Abstract
Homologous recombination mediated by the RAD51 recombinase helps eliminate chromosomal lesions, such as DNA double-stranded breaks induced by radiation or arising from injured DNA replication forks. The tumor suppressors BRCA2 and PALB2 act together to deliver RAD51 to chromosomal lesions to initiate repair. Here we document a new function of PALB2 in the enhancement of RAD51's ability to form the D-loop. We show that PALB2 binds DNA and physically interacts with RAD51. Importantly, while PALB2 alone stimulates D-loop formation, a co-operative effect is seen with RAD51AP1, an enhancer of RAD51. This stimulation stems from PALB2's ability to function with RAD51 and RAD51AP1 to assemble the synaptic complex. Our results help unveil a multi-faceted role of PALB2 in chromosome damage repair. Since PALB2 mutations can cause breast and other tumors or lead to Fanconi anemia, our findings are important for understanding the mechanism of tumor suppression in humans.
BRCA2 protein plays a key role in mediating homologous recombination (HR) and the repair of damaged chromosomes via HR1 in humans. BRCA2 fulfills these biological roles by regulating the function of the RAD51 recombinase protein2,3. Recently, a novel protein called PALB2 (Partner and Localizer of BRCA2) was found to associate with BRCA2. Importantly, BRCA2 and PALB2 mutations lead to destabilization of the genome4-6 and engender cancer risk. While monoallelic mutations predispose affected individuals to breast, pancreatic and other cancers7,8, biallellic mutations cause Fanconi anemia subtypes -N and -D1 respectively9-11. PALB2 is thought to stabilize BRCA2 by promoting its chromatin association. Recent studies have also demonstrated an architectural function of PALB2 in linking BRCA2 with the BRCA1 tumor suppressor12,13.
To better understand the biological functions of PALB2, we have purified full length PALB2 and several PALB2 fragments and subjected them to a series of biochemical analysis. We demonstrate that PALB2 binds DNA and associates with the RAD51 recombinase in a species-specific fashion. Importantly, PALB2 exerts a stimulatory effect on the ability of RAD51 to make D-loops. Moreover, PALB2 physically interacts with RAD51AP1, a key accessory factor of RAD5114,15, and functionally co-operates with RAD51AP1 in the enhancement of RAD51's recombinase attribute. These results help define the multi-faceted role that PALB2 protein fulfills in HR and chromosome damage repair.
Results
Purification of PALB2 and protein fragments
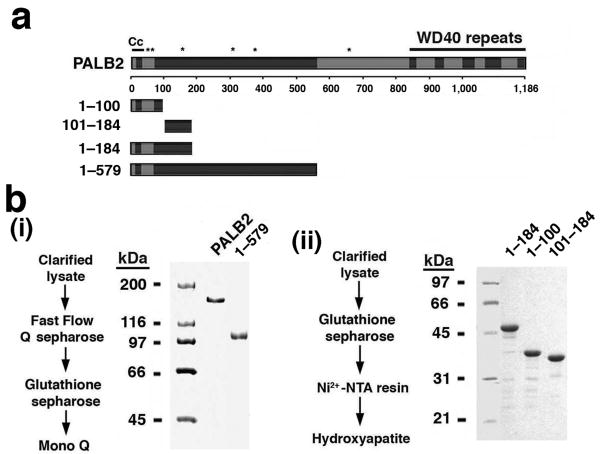
To obtain highly purified PALB2 and protein fragments for functional analyses, full-length GST-tagged PALB2 and PALB2 fragments were expressed in insect cells with the use of recombinant bacculoviruses or in the bacterium Escherichia coli (E. coli). All the PALB2 species were found to be soluble, and multi-step procedures were devised for purifying them to near homogeneity (Fig. 1a,b and Supplementary Fig. 1a,b).
Figure 1. PALB2 purification.
(a) Schematic summarizing the known features of PALB2 and the PALB2 species used in this work. (b) Protein purification procedures and SDS-PAGE analysis of the purified PALB2 species (1 μg each in (i) and 2 μg each in (ii)).
Demonstration of PALB2's DNA binding function
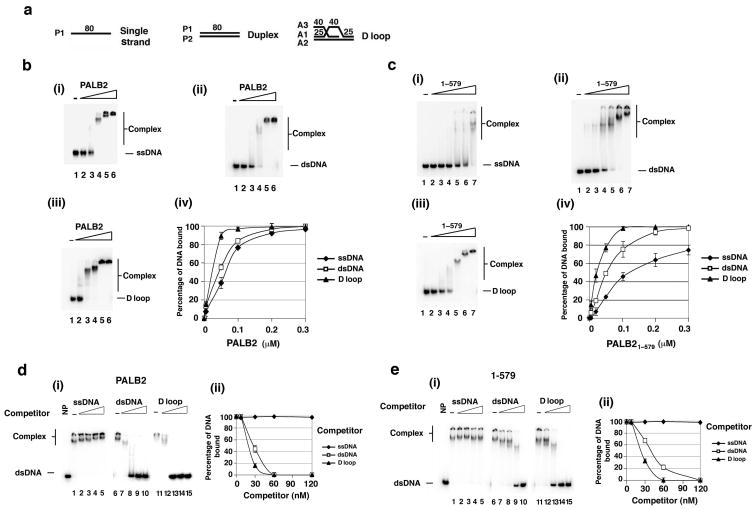
Since PALB2 mediates the chromatin association of BRCA2 in cells5, we were interested in testing whether it has DNA binding activity. A DNA electrophoretic mobility shift assay with 32P-labeled ssDNA, dsDNA, or D-loop as substrate (Fig. 2a) was therefore used to test various purified PALB2 species. Full length PALB2 bound all three species of DNA with the following preference: D-loop > dsDNA > ssDNA (Fig. 2b). Importantly, while the PALB2 amino-terminal fragment encompassing residues 1-579 also bound DNA with the same preference and a similar affinity as the full length protein (Fig. 2c), the PALB2 fragment that harbors residues 611-764 or the C-terminal region (residue 850-1186) did not shift any of the DNA substrates tested (Supplementary Fig. 1c and data not shown).
Figure 2. DNA binding by PALB2.
(a) The DNA substrates used. The prefix denotes the identity of the oligonucleotide. (b) Full length PALB2 (5, 50, 100, 200, and 300 nM) or (c) PALB2 1-579 (5, 20, 50, 100, 200, and 300 nM) was incubated with different DNA substrates (30 nM) and then analyzed (i, ii, and iii). The results were quantified and graphed (iv). (d) and (e) PALB2 (150 nM) or PALB2 1-579 (200 nM) was incubated with radiolabeled dsDNA (30 nM) and then the nucleoprotein complex was challenged with an increasing concentration of unlabeled ssDNA, dsDNA, or D-loop (30, 60, 90, and 120 nM), as indicated. The reaction mixtures were analyzed (i), and the levels of radiolabeled nucleoprotein complex were quantified and graphed (ii). Error bars represent the standard deviation (±s.d.) calculated based on at least three independent experiments.
In order to establish the DNA binding specificity of PALB2 more definitively, we conducted a series of competition experiments wherein a preformed nucleoprotein complex of either full length PALB2 or PALB2 1-579 with radiolabeled dsDNA was challenged with increasing amounts of unlabeled ssDNA, dsDNA, or D-loop. The results revealed that, with either PALB2 species, the D-loop was the most effective competitor, followed by dsDNA (Fig. 2d,e). By contrast, ssDNA was less able to reduce the level of the preformed radiolabeled nucleoprotein complex (Fig. 2d,e).
Taken together, the results from the experiments documented herein have helped establish that (1) PALB2 is a DNA binding protein, (2) the N-terminal half of PALB2 harbors a DNA binding domain, and (3) both full length PALB2 and the isolated N-terminal DNA binding domain bind the D-loop avidly. PALB2 clearly also binds dsDNA, and we note that results reported by Jean Yves Masson's group (Buisson et al; personal communication) suggest that PALB2 has high affinity for certain sequences or secondary structures in ssDNA as well.
Association of purified PALB2 with RAD51
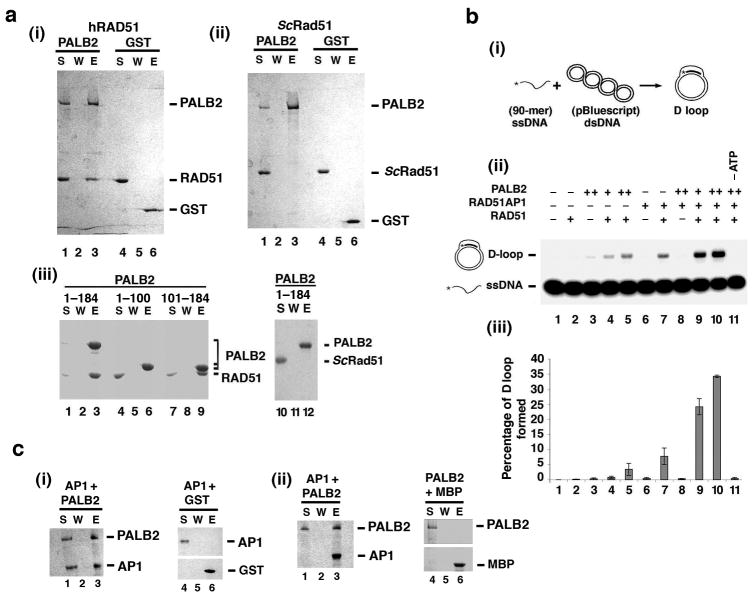
Since PALB2 affects RAD51-mediated chromosome damage repair in cells5, we employed an affinity pulldown assay to query whether PALB2 directly interacts with RAD51. For this, purified PALB2 and RAD51 were incubated together, followed by the capture of the assembled protein complex via the GST tag on PALB2 using glutathione Sepharose beads, elution of proteins by SDS treatment, and SDS-PAGE analysis. The results clearly showed an association of RAD51 with PALB2 (Fig. 3a, (i)). As expected, RAD51 did not associate with purified GST (Fig. 3a, (i)). Importantly, no interaction of PALB2 with the budding yeast Saccharomyces cerevisiae Rad51 (ScRad51) was observed (Fig. 3a, (ii)), indicating that the PALB2-RAD51 complex is species-specific. By testing purified GST-tagged fragments of PALB2 that encompass different portions of the protein (Fig. 1b, (ii)), we were able to map the RAD51 interaction domain to residues 101-184 of PALB2 (Fig. 3a, (iii)). Again, no interaction of the PALB2 1-184 fragment with ScRad51 was seen (Fig. 3a, (iii)).
Figure 3. Enhancement of the RAD51-mediated D-loop reaction by PALB2 and RAD51AP1.
(a) In (i), GST-PALB2 or GST (2 μg) was incubated with RAD51 (3 μg) and the PALB2-RAD51 complex was captured on glutathione resin and analyzed. In (ii), ScRad51 (3 μg) was used in place of hRAD51. In (iii), the indicated GST-tagged PALB2 fragments (4 μg) were examined for complex formation with RAD51 or ScRad51 (3 μg). S: supernatant containing unbound proteins; W: wash; E: SDS eluate of the glutathione resin. (b) Schematic of the D-loop assay is shown in (i). Panel (ii) shows D-loop reactions conducted with RAD51, PALB2, RAD51AP1, or combinations of them. ATP was omitted from the reaction in lane 11. The results were quantified and graphed in (iii). Error bars represent the standard deviation (±s.d.) calculated based on at least three independent experiments. (c) In panel (i) GST-tagged PALB2 or GST (2 μg) was incubated with MBP-RAD51AP1 (2 μg), and the PALB2-RAD51AP1 complex was captured on glutathione resin. In panel (ii), MBP-tagged RAD51AP1 (2 μg) or MBP was incubated with GST-tagged PALB2 and the PALB2-RAD51AP complex was captured on amylose resin. Elution of the protein complex from the affinity resin and subsequent analysis were conducted as in (a).
Enhancement of RAD51 activity by PALB2 and RAD51AP1
We noted that the DNA binding properties of PALB2 are reminiscent of RAD51AP114,15, which interacts with and enhances the ability of RAD51 to make D-loops14,15. Given this, and the fact that PALB2 associates with RAD51, we wanted to test whether PALB2, alone or in combination with RAD51AP1, would enhance the recombinase function of RAD51. A well-established D-loop assay (Fig. 3b, (i)) was used for this purpose. The addition of PALB2 alone to the D-loop reaction increased the level of product several fold, from a barely detectable amount in PALB2's absence to ∼5-8% of D-loop with PALB2 being present (Fig. 3b, (ii) and (iii), and Supplementary Fig. 2a). The increase in D-loop formation stemmed from a stimulation of the RAD51 recombinase activity by PALB2, as PALB2 alone was not capable of D-loop formation (Fig. 3b, (ii) and (iii)). As expected14,15, the efficiency of the D-loop reaction could be enhanced by RAD51AP1 alone (Fig. 3b, (ii) and (iii), and Supplementary Fig. 2b). We then conducted an extensive series of experiments in which we tested a wide range of reactant concentrations to ask whether PALB2 and RAD51AP1 would synergize in the promotion of D-loop formation. Importantly, at PALB2 and RAD51AP1 concentrations that are suboptimal on their own, a clear synergy was seen (Fig. 3b, (ii) and (iii)). ATP was always required for D-loop formation, regardless of whether PALB2, RAD51AP1, or the combination of these two recombinase co-factors was used (Fig. 3b, (ii) and (iii), and data not shown). We note that the stimulatory effect of PALB2 and RAD51AP1 on RAD51 is specific, as we did not find any enhancement of the D-loop reaction catalyzed by ScRad51 by PALB2 or RAD51AP1 alone or the two factors in combination (Supplementary Fig. 3).
Interactions of PALB2 with RAD51AP1
Based on the observed synergy between PALB2 and RAD51AP1 in the D-loop reaction (Fig. 3b), we wished to determine whether these two HR factors physically interact. With the aid of affinity pulldown using either the GST-tag on PALB2 and glutathione resin, or the MBP-tag on RAD51AP1 and amylose resin, we were able to show a direct interaction of PALB2 with RAD51AP1 (Fig. 3c). The PALB2-RAD51AP1 complex is destabilized by KCl concentrations above 100 mM (data not shown). To further map this interaction, we used an N-terminally truncated variant of RAD51AP1, NΔ120 that lacks the first 120 residues of the protein. With the aid of MBP-pulldown by amylose resin, we observed that MBP-tagged RAD51AP1 NΔ120 is deficient in PALB2 interaction (Supplementary Fig. 4a, (i)). We note that, since the RAD51 interaction domain resides within the extreme C-terminus of RAD51AP114,15, the RAD51AP1 NΔ120 variant retains the ability to interact with RAD51 (Supplementary Fig. 4a, (ii)). Importantly, while RAD51AP1 NΔ120 could enhance the RAD51-mediated D-loop reaction, it failed to synergize with PALB2 in this regard (Supplementary Fig. 4b). Thus, it appears that the interaction between PALB2 and RAD51AP1 is required for their functional synergy in the enhancement of the D-loop reaction.
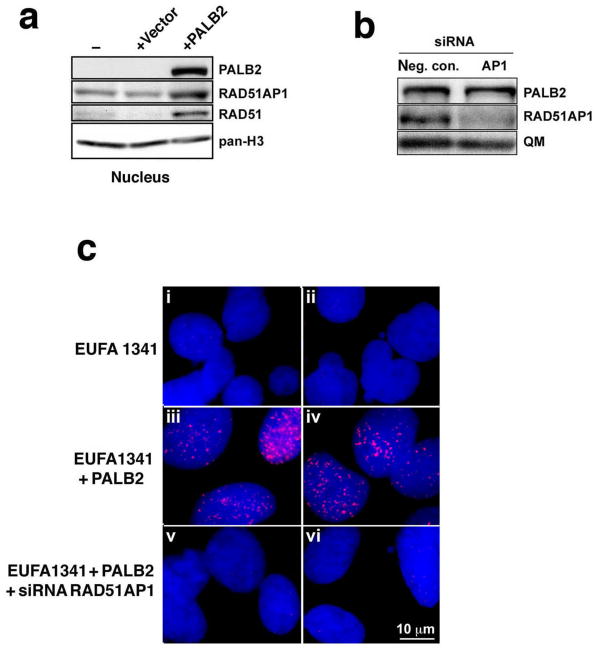
To investigate the functional relationship between PALB2 and RAD51AP1 in vivo, we analyzed protein foci formation in HeLa and U2OS cells after DNA damaging treatment. Whereas mock-treated cells showed dispersed nuclear staining of RAD51AP1 and PALB2 but no significant co-localization (Supplementary Fig. 5a,b and i,j), exposure to ionizing radiation induced distinct PALB2 and RAD51AP1 foci in both HeLa (Supplementary Fig. 5e,f) and U2OS cells (Supplementary Fig. 5m,n) that frequently co-localized (Supplementary Fig. 5g,o, for HeLa and U2OS cells respectively). Similar results were obtained in U2OS cells with mitomycin C (MMC) treatment (data not shown). Interestingly, we found that even though PALB2 deficient fibroblasts express RAD51AP1 protein (Fig. 4a), they are unable to assemble RAD51AP1 foci upon treatment with MMC (Fig. 4b). As expected, complementation of these cells with PALB2 restored the ability to form RAD51AP1 foci in response to MMC (Fig. 4b), and siRNA treatment against RAD51AP1 could diminish these foci (Fig.4b,c). These results indicate that PALB2, either directly or through an intermediary, such as RAD51, plays a key role in the recruitment of RAD51AP1 to DNA damage.
Figure 4. Effect of PALB2 on DNA damage-induced RAD51AP1 foci formation.
(a) Western blots of nuclear extracts from PALB2-deficient fibroblasts (-) transduced with empty pOZC vector or with PALB2-expressing pOZC vector (+PALB2). (b) Western blots to show RAD51AP1 knockdown in the PALB2-complemented EUFA1341 cells. The signal for QM serves as a loading control. (c) Shown are representative micrographs obtained by superimposing the RAD51AP1 signal (red) of PALB2-deficient (i-ii) and PALB2-complemented (iii-vi) EUFA1341 fibroblasts onto the DAPI counter stain (blue). PALB2-deficient EUFA1341 fibroblasts are greatly impaired in RAD51AP1 foci formation after MMC treatment (i, ii), whereas PALB2-complemented EUFA1341 cells form RAD51AP1 foci (iii, iv) in response to MMC. RAD51AP1 foci can be abrogated in PALB2-complemented EUFA1341 cells by treatment with RAD51AP1 siRNA (v, vi).
In co-immunoprecipitation experiments using anti-PALB2 and anti-RAD51AP1 antibodies, only a negligible amount of PALB2 and RAD51AP1 could be co-precipitated from cell extracts before and after treatment of cells with camptothecin (data not shown). Considering that the PALB2-RAD51 complex is salt-sensitive, the failure to co-immunoprecipitate these proteins could stem from the relatively high concentration of salt (120 mM) and extensive washing steps designed to reduce non-specific protein binding to the anti-mouse or anti-rabbit IgG resin. Alternatively, or in addition, the antibodies used might have a destabilizing effect on any PALB2-RAD51AP1 complex present in the extracts.
Enhancement of synaptic complex assembly by PALB2-RAD51AP1
In the RAD51-mediated D-loop reaction, the presynaptic filament captures the duplex DNA partner, followed by a DNA homology search process that leads to the formation of a nucleoprotein intermediate, the synaptic complex, in which the DNA molecules are homologously aligned and base switching has occurred16,17.
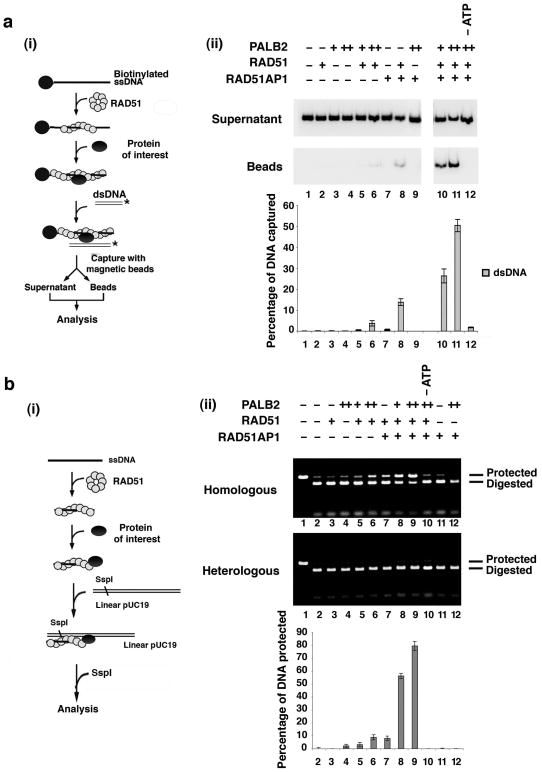
We employed two separate assay systems to verify whether PALB2 and RAD51AP1 promote the formation of the synaptic complex with the RAD51 presynaptic filament. First, the duplex capture step (Fig. 5a, (i)) was examined by mixing the RAD51 presynaptic filaments assembled on ssDNA linked to magnetic beads with a radiolabeled duplex in the presence of PALB2, RAD51AP1, or both of these recombinase cofactors. As had been reported before, the RAD51 presynaptic filament alone exhibited only a weak ability to engage dsDNA18. PALB2 alone enhanced the ability of the presynaptic filament to capture duplex DNA (Fig. 5a, (ii)). RAD51AP1 also worked with the RAD51 presynaptic filament in duplex capture (Fig. 5a, (ii)). Then, we tested PALB2 and RAD51AP1 together to see if they would act synergistically in the duplex capture reaction. Numerous reactant concentrations were examined in this endeavor. As in the D-loop reaction, at certain PALB2 and RAD51AP1 concentrations, these two factors clearly synergized in duplex capture in a manner that required the RAD51 presynaptic filament and ATP (Fig. 5a, (ii)). As expected, no dsDNA capture occurred when magnetic beads without ssDNA were first incubated with RAD51, PALB2, and RAD51AP1 and then with the radiolabeled dsDNA (Supplementary Fig. 6a). Importantly, the combination of PALB2, RAD51AP1, and the RAD51 presynaptic filament failed to capture ssDNA (Supplementary Fig. 6b).
Figure 5. PALB2 enhances synaptic complex assembly.
(a) Shown in (i) is the schematic of the duplex capture assay18. Panel (ii) shows reactions carried out with the RAD51 presynaptic filament, RAD51AP1, PALB2, or their combinations. ATP was omitted from the reaction in lane 12. The results were quantified and graphed. (b) The basis for the protection against restriction digest is explained in (i)19. Panel (ii) shows reactions carried out with the RAD51 presynaptic filament, RAD51AP1, PALB2, or their combinations. No restriction enzyme was added to the reaction in lane 1, and ATP was omitted from the reaction in lane 10. The results were quantified and graphed; the background of 5% (lane 2) had been subtracted from all the values. Error bars represent the standard deviation (±s.d.) calculated based on at least three independent experiments.
We next used protection of linear dsDNA against digestion by the restriction enzyme SspI (Fig. 5b, (i)) to directly examine synaptic complex formation19. The results from testing numerous reactant concentrations again provided clear evidence for an enhancement of RAD51-dependent synaptic complex formation by PALB2 and RAD51AP1 individually, and for a co-operative action of these recombinase co-factors (Fig. 5b, (ii)). Again, the reaction mediated by PALB2 and RAD51AP1 in conjunction with the RAD51 presynaptic filament showed a strict dependence on ATP (Fig. 5b, (ii)).
Discussion
In cells, ssDNA arising from the nucleolytic processing of DSBs and other chromosomal lesions is first bound by RPA. The utilization of the RPA-bound ssDNA as substrate for HR necessitates RPA's replacement by RAD51, which is promoted by the BRCA2 protein1,20. This attribute of the BRCA2 protein qualifies it as a recombination mediator1. Recent work5 has shown that the proper localization of BRCA2 and the targeting of BRCA2 to damaged chromosomes are dependent on PALB2. Thus, in PALB2-deficient cells, as in BRCA2-deficient cells21, the assembly of DNA damage-induced RAD51 foci is impaired5. Based on these properties of PALB2, it has been generally assumed that its primary function is to help promote the delivery of RAD51 to DNA lesions via BRCA25 (Supplementary Fig. 7a). Here, our biochemical studies and those reported by Buisson et al (personal communication) have revealed a direct interaction of PALB2 with RAD51 and the RAD51 presynaptic filament. Importantly, our results have shown that PALB2 binds D-loop and also dsDNA, and the results of Buisson et al (personal communication) suggest that PALB2 also has high affinity for certain sequences or secondary structures in ssDNA. These DNA binding properties distinguish PALB2 from the isolated DNA binding domain of the human or mouse BRCA2 protein described until now, which appear to have little affinity for dsDNA3,20. We have carried out biochemical analyses to test the hypothesis that PALB2 functions with RAD51 after presynaptic filament assembly. The results have provided evidence for an ability of PALB2 to synergize with the RAD51AP1 protein in the promotion of D-loop formation by RAD51 (Supplementary Fig. 7a,b). In addition to facilitating the assembly of the synaptic complex, given the affinity of PALB2 (personal communication from Buisson et al and this work) and RAD51AP114 for the D-loop structure, they could also stabilize the nascent D-loop made by RAD51. We note that since RAD51AP1 plays little or no role in the assembly of DNA damage-induced RAD51 foci in cells14,15, its function in HR is probably limited to the DNA strand invasion stage of the HR reaction.
Like BRCA2, PALB2 is indispensable for genome maintenance and cancer suppression in humans7,9,10,22,23. Our results showing an ability of PALB2 to stimulate the RAD51-mediated D-loop reaction speak to the multi-faceted role that this tumor suppressor fulfills in DNA homology-directed chromosome damage repair and should be valuable for understanding the tumor suppressor attribute of this recombinase accessory factor.
Methods
DNA binding assays
PALB2 was incubated with the indicated radiolabeled DNA substrate (30 nM each) and amounts of PALB2 or PALB2 fragment in 5 μl buffer (50 mM Tris-HCl, pH 7.5, 100 mM KCl, 1 mM MgCl2, 1 mM DTT, 100 μg.ml-1 BSA) at 37°C for 10 min. The reaction mixtures were resolved in 4% polyacrylamide gels in Tris-Glycine buffer (12.5 mM Tris-HCl, 100 mM glycine pH 8.0). For the competition experiments in Fig. 2, d & e, dsDNA (30 nM) was incubated with 150 nM of PALB2 or 200 nM of PALB2 1-579 at 37°C for 10 min. The preformed nucleoprotein complex was then challenged with the indicated concentrations of unlabeled DNA competitor (added in 1 μl) for 5 min at 37°C. The reaction mixtures were analyzed as above.
Affinity pulldown
The indicated GST-tagged, MBP-tagged, or untagged protein species were incubated in 30 μl of buffer (25 mM Tris-HCl, pH 7.5, 0.01% Igepal, 1 mM 2-mercaptoethanol) containing 100 mM KCl for 30 min at 4°C and then mixed with 7 μl glutathione resin (GE Healthcare) or amylose resin (New England Biolabs) for 30 min at 4°C to capture the tagged protein and its partner. The resin was washed three times with 30 μl of buffer containing 70 mM KCl and then treated with 20 μl of 2% SDS to elute proteins. The supernatant (S), last wash (W), and SDS eluate (E) were analyzed by SDS-PAGE.
D-loop reaction
The 32P-labeled 90-mer oligonucleotide substrate (2.4 μM nucleotides) was incubated for 5 min at 37°C with RAD51 (0.8 μM) in 10.5 μl buffer (35 mM Tris-HCl, pH 7.5, 1 mM DTT, 50 mM KCl, 2 mM MgCl2, 2 mM ATP), followed by the addition of PALB2 (200 or 300 nM), RAD51AP1 (50 nM), or the combination of the two proteins in 1 μl and a 5-min incubation at 37°C. pBlueScript replicative form I DNA (35 μM base pairs) was then incorporated in 1 μl, and the reactions were incubated for 12 min at 37°C and analyzed.
Duplex DNA capture
Streptavidin magnetic beads (4 μl; Roche) containing 5′-biotinylated 83-mer oligo dT (9 μM nucleotides) were incubated with RAD51 (3 μM) in 20 μl buffer (35 mM Tris-HCl, pH 7.5, 1 mM DTT, 100 μg.ml-1 BSA, 50 mM KCl, 2 mM MgCl2, 2 mM ATP) for 5 min at 37°C. The beads were captured with a magnet, washed with 20 μl buffer, and then resuspended in 19 μl buffer. Following the incorporation of PALB2 (200 or 400 nM), RAD51AP1 (300 nM), or the combination of the two proteins in 1 μl and a 5-min incubation at 37°C, the beads were again captured, washed with 20 μl buffer, and resuspended in 19 μl buffer. The reaction was completed by adding radiolabeled 80-mer dsDNA (4 μM base pairs) in 1 μl. After a 10-min incubation at 37°C, the beads were captured, and the supernatant was set aside. After washing the beads twice with 20 μl buffer, 20 μl 2% SDS was used to elute bound proteins and radiolabeled DNA, followed by analysis of the supernatant and eluate.
Synaptic assay
RAD51 (4 μM) was incubated with a 60-mer oligonucleotide (12 μM nucleotides) homologous to the region of pUC19 DNA spanning positions 2471 to 2531 (that covers the SspI restriction site and serves as the homologous DNA substrate) or positions 776 to 836 (that covers the AflIII restriction site and serves as the heterologous control) in 8 μl buffer (35 mM Tris-HCl, pH 7.5, 1 mM DTT, 2 mM MgCl2, 50 mM KCl, 2 mM ATP) for 5 min at 37°C to assemble the presynaptic filament, which was further incubated for 5 min with PALB2 (200 or 400 nM), RAD51AP1 (400 nM), or both proteins added in 1 μl of buffer. Following the addition of linear pUC19 DNA (83 μM nucleotides) in 1 μl and another 5-min incubation at 37°C, the reaction was treated with 2.5 units of SspI for 10 min at 37°C and then analyzed.
Immunofluorescence
EUFA1341 (PALB2-/-) fibroblasts and their derivatives (+ vector, + PALB2), a kind gift from Dr. Bing Xia, were maintained as described24. U2OS and HeLa cells were grown in Dulbecco's Modified Eagle's Medium and Minimum Essential Medium, respectively, with 10% fetal bovine serum. Cells were exposed to 160 kVp X-rays filtered through 0.5 mm copper at a dose rate of ∼1 Gy.min-1 or to 6 μM MMC for 1 h in regular growth medium at 37°C. The treated cells were kept in the regular growth medium at 37°C for 16 h before fixation and permeabilization as described12. For detection of RAD51AP1 and PALB2 foci, cells were incubated with mouse polyclonal α-RAD51AP1 antibody (SAB1400398; Sigma; 1:2000) and affinity-purified rabbit polyclonal α-PALB2F4 antibody5 (1:600) in 2% bovine serum albumin (BSA) in Dulbecco's Phosphate Buffered Saline (DPBS; 137.9 mM NaCl, 8.1 mM Na2HPO4, 2.7 mM KCl, 1.5 mM KH2PO4, pH 7.5) at 4°C overnight. After 5 washes with DPBS, the cells were incubated for 1 h with Alexa Fluor 594 goat anti-mouse and Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen; 4 μg.ml-1) in 2% BSA/DPBS, and DNA was stained by DAPI (50 ng.ml-1). The slides were viewed at 2,000 × magnification with a Zeiss Axioskop microscope.
Immunoblotting
Western blot analysis was conducted as described24. The following primary antibodies were used: α-PALB2 (A301-246A, Bethyl Lab., 1:3,000), α-RAD51 (PC-130; Calbiochem; 1:3,000), α-pan-H3 (ab1791; Abcam; 1:5,000), and α-QM (C-17; Santa Cruz Biotechnology; 1:3,000). Rabbit polyclonal α-RAD51AP1 antibody was raised against a RAD51AP1 fragment encompassing residues 21-166. HRP-conjugated goat α-rabbit IgG (Jackson ImmunoResearch Laboratories; 1:10,000) was used as secondary antibody.
siRNA and transfections
The lead siRNA sequences used were r(GCAGUGUAGCCAGUGAUUA)d(TT) for RAD51AP115 and r(UUCUCCGAACGUGUCACGU)d(TT) for the negative control siRNA (Qiagen). Transfections were carried out using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions.
Preparation of all figures
Figures were prepared by recording protein gels in a BioRad gel documentation station, scanning and quantifying radioactive gels with a Biorad phospho imager and all figures were assembled in Adobe software (Photoshop or Illustrator).
Supplementary Material
Acknowledgments
We are grateful to Rémi Buisson and Jean-Yves Masson (CRCHUQ, Quebec) for the communication of results prior to publication, to Sead Begovic, Simonne Longerich and Yong-Chul Kim (Yale University) for assistance, to Youngho Kwon (Yale University) for providing ScRad51 protein, and to Bing Xia (UMDMJ, New Jersey) for providing PALB2-deficient and PALB2-complemented cells, as well as for generously providing α-PALB2 antibody. This study was supported by research and program project grants RO1CA120315, RO1ES07061, RO1ES015252, RO1ES015632, PO1CA129186 and PO1CA92584 from the US National Institutes of Health, and by postdoctoral fellowship PDF0706844 from the Susan G. Komen for the Cure Foundation.
Footnotes
Author Contributions: E.D., J.E., and C.W., conceived and carried out experiments; E.D. and C.W also wrote the paper. D.Sa., G.J.W, M.H., X.Y, V.E.G, C.W., D.L., M.S.T., and S.M-H.S. contributed key material. D.Sc., E.E, J.C. and P.S. contributed expertise. P.S. provided guidance throughout, wrote and edited the paper.
Note: Supplementary information is available online.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–57. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 2.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–72. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 3.San Filippo J, et al. Recombination mediator and Rad51 targeting activities of a human BRCA2 polypeptide. J Biol Chem. 2006;281:11649–57. doi: 10.1074/jbc.M601249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tutt A, et al. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr Biol. 1999;9:1107–10. doi: 10.1016/s0960-9822(99)80479-5. [DOI] [PubMed] [Google Scholar]
- 5.Xia B, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–29. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 6.Yu VP, et al. Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 2000;14:1400–6. [PMC free article] [PubMed] [Google Scholar]
- 7.Erkko H, et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446:316–9. doi: 10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- 8.Foulkes WD, et al. Identification of a novel truncating PALB2 mutation and analysis of its contribution to early-onset breast cancer in French-Canadian women. Breast Cancer Res. 2007;9:R83. doi: 10.1186/bcr1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahman N, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–7. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reid S, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–4. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 11.Hirsch B, et al. Association of biallelic BRCA2/FANCD1 mutations with spontaneous chromosomal instability and solid tumors of childhood. Blood. 2004;103:2554–9. doi: 10.1182/blood-2003-06-1970. [DOI] [PubMed] [Google Scholar]
- 12.Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang F, et al. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19:524–9. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Modesti M, et al. RAD51AP1 is a structure-specific DNA binding protein that stimulates joint molecule formation during RAD51-mediated homologous recombination. Mol Cell. 2007;28:468–81. doi: 10.1016/j.molcel.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 15.Wiese C, et al. Promotion of homologous recombination and genomic stability by RAD51AP1 via RAD51 recombinase enhancement. Mol Cell. 2007;28:482–90. doi: 10.1016/j.molcel.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 recombinase and recombination mediators. J Biol Chem. 2003;278:42729–32. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 17.Gupta RC, Folta-Stogniew E, O'Malley S, Takahashi M, Radding CM. Rapid exchange of A:T base pairs is essential for recognition of DNA homology by human Rad51 recombination protein. Mol Cell. 1999;4:705–14. doi: 10.1016/s1097-2765(00)80381-0. [DOI] [PubMed] [Google Scholar]
- 18.Chi P, San Filippo J, Sehorn MG, Petukhova GV, Sung P. Bipartite stimulatory action of the Hop2-Mnd1 complex on the Rad51 recombinase. Genes Dev. 2007;21:1747–57. doi: 10.1101/gad.1563007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pezza RJ, Voloshin ON, Vanevski F, Camerini-Otero RD. Hop2/Mnd1 acts on two critical steps in Dmc1-promoted homologous pairing. Genes Dev. 2007;21:1758–66. doi: 10.1101/gad.1562907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–7. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 21.Yuan SS, et al. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–51. [PubMed] [Google Scholar]
- 22.Walsh T, King MC. Ten genes for inherited breast cancer. Cancer Cell. 2007;11:103–5. doi: 10.1016/j.ccr.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 23.Xia B, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–61. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 24.Wiese C, et al. Interactions involving the Rad51 paralogs Rad51C and XRCC3 in human cells. Nucleic Acids Res. 2002;30:1001–8. doi: 10.1093/nar/30.4.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.