. [DOI] [PubMed] [Google Scholar]
- Filatov DA, Moneger F, Negrutiu I, Charlesworth D. Low variability in a Y-linked plant gene and its implications for Y-chromosome evolution. Nature. 2000;404:388–390. doi: 10.1038/35006057. [DOI] [PubMed] [Google Scholar]
- Gordo I, Charlesworth B. Genetic linkage and molecular evolution. Curr Biol. 2001;11:R684–R686. doi: 10.1016/s0960-9822(01)00408-0. [DOI] [PubMed] [Google Scholar]
- Grant PR, Grant BR. Hybridization of bird species. Science. 1992;256:193–197. doi: 10.1126/science.256.5054.193. [DOI] [PubMed] [Google Scholar]
- Graves JAM. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Haddrill PR, Halligan DL, Tomaras D, Charlesworth B. Reduced efficacy of selection in regions of the Drosophila genome that lack crossing over. Genome Biol. 2007;8:R18. doi: 10.1186/gb-2007-8-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL. Genetic dissection of segregation distortion II. Mechanism of suppression of distortion by certain inversions. Genetics. 1975;80:539–547. doi: 10.1093/genetics/80.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson E, Eanes WF. Contrasting histories of three gene regions associated with In(3L)Payne of Drosophila melanogaster. Genetics. 1996;144:1565–1575. doi: 10.1093/genetics/144.4.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellborg L, Ellegren H. Low levels of nucleotide diversity in mammalian Y chromosomes. Mol Biol Evol. 2004;21:158–163. doi: 10.1093/molbev/msh008. [DOI] [PubMed] [Google Scholar]
- Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;8:269–294. [PubMed] [Google Scholar]
- Hoffmann AA, Sgro CM, Weeks AR. Chromosomal inversion polymorphisms and adaptation. TREE. 2004;19:482–488. doi: 10.1016/j.tree.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- ICGSC Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Ironside JE, Filatov DA. Extreme population structure and high interspecific divergence of the Silene Y chromosome. Genetics. 2005;171:705–713. doi: 10.1534/genetics.105.041210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MA, Charlesworth B, Kreitman M. Patterns of genetic variation at a chromosome 4 locus of Drosophila melanogaster. Genetics. 2002;160:493–507. doi: 10.1093/genetics/160.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JK. A test of neutrality based on interlocus associations. Genetics. 1997;146:1197–1206. doi: 10.1093/genetics/146.3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennington JW, Partridge L, Hoffmann AA. Patterns of diversity and linkage disequilibrium within the cosmopolitan inversion In(3R)Payne in Drosophila melanogaster are indicative of coadaptation. Genetics. 2006;172:1655–1663. doi: 10.1534/genetics.105.053173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Barton N. Chromosome inversions, local adaptation and speciation. Genetics. 2006;173:419–434. doi: 10.1534/genetics.105.047985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Hornung U, Nanda I, Imai S, Sasaki T, Shimizu A, et al. Genomic organization of the sex-determining and adjacent regions of the sex chromosomes of medaka. Genome Res. 2006;16:815–826. doi: 10.1101/gr.5016106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacevic M, Schaeffer SW. Molecular population genetics of X-linked genes in Drosophila pseudoobscura. Genetics. 2000;156:155–172. doi: 10.1093/genetics/156.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Lowther JK. Polymorphism in the white-throated sparrow, Zonotrichia albicollis Gmelin. Can J Zool. 1961;39:281–292. [Google Scholar]
- Lyon MF. Transmission ratio distortion in mice. Annu Rev Genet. 2003;37:393–408. doi: 10.1146/annurev.genet.37.110801.143030. [DOI] [PubMed] [Google Scholar]
- Mallet J. Hybridization as an invasion of the genome. TREE. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Marais GAB. Sex chromosomes and mitochondrial DNA polymorphism in birds: the Hill-Robertson effects extend from nucleus to mitochondria. Heredity. 2007;99:357–358. doi: 10.1038/sj.hdy.6801034. [DOI] [PubMed] [Google Scholar]
- Michopoulos V, Maney DL, Morehouse CB, Thomas JW. A genotyping assay to determine plumage morph in the white-throated sparrow, Zonotrichia albicollis. Auk. 2007;124:1330–1335. [Google Scholar]
- Montell H, Fridolfsson A-K, Ellegren H. Contrasting levels of nucleotide diversity on the avian Z and W sex chromosomes. Mol Biol Evol. 2001;18:2010–2016. doi: 10.1093/oxfordjournals.molbev.a003742. [DOI] [PubMed] [Google Scholar]
- Munté A, Rozas J, Aguadé M, Segarra C. Chromosomal inversion polymorphism leads to extensive genetic structure: a multilocus survey in Drosophila pseudoobscura. Genetics. 2005;39:1573–1581. doi: 10.1534/genetics.104.032748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagylaki T. The inbreeding effective population number in dioecious populations. Genetics. 1995;148:1245–1255. doi: 10.1093/genetics/139.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, Betrán E, Barbadilla A, Ruiz A. Recombination and gene flux caused by gene conversion and crossing over in inversion heterozygotes. Genetics. 1997;146:695–709. doi: 10.1093/genetics/146.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MAF, Garfield DA, Schaeffer SW, Machado CA. Divergence between the Drosophila pseudoobscura and D. persimilis genome sequences in relation to chromosomal inversions. Genetics. 2007;177:1417–1428. doi: 10.1534/genetics.107.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MAF, Grams KL, Bertucci LA, Reiland J. Chromosomal inversions and the reproductive isolation of species. Proc Natl Acad Sci USA. 2001;98:12084–12088. doi: 10.1073/pnas.221274498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitski E, Braver G. An analysis of crossing over within a heterozygous inversion in Drosophila melanogaster. Genetics. 1954;39:197–209. doi: 10.1093/genetics/39.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panithanarak T, Hauffe HC, Dallas JF, Glover A, Ward RG, Searle JB. Linkage-dependent gene flow in a house mouse chromosomal hybrid zone. Evolution. 2004;58:37–47. doi: 10.1111/j.0014-3820.2004.tb01585.x. [DOI] [PubMed] [Google Scholar]
- Peichel CL, Ross JA, Matson CK, Dickson M, Grimwood J, Schmutz J, et al. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr Biol. 2004;14:1416–1424. doi: 10.1016/j.cub.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Piper WH, Wiley RH. Distinguishing morphs of the white-throated sparrow in basic plumage. J Field Ornithol. 1989;60:73–83. [Google Scholar]
- Price TD, Bouvier MM. The evolution of F1 postzygotic incompatibilities in birds. Evolution. 2002;56:2083–2089. [PubMed] [Google Scholar]
- Rice WR. Degeneration of a nonrecombining chromosome. Science. 1994;263:230–232. doi: 10.1126/science.8284674. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Whitton J, Gardner K. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics. 1999;152:713–727. doi: 10.1093/genetics/152.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov MN, Tuttle EM, Houck ML, Modi WS, Chemnick LG, Korody ML, et al. The value of avian genomics to the conservation of wildlife. BMC Genomics. 2009;10:S10. doi: 10.1186/1471-2164-10-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Schaeffer SW, Anderson WW. Mechanisms of genetic exchange within the chromosomal inversions of Drosophila pseudoobscura. Genetics. 2005;171:1729–1739. doi: 10.1534/genetics.105.041947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer SW, Goetting-Minesky MP, Kovacevic M, Peoples JR, Graybill JL, Miller JM, et al. Evolutionary genomics of inversions in Drosophila pseudoobscura: evidence for epistasis. Proc Natl Acad Sci USA. 2003;100:8319–8324. doi: 10.1073/pnas.1432900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, et al. Evolutionarily conserved elements in vertebrate, insect, worm and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawson EE, Shaffer CD, Malone CD, Leung W, Kellmann E, Shevchek RB, et al. Comparison of dot chromosome sequences from D.melanogaster and D. virilis reveals an enrichment of DNA transposon sequences in heterochromatic regions. Genome Biol. 2006;7:R15. doi: 10.1186/gb-2006-7-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, Barnard J, et al. A common inversion under selection in Europeans. Nat Genet. 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith N, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stump AD, Fitzpatrick MC, Lobo NF, Traoré S, Sagnon N, Costantini C, et al. Centromere-proximal differentiation and speciation in Anopheles gambiae. Proc Natl Acad Sci USA. 2005;102:15930–15935. doi: 10.1073/pnas.0508161102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH. A case of rearrangement of genes in Drosophila. Proc Natl Acad Sci USA. 1921;7:235–237. doi: 10.1073/pnas.7.8.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH, Beadle GW. The relations of inversions in the X chromosome of Drosophila melanogaster to crossing over and disjunction. Genetics. 1936;21:554–604. doi: 10.1093/genetics/21.5.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics. 1993;135:599–607. doi: 10.1093/genetics/135.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry A, Bucciarelli G, Bernardi G. Restricted gene flow and incipient speciation in disjunct Pacific Ocean and Sea of Cortez populations of a reef fish species, Girella nigricans. Evolution. 2000;54:652–659. doi: 10.1111/j.0014-3820.2000.tb00066.x. [DOI] [PubMed] [Google Scholar]
- Thomas JW, Cáceres M, Lowman JJ, Morehouse CB, Short ME, Baldwin EL, et al. The chromosomal polymorphism linked to variation in social behavior in the white-throated sparrow (Zonotichia albicollis) is a complex rearrangement and a suppressor of recombination. Genetics. 2008;179:1455–1468. doi: 10.1534/genetics.108.088229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorneycroft HB. Chromosomal polymorphism in the white-throated sparrow, Zonotrichia albicollis (Gmelin) Science. 1966;154:1571–1572. doi: 10.1126/science.154.3756.1571. [DOI] [PubMed] [Google Scholar]
- Thorneycroft HB. A cytogenetic study of the white-throated sparrow, Zonotrichia albicollis Gmelin. Evolution. 1975;29:611–621. doi: 10.1111/j.1558-5646.1975.tb00855.x. [DOI] [PubMed] [Google Scholar]
- Tuttle EM. Alternative reproductive strategies in the white-throated sparrow: behavioral and genetic evidence. Behav Ecol. 2003;14:425–432. [Google Scholar]
- Wallace LT, Erhart MA. Recombination within mouse t haplotypes has replaced significant segments of t-specific DNA. Mamm Genome. 2008;19:263–271. doi: 10.1007/s00335-008-9103-3. [DOI] [PubMed] [Google Scholar]
- Watt DJ. Plumage brightness index for white-throated sparrows. J Field Ornithol. 1986;57:105–115. [Google Scholar]
- White BJ, Hahn MW, Pombi M, Cassone BJ, Lobo NF, Simard F, et al. Localization of candidate regions maintaining a common polymorphic inversion (2La) in Anopheles gambiae. PloS Genet. 2007;3:e217. doi: 10.1371/journal.pgen.0030217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wheeler DA, Yakub I, Wei S, Sood R, Rowe W, et al. SNPdetector: a software tool for sensitive and accurate SNP detection. PloS Comput Biol. 2005;5:e53. doi: 10.1371/journal.pcbi.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Wang J, Huang L, Nie W, Wang J, Liu Y, et al. Neo-sex chromosomes in the black muntjac recapitulate incipient evolution of mammalian sex chromosomes. Genome Biol. 2008;9:R98. doi: 10.1186/gb-2008-9-6-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.