Abstract
Mammalian sperm are transcriptionally and translationally inactive. To meet changing needs in the epididymis and female tract, they rely heavily on post-translational modifications and protein acquisition/degradation. Membrane rafts are sterol and sphingolipid-enriched micro-domains that organize and regulate various pathways. Rafts have significance in sperm by transducing the stimulus of sterol efflux into changes in intracellular signaling that confer fertilization competence. We recently characterized 3 biochemically distinct sub-types of sperm rafts, and now present profiles for proteins targeting to and associating with these sub-types, along with a fraction largely comprised of “non-raft” domains. Proteomics analysis using a gel-based LC-MS/MS approach identified 190 strictly validated proteins in the raft sub-types. Interestingly, many of these are known to be expressed in the epididymis, where sperm membrane composition matures. To investigate potential roles for rafts in epididymal protein acquisition, we compared the expression and localization of 2 different sterol-interacting proteins, apolipoprotein-A1 and prominin-1 in sperm from different zones. We found that apolipoprotein-A1 was gradually added to the plasma membrane overlying the acrosome, whereas prominin-1 was not, suggesting different mechanisms for raft protein acquisition. Our results define raft-associating proteins, demonstrate functional similarities and differences among raft sub-types, and provide insights into raft-mediated epididymal protein acquisition.
Keywords: Membranes, membrane rafts, proteins, proteomics, sperm
1 Introduction
After leaving the testis, mammalian sperm must undergo two distinct maturational processes in order to become fertilization competent. In most mammals, the first step occurs when sperm pass through the caput, corpus, and cauda regions of the epididymis. During epididymal transit, complex changes take place in the lipid and protein compositions of the sperm membranes [1, 2]. After storage in the cauda and then ejaculation, sperm are still unable to fertilize an oocyte until they mature in the female tract in response to external stimuli in the process of “capacitation” [3]. This second, functional maturation is associated with multiple physiological events taking place at the level of the plasma membrane, including a requirement for sterol efflux.
The central role of lipids in the regulation of capacitation suggests functional connections between individual lipid species and/or regions of membrane with intracellular signaling events. Membrane rafts are a specific type of membrane micro-domain enriched in sterols and sphingolipids relative to phospholipids, and are involved in diverse cellular functions such as scaffolding intracellular signaling pathways [4, 5]. Because of their importance, several approaches have been taken to isolate rafts and define their proteomes. The newest method, stable isotope labeling by amino acids in cell culture (SILAC; [6]), is not relevant for studies of sperm because no culture system exists that practically supports spermatogenesis; therefore, protein translation with controlled labeling can’t be performed. The most commonly used approach has been fractionation to partition detergent-resistant membranes (DRM), but it is accepted that this strategy is insufficient for this purpose [7–9]. Because true rafts are only a subset of what might partition with DRM (as they can artifactually cause disparate molecules to coalesce), and because resistance to solubilization is detergent-dependent and doesn’t correspond with known physiological entities, identification of proteins that partition to DRM will at best yield candidates for targeting to or associating with, true rafts [7]. Despite this limitation, DRM from sperm have shown interesting capabilities, such as possessing components that bind to the zona pellucida [10, 11], making the further characterization of rafts of great interest.
Non-detergent-based isolation of membrane rafts is designed to partition these domains in a manner that better mirrors pre-existing rafts [5]. However, this approach has its own possible drawbacks, including the potential for cytoplasmic proteins that peripherally or indirectly associate with rafts to partition with these domains. Motivated by our desire to understand the mechanism of sterol efflux and its intracellular sequelae, we set out to define membrane rafts and raft-associating proteins in sperm using this non-detergent approach. This methodology could identify both resident raft proteins and intracellular interactors, potentially shedding light on how sterol efflux might be transduced into changes in cell function.
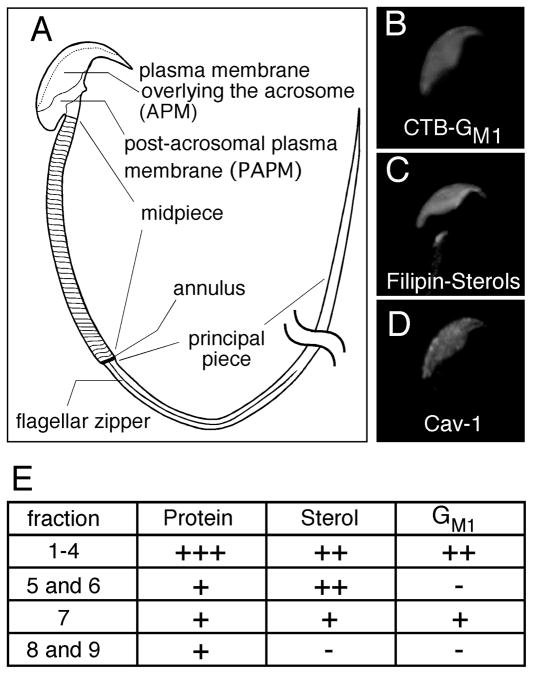
By utilizing a linear density gradient separation of membrane vesicles, we were recently able to demonstrate that murine sperm possess at least 3 distinct sub-types of raft, each with predictable molar ratios of sterols and ganglioside GM1 (GM1) to phospholipids, and mass amounts of protein relative to total lipid [9]. Existence of raft sub-types is supported by evidence of both large scale lipid segregation as well as micro-heterogeneities in the distribution of sterols and GM1 in the plasma membrane overlying the acrosome (APM) [12]. In terms of macro-domains, GM1 is highly enriched in the APM and, but not in the post-acrosomal plasma membrane (PAPM) ([9, 12, 13]: Fig. 1). Caveolin-1 and filipin-sterol complexes are restricted to the APM in a similar fashion, although focal enrichments of sterols are distributed unevenly in the APM (Fig. 1 and [12, 14–16]). We have also shown that the membrane enclosing the acrosome is highly enriched in GM1 but does not have the same focal enrichments of sterols as the APM [9, 12, 16]. Regional and organelle-specific differences in membrane composition/appearance have also been noted at the annulus and flagellar zipper, as well as within the mitochondrial membranes that face other membranes versus cytoskeletal elements [17–21]. Together, these biochemical and cell biological data suggest that the membranes of sperm are not uniform, but rather are dynamic and possess diverse micro-domains [9, 12, 22].
Fig. 1.
Membrane organization and biochemical characteristics of raft sub-types in murine sperm. A schematic diagram of murine sperm shows the high degree of cell polarization and compartmentalization of the plasma membrane (A). The plasma membrane overlying the acrosome (APM) is highly enriched in GM1, detected by FITC-CTB (B). Sterols and caveolin-1 are also segregated into the same area (C and D, [12, 14]). Biochemical analysis of membrane raft fractions demonstrated the presence of 3 different raft sub-types (fx 1-4, 5 and 6, and 7) characterized by reproducibly distinct lipid and protein compositions [summarized in panel E; “+” signs denote relative molar ratios (sterols or GM1 versus phospholipid) or mass ratios (total protein:total lipid) in the sub-types [9]].
Using fractions obtained from our non-detergent methodology, we previously performed an iTRAQ-based proteomic comparison, combined with in-solution tryptic digestion, in order to quantify differences in protein composition among the 3 raft sub-types. For a true ratiometric comparison, we were limited to the 11 proteins found in each of the 3 raft sub-types as well as in a fraction largely comprised of non-raft domains [9]. However, during those experiments, we noted many examples of proteins that were enriched in one or two specific membrane raft sub-types. These proteins had to be excluded from ratiometric comparisons because lack of quantitative data from a sample precludes a relative abundance analysis of that protein. As mentioned earlier, other approaches at quantitative proteomics, such as SILAC, cannot be used with sperm because they do not make new proteins and there is no culture system that supports full spermatogenesis. Because the understanding of sperm membrane proteomes and sterol efflux is so limited, we have now pursued a complementary, qualitative characterization of the proteomes of the distinct raft sub-types. This approach describes all the proteins associated with each membrane sub-type, precisely because proteins that associate with a specific subtype might provide it with a unique function, which in turn might shed light on the cell biological need for the sperm’s complex membrane organization. For this, we used 1-D SDS-PAGE to enhance solubilization and separation of membrane proteins. Using strict criteria, we identified 190 proteins in the different raft fractions. Our results indicate that there are significant functional distinctions among the different raft sub-types, and also suggest important roles for membrane rafts in the processes of epididymal maturation and sterol efflux.
2 Materials and methods
2.1 Reagents and animals
All reagents were purchased from Sigma (St. Louis, MO), unless otherwise noted. Antisera to apolipoprotein-A1 (apoA1) and prominin-1 (prom1) were purchased from Abcam Inc. (Cambridge, MA) and eBioscience, Inc. (San Diego, CA), respectively. Caveolin-1 antiserum was from BD Biosciences (San Diego, CA). A monoclonal antibody against murine erythroid cells was from eBioscience (TER-119, San Diego, CA). All animal work was performed with the approval of Cornell University’s Institutional Animal Care and Use Committee, in accordance with the NIH guidelines for the Care and Use of Laboratory Animals.
2.2 Preparation of membrane raft sub-types
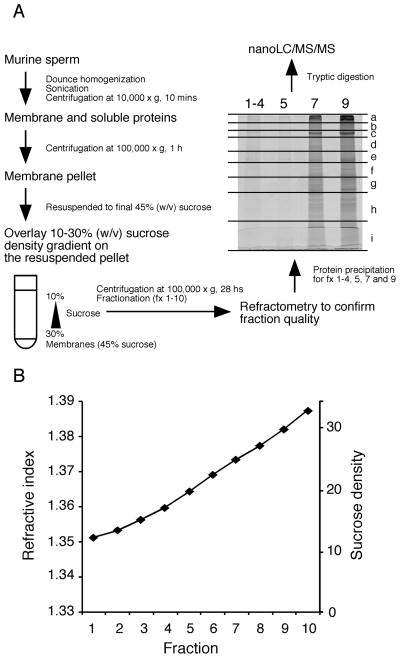
Preparation of cauda epididymal sperm and membrane raft fractions were performed as described previously [9]. Briefly, sperm were washed by sequential differential centrifugation steps to remove contaminants such as epithelial cells and blood cells. The purity of the final sperm pellet was confirmed by both visual examination using a phase-contrast microscope and by specific testing for the lack of immunoreactivity for TER-119, an erythrocyte-specific membrane protein (data not shown; whole blood proteins were used as a positive control). Membranes were isolated as vesicles from sperm (1.2 × 109 cells) using homogenization, sonication, and centrifugation according to standard methods (Fig. 2A). The resultant membrane pellet was mixed with a sucrose solution [45% (v/v) final], and overlaid with a 10–30 % continuous sucrose density gradient. Centrifugation for 28 hours allowed membrane vesicles to partition by buoyancy, and fractions were analyzed by refractometry to ensure their nature prior to protein extraction and proteomic profiling. The most buoyant fractions (1–4) were pooled to match our previous biochemical analyses and to provide a sufficient amount of protein for profiling.
Fig. 2.
Non-detergent based separation of membrane rafts and sample preparation for mass spectrometry. Sperm membranes were isolated by dounce homogenization and sonication and then spun down at 10,000 × g for 10 minutes. The supernatant was then centrifuged at 100,000 × g to yield a membrane pellet. This was mixed with 80% (w/v) sucrose to obtain a final sucrose concentration of 45%. The lysate was overlaid with a 10–30% linear sucrose density gradient and centrifuged at 100,000 × g for 28 hours. The supernatant was divided into 10 fractions from the top. Reproducibility of the raft separation was confirmed with refractometry (B) [9]. 1-D SDS-PAGE was utilized to separate proteins and the gel excised into 9 gel pieces (a–i) for tryptic digestion followed by mass spectrometry.
2.3 Protein extraction and in-gel tryptic digestion/extraction
The proteins for fractions 1–4, 5, 7 and 9 were extracted in 10% (v/v) trichloroacetic acid at 4°C for 2 hours and centrifuged at 100,000 × g for 3 hours. The precipitated proteins were separated by 10% SDS-PAGE, and then visualized with SYPRO Ruby (Fig. 2A) (Invitrogen, Carlsbad, CA). Each lane of the gel was excised in parallel into 9 discrete pieces, and subjected to in-gel digestion and tryptic peptide extraction following a protocol modified slightly from Shevchenko et al. [23]. The gel pieces were destained, reduced with DTT and alkylated by treatment with iodoacetamide. Samples were treated overnight with 0.2 μg trypsin, and the resultant tryptic-digested peptides were removed by centrifugation for 2 min at 4,000 ×g. The remaining peptides in the gel were then extracted by sonication in 50 μL of 5% formic acid in 50% ACN and collected similarly. All gel-extracted supernatants were combined and evaporated to dryness in a Speedvac SC110 (Thermo Savant, Milford, MA).
2.4 Protein identification by nanoLC/MS/MS analyses
The digested samples were reconstituted in 15 μL of 2% ACN with 0.5% formic acid and injected using a Famous auto sampler onto a C18 column (5 μm, 300 μm × 5 mm, Dionex, Sunnyvale, CA) for on-line desalting. They were then separated on a C-18 RP nano column (3 μm, 75 μm × 15 cm, Dionex) connected in-line to a hybrid triple quadrupole linear ion trap mass spectrometer, 4000 Q Trap, equipped with a Micro Ion Spray Head II ion source (Applied Biosystems/MDS SCIEX, Framingham, MA).
MS data acquisition was performed using Analyst 1.4.1 software (Applied Biosystems) for information dependent acquisition (IDA) analysis. The nanospray voltage was 2.0 kV, and was used in positive ion mode for all experiments. The declustering potential was set at 50 eV and nitrogen was used as the collision gas. In IDA analysis, after each survey scan for m/z 400 to m/z 1550 and an enhanced resolution scan, the three highest intensity ions with multiple charge states were selected for tandem MS (MS/MS) with rolling collision energy applied for detected ions based on different charge states and m/z values.
2.5 MS data analysis
The MS/MS data were submitted to Mascot 2.2 (Matrix Science, London, UK) for NCBInr database searching (downloaded in July 2007), with one missed cleavage site by trypsin allowed and with decoy database searching on. The following variable modifications were accepted: methionine oxidation, carbamidomethyl cysteine, peptide mass assignment ± 1.5 Da, fragment mass assignment ± 0.6 Da. All detected peptides were also strictly validated by two criteria: Mascot expectation value < 0.05 and Mascot ion score > 35. For protein validation, peptides corresponding to keratin contaminants were manually excluded and then the proteins with 2 or more unique peptides were classified as “identified proteins”. The decoy database search in Mascot search engine allows us to estimate false discovery rate (FDR) for detected tryptic peptides, which yielded about 1% for each of the fractions. After the additional filters described above were applied, the peptide FDR decreased significantly down to 0.1–0.2%. In each case, all proteins identified in the FDR analysis (in decoy database search) were inferred by a single peptide hit. Therefore, using these strict filter criteria with 2 or more unique peptides, the identifications of the proteins reported here are highly confident.
2.6 Localization of lipids and proteins in sperm
Mouse caput, corpus and cauda epididymides were incised with scissors in separate volumes of a modified Whitten’s medium [24] to free the sperm from those regions. The sperm suspensions were centrifuged at 100 × g at 37°C for 1 minute to pellet epithelial cells. Sperm were fixed and permeabilized as described previously [9].
For localization of proteins, the sperm were blocked with 10% rabbit serum overnight at 4°C. Samples were incubated overnight at 4°C with the primary antibodies, washed, and then incubated with appropriate fluorophore-conjugated secondary antisera for 2 hours at 37°C. Localization and observation of sterols and caveolin-1 in fixed sperm and GM1 in live sperm were performed using filipin, anti-caveolin-1, and FITC-cholera toxin B (CTB) as described previously [13, 14]. Consistency in results was confirmed from 3 replicate trials.
2.7 Immunoblot
Caput, corpus and cauda epididymal sperm were collected and washed to remove gross cellular debris as described above. Sperm suspensions were layered on 20 and 30% discontinuous percoll density gradients and centrifuged at 1,000 × g for 10 minutes. After separation from cell contaminants, the protein content for each sperm suspension was measured with a bicinchoninic acid protein assay according to the manufacturer’s instructions (Micro BCA Protein Assay Kit, Thermo Scientific/Pierce, Rockford, IL). Equivalent amounts of sperm protein (0.4 mg) from the three regions were used for SDS-PAGE. Transfer and immunodetection of the specific proteins were performed essentially as described previously [25], except that 5% BSA in TBS containing 0.1 % tween 20 (TTBS) was used as a blocking solution. The dilution used for anti-apoA1 or anti-prom1 was 1:2,000 in the blocking solution. Secondary antiserum was also used at 1:2000 in TTBS. Chemiluminescence (Super Signal West Pico Chemiluminescent kit, Thermo Scientific/Pierce) was used for visualization.
3 Results
3.1 Preparation of membrane rafts without detergent
The procedures we employed for separation of membrane rafts and extraction of their proteins are schematized in Fig. 2A. Previously, we showed that this non-detergent based method separates membrane raft sub-types with high purity and reproducibility by means of their buoyancy [9]. To confirm the quality and identity of fractions obtained from the linear density gradient, we analyzed them using refractometry to ensure that the collected fractions represented the same sucrose densities as those we had described (Fig. 2B; note that we performed the fractionation of the sperm membranes 10 times, with very high reproducibility in terms of the biochemical characterization and the refractive indices between replicates [9]). Our previous biochemical characterization of lipids and mass amounts of protein contained within these membrane fractions revealed that murine sperm have at least 3 distinct raft sub-types (Fig. 1). Fx1-4 had high molar ratios of sterols:PL and GM1:PL, and high mass amount of total protein to total lipid (estimated by PL plus sterols). Fx5 and fx6 had a high molar ratio of sterols:PL but had a lower molar ratio of GM1:PL. Fx7 had a high molar ratio of GM1:PL but had a molar ratio of sterols:PL that was close to 1:1, putting it near the minimal limit of what is customarily defined as a raft. Fx 5, 6, 7, 8, and 9 all had relatively similar estimated ratios of total protein:total lipid [9]. Based on these findings, we analyzed the protein profiles of fx1-4, fx5, and fx7 to cover the three raft sub-types, and fx9, which is largely comprised of non-raft membranes.
3.2 Mass spectrometric analysis of protein composition of the different raft sub-types
Because the fractions would be predicted to have complex profiles with a high relative abundance of hydrophobic proteins, we utilized 1-D SDS-PAGE and divided each lane into 9 sections to facilitate the detection of relatively lower abundance proteins in the wide dynamic range of complex samples (Fig. 2A). Resultant total ion spectra, unique peptides and identified proteins are summarized in Table 1. Our strict validation allowed us to identify 14, 12, 188 and 247 proteins in fx1-4, 5, 7, and 9 (supplemental data 1 and 2).
Table 1.
Summary of proteomic analysis of different types of membrane fractions1.
| Fraction | 1–4 | 5 | 7 | 9 |
|---|---|---|---|---|
| Ion spectra | 284 | 1765 | 4500 | 6075 |
| Unique peptides (2 exp < 0.05) | 51 | 57 | 837 | 1247 |
| Identified proteins (unique pep ≥ 2) | 14 | 12 | 188 | 247 |
Membrane fractions were prepared by a non-detergent based method as described in the Materials and Methods. After confirming the quality of the partitioning by comparing the refractive index of each fraction with our previous sucrose density data [9], proteins for fractions 1-4, 5, 7 and 9 were extracted by TCA precipitation and were separated by SDS-page. Each lane was cut into 9 pieces and utilized for GeLC-MS/MS analysis after “in-gel digestion” using trypsin.
Mascot expectation value.
3.3 Characterization of protein profiles
A functional characterization was performed based on the known biological processes and molecular functions with which identified proteins are associated using a PANTHER analysis (supplemental data 3) [http://www.pantherdb.org/] ([26, 27]). For this analysis, GI accession numbers provided from the Mascot database were converted into each corresponding gene ID manually. Fx 1-4 and fx 5 shared 2 major biological processes: cell structure and motility and intracellular protein traffic. Proteins involved in protein metabolism and modification were found in all fractions, with particular enrichment in fx 5, 7, and 9. The distribution of proteins across the major biological processes was very similar between fx 7 and 9. Proteins involved in unclassified biological processes, transport, and protein metabolism and modification were most prevalent in these fractions as well.
When looking at proteins individually by their molecular functions, some sharper differences emerged among raft sub-types. For example, proteases and cytoskeletal proteins were greatly enriched in fx 1-4 and fx 5, versus fx 7 or fx 9. Hydrolases were found to comprise large percentages of all 3 raft sub-types, but represented only a minimal percentage of the proteins in non-raft fx 9. Transporters and proteins with unclassified molecular functions were greatly enriched in fx 7 and fx 9. Oxidoreductases were found to comprise a relatively high percentage of proteins in all fractions, underscoring their important roles in sperm metabolism and protection against oxidative stress.
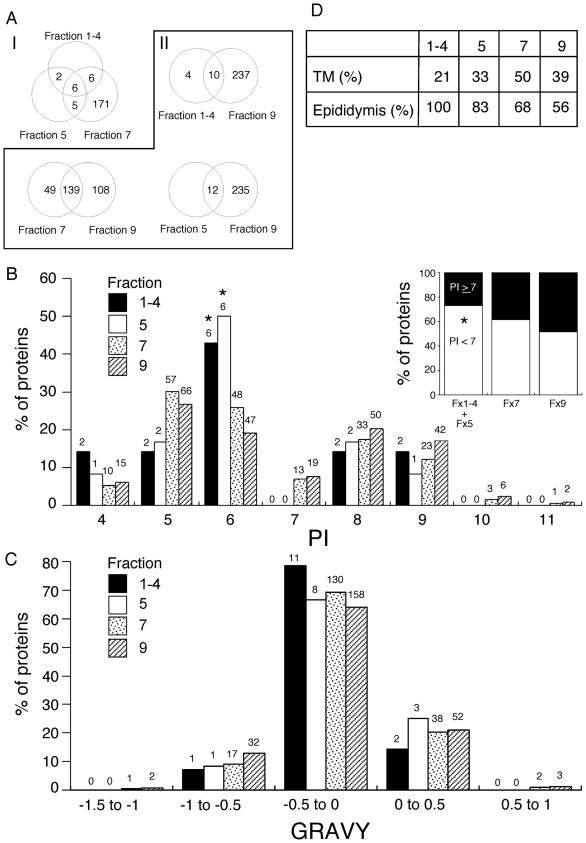
Similarity analysis of the protein profiles among raft sub-types showed that all proteins identified in fx 1-4 and fx 5 were shared between two or more sub-types (Fig. 3A–I). Although based on small numbers of proteins identified from the most buoyant (and lowest abundance) raft sub-types, our data are consistent with brief temporal associations of these proteins with specific raft sub-domains, in line with the dynamic definition of membrane rafts [28]. This was also supported by comparison with the non-raft fraction (fx 9), which showed that all proteins in fx 5 were also identified in fx 9. Finding raft resident or raft-associated proteins in non-raft fractions is not surprising because of the dynamic nature of rafts. Also, it should be stressed that our methodology was designed to isolate membrane raft subtypes specifically and with high purity, but it was not designed to isolate non-raft fractions with purity, nor to distinguish the potential diversity of non-raft sub-types that has been suggested [29]. Therefore, fx 9 likely possesses contaminating raft micro-domains. Despite these caveats, 4 and 49 proteins of fx 1-4 and fx 7 were not present in the profile of fx 9 (Fig. 3A–II), demonstrating some degree of selectivity of protein targeting and/or association.
Fig. 3.
Characterization of protein profiles among raft fractions. A Venn diagram showing overlap of identified proteins among three raft fractions (A–I), and raft versus non-raft fractions (A–II). Similarities of protein profiles were characterized manually among raft fractions (fx 1-4, 5 and 7) and between each raft sub-type (fx 1-4, 5 and 7) and non-raft fractions (fx 9). The distributions of isoelectric points (pI) and GRAVY (http://www.expasy.ch/tools/protparam.html) in raft sub-types and non-rafts are portrayed by use of a relative frequency histogram (B) and (C). The numbers of proteins categorized in the ranges are indicated above the bars. The proportions of negatively or positively charged proteins (pI < 7 or ≥ 7) at physiological pH were compared (B inset). Statistical analysis among groups was performed with chi-square test for independence or Fisher’s exact probability test when sample sizes smaller than 5 were analyzed. The asterisks denote significant difference for the raft sub-types when compared with fx 9 in the respective compartments (P < 0.05). Percentages of proteins with at least one transmembrane domain (TM) and protein expression in murine epididymides (D).
Because electric charge and hydrophobicity are important determinants of lipid-protein and protein-protein interactions, analyses of the biochemical characteristics of identified proteins were performed using a relative frequency histogram. As expected, the isoelectric points (pI) tended away from physiological pH in all fractions (Fig. 3B). Bimodal pI distribution flanking the physiological pH was observed throughout fractions, similar to other analyses of proteomes from a variety of cell types [30, 31]. Although potentially influenced by the relatively small number of proteins identified in fx 1-4 and fx 5, the relative frequencies of proteins with pI = 6 in fx 1-4 and fx 5 were significantly higher than that of fx 9 (Fig. 3B, P < 0.05). Because of the high similarity of pI frequencies in fx 1-4 and fx 5, the data of these fractions were combined to analyze the relative frequency of negatively charged proteins (i.e. those with lower pI). Interestingly, more proteins in the raft fractions were negatively charged than proteins in the non-raft fraction (fx 1-4 + fx 5 vs. fx 9, P < 0.05; fx 7 vs. fx 9, P=0.05) at physiological pH (Fig. 3B inset). Bias in pI distribution has been reported to differ between organisms and with sub-cellular protein localization, although the exact reasons are often unknown [31–33]. The impact of specific membrane micro-domains being associated with proteins with a more acidic pI might have relevance for sperm biology and will be discussed further below.
Grand average of hydropathicity (GRAVY) was compared and no statistical differences in the relative frequencies between the sub-types (Fig. 3C), or rafts and non-rafts (data not shown) were found. Analysis for transmembrane domains revealed that 21, 33, 50 and 39% of identified proteins in fx 1-4, 5, 7, and 9 respectively, were integral membrane proteins (Fig. 3D), suggesting that many of the proteins were membrane-associated as opposed to membrane-resident proteins.
Sperm are known to acquire GPI-anchored proteins, as well as integral and peripheral membrane proteins during epididymal transit [34–39]. Therefore, we analyzed identified proteins for potential expression in the epididymis using a database of transcriptome analysis (http://mrg.genetics.washington.edu). Though not demonstrative of origin, this analysis indicated that proteins identified from the raft sub-types were relatively more enriched in epididymal proteins than non-rafts (Fig. 3D)[100%, 83.3%, 67.6% vs. 55.8%]. These results suggested the possibility that protein acquisition during epididymal passage is associated with targeted transfer into specific membrane domains.
3.4 Expression of raft proteins in epididymis
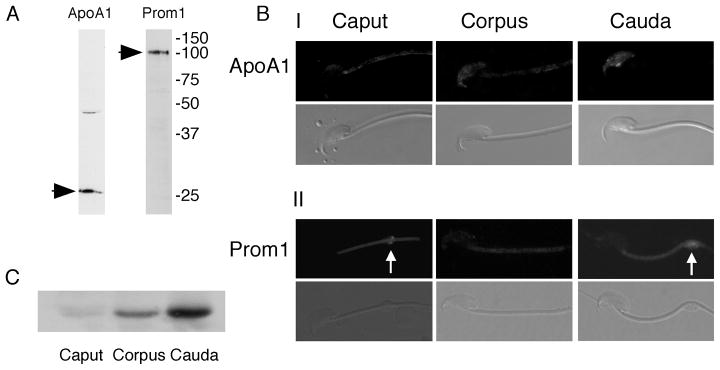
To begin investigation of this possibility, we compared the expression and potential incorporation of two of these proteins, prom1 and apoA1, which both bind membrane sterols. Immunoblotting confirmed apoA1 and prom1 were present in cauda epididymal sperm (28 kDa and 100 kDa, respectively; Fig. 4A). Interestingly, our preliminary immunohistochemistry experiments on testis sections and our searches of EST databases (http://www.ncbi.nlm.nih.gov/Unigene/) and high-throughput gene expression (http://symatlas.gnf.org/) suggested that apoA1 was not expressed in the murine testis (data not shown), consistent with a previous report [40]. This was confirmed by immunoblotting of mixed germ cell proteins, which were also negative (data not shown). Together, our data, the expression databases, and other reports suggested that sperm might be acquiring apoA1 during epididymal maturation. Therefore, we examined the localization of apoA1 and prom1 in sperm from the caput, corpus and cauda epididymis. In caput epididymal sperm, minor labeling of apoA1 was observed in the caudal APM and the midpiece (Fig. 4B–I). The signal in the APM increased in intensity as sperm progressed to the cauda. Immunoblotting of apoA1 confirmed increasing amounts of epididymal apoA1 binding to sperm during epididymal transit (Fig. 4C). Conversely, prom1 localization was faintly observed in the midpiece and cytoplasmic droplet (when present) in all regions of the epididymis, and there was no difference in the amount of prom1 expression (Fig. 5B–II and data not shown). These results are consistent with a previous report [41]. Our results demonstrate that apoA1 was transferred to raft micro-domains of the APM in murine sperm during epididymal transit.
Fig. 4.
Expression and localization of apoA1 and prom1 in murine sperm. Immunoblots of proteins from cauda epididymal sperm confirmed that apoA1 and prom1 were expressed at the appropriate molecular weights (A). ApoA1 was transferred to the sperm plasma membrane during epididymal transit (B–I), consistent with the results of immunoblots for this protein (C). However, localization of prom1 was conserved between different epididymal regions, showing the midpiece and the cytoplasmic droplet (arrows) (B–II).
4 Discussion
4.1 Non-detergent based preparation of membrane rafts
Mass spectrometry-based proteomic analysis has been used to reveal the protein composition of membrane rafts in several cell types. We recently reported using a non-detergent approach that murine sperm possess at least 3 different raft sub-types, each reproducibly possessing a characteristic lipid and protein composition (Fig. 1E) [9]. This approach offers several advantages, including: 1). allowing potential identification of cytoplasmic interactors with raft resident proteins, 2). avoiding the artifactual coalescence of disparate membrane components that is caused by detergents, and 3). avoiding the potential interference with the chromatographic separation of peptides and/or detection of ion spectra that can be caused by detergents [42]. Perhaps most importantly, partitioning based on detergent insolubility promotes the binary division of membranes into “raft” versus “non-raft,” masking the presence of the multiple membrane domain sub-types that occur in nature [9]. In part because of these methodological differences, our combined total of 190 raft-associated proteins that were identified using strict criteria is significantly larger and has important differences versus previous DRM-based studies [11, 43].
Although methodological differences between the present study and our previous quantitative proteomics make comparison imprecise, our current profiles closely matched our previously identified proteins, with the single exception being that we did not currently identify epidermal growth factor receptor precursor (previously found to be relatively enriched in fx 9 [9]). However, other proteins that we had shown to be enriched in that non-raft fraction (e.g. facilitated glucose transporter 3, basigin, hexokinase type 1, and the sodium potassium transporting ATPase alpha 4 and beta 3 subunits) were all currently identified in fx 9. In addition, although we note that the current method is non-quantitative, these proteins tended to be identified on the basis of more unique peptides in the non-raft fraction than were identified in the less buoyant fractions. The concordance of our findings with the previous iTRAQ both supports the highly reproducible fractions produced by our non-detergent method, and the rigor of the mass spectrometric approaches.
4.2 Functional differences between raft sub-types
Previously, our biochemical characterization of the lipid molar ratios of raft fractions and our cell biological data showing enrichment of GM1 in acrosomal membranes, together suggested that membranes of the acrosome constitute a large portion of the membranes found in fx 7 [9]. A major finding of our present study is that fx 7 contains a variety of calcium channels and SNARE (soluble N-ethylmaleimide sensitive factor attachment protein receptors) proteins. Because an increase in intracellular Ca2+ concentration is critical in several signaling pathways leading to the induction of acrosomal exocytosis [44], and SNARE proteins have been demonstrated to be involved in acrosomal exocytosis [45], our data suggest that the membrane micro-domain sub-type partitioning to this fx is heavily involved in acrosomal exocytosis. To identify whether other specific biological functions or pathways were associated with specific fractions, we performed a database analysis (supplemental data 3). Because of the low number of proteins identified in the most buoyant fractions, caution should be exercised when comparing percentages between fractions. However, the high percentages of proteins involved in post-translational modifications and proteins whose functions are as of yet unclassified, offer attractive targets for future studies.
Based on similarity analyses of protein profiles among the different raft fractions, we found that several proteins selectively associated with specific raft sub-types. This is consistent with our previous, highly limited quantitative proteomic comparison of the same four fractions [9]. For example, alkaline phosphatase 2 (liver) was found in only fx 1-4 and fx 7 out of 4 membrane fractions. The presence of this GPI-anchored protein in sperm is consistent with previous reports of high alkaline phosphatase activity in the plasma membrane fraction [46, 47]. GPI-anchored proteins such as this enzyme have been shown to be spatially and biochemically associated with raft micro-domains enriched in GM1 [48–51]. Considering that fx 1-4 and fx 7 were enriched in GM1 but fx 5 was not, it is possible that for proteins such as this form of alkaline phosphatase, functional discrimination between raft sub-types might directly or indirectly be associated with degree of enrichment in GM1.
Comparison of the pI of proteins partitioning to the different fractions revealed some intriguing biochemical characteristics as well. A bi-modal distribution of pI away from neutral was confirmed for all fractions, as expected. This supports the notion that electrostatic interactions between proteins and other membrane components play important roles in a variety of cellular processes [52]. Although membranes used for our study were never exposed to seminal plasma, it is known that basic proteins are highly abundant in seminal plasma [53], and at least several of these interact directly with components of the sperm plasma membrane after ejaculation [54, 55]. These interactions have physiological importance in terms of acquisition and maintenance of motility [56–58], or prevention of activation of signaling cascades involved in capacitation [55, 59]. In agreement with this, our data demonstrated that acidic, negatively charged proteins are enriched particularly in the most buoyant raft fractions. Differences in charge between the components of different membrane micro-domains might then facilitate interactions between those specific domains with epididymal and/or seminal plasma proteins. This hypothesis will need to be investigated by studying the nature of the specific interactions between proteins secreted from accessory sex glands and their binding partners on the sperm plasma membrane.
4.3 Detection of mitochondrial proteins
We detected several mitochondrial proteins in raft fractions. In itself, this finding is not surprising because our non-detergent methodology was designed to obtain membrane rafts from all sub-cellular compartments of sperm. However, in somatic cells, the presence of mitochondrial rafts is controversial. There are several reports suggesting that intracellular organelles such as mitochondria might possess raft domains [60–62]. In contrast to these, recent quantitative proteomic studies of specific cultured cells demonstrated that mitochondrial proteins often observed in DRM were insensitive to sterol efflux, suggesting that those proteins might be contaminants that simply co-purify with rafts during separation process [6]. One possibility underlying this controversy is difference in mitochondrial membranes between cell types. Sperm are well known to have mitochondria that differ from their somatic counterparts in terms of morphology, localization, interaction with the plasma membrane, and their own enzyme compositions. Indeed, sperm mitochondria have evolved species-specific differences giving them the ability to adapt their metabolism to varying oviductal environments [63]. Rather than over-interpret or generalize the findings of these particular mitochondrial proteins, we instead view them as a starting point for future investigations of the composition of sperm mitochondrial membranes.
4.4 Acquisition of raft proteins during epididymal maturation
Because many of the epididymal proteins that are added to sperm play important roles in capacitation and sperm-oocyte interactions, knowledge of the species and functions of these proteins are of considerable interest as targets for male contraception [64–67]. The importance of membrane protein function in fertilization is reflected in the strong evolutionary pressures that shape them; indeed, combinations of proteomic and genomic investigations reveal their use as tools to investigate evolutionary history and possibly gain insight into protein-protein interactions [68, 69]. Our proteomic analysis revealed that many of the proteins in raft fractions are highly expressed in the epididymis. These results corroborate the notion [70] that an important function of sperm raft micro-domains is acting as targets for the selective acquisition of proteins during epididymal maturation.
In the current study, we found that apoA1 and prom1 were present in raft fx 7. Our preliminary analyses confirmed that apoA1 does not appear to be synthesized in the testis [71] unlike prom1, despite the sterol-binding nature of both proteins. These important similarities and differences made us choose these proteins for comparative studies on targeting to raft micro-domains. Our combined data suggest that sperm rafts already possess prom1, but then acquire apoA1 during epididymal transit. In support of this, another proteomic analysis showed apoA1 in cauda epididymal fluid [72].
Interestingly, the major site for apoA1 transfer was the APM. Recently, we demonstrated in live sperm that the APM is the site of multiple focal enrichments of sterols [12], which is consistent with this region being the site of one or more raft sub-types. A recent report that apoA1-binding protein is enriched in the APM region provides corroborative support for our finding regarding the localization of apoA1. The same report showed that apoA1-binding protein undergoes tyrosine phosphorylation during capacitation and is shed into the extracellular space, which is consistent with sterol efflux [73]. However, this protein is not known to have a binding interaction with sterols, suggesting that it might function in a complex with apoA1. Our present data showing different spatial and temporal patterns of apoA1 and prom1 acquisition highlight the need for future investigations of the physiological roles of membrane rafts in selective acquisition of new proteins in sperm during epididymal transit.
Supplementary Material
Acknowledgments
We thank Dr. Colin Parrish for his generous gift of a secondary antibody and Ms. Sophie Kay for technical assistance. This study was supported by NIH R01-HD-045664 and 5DP1-OD-006431 (A.J.T), and the Baker Institute for Animal Health.
Abbreviations
- APM
plasma membrane overlying the acrosome
- PAPM
post-acrosomal plasma membrane
- DRM
detergent-resistant membranes
- prom1
prominin 1
- apoA1
apolipoprotein A1
- GM1
ganglioside GM1
- IDA
information dependent acquisition
- FDR
false discovery rate
- TTBS
TBS containing 0.1 % tween 20
- CTB
cholera toxin B
- fx
fraction
- PL
phospholipids
- GPI
glycerophosphatidylinositol
- pI
isoelectric point
Footnotes
Conflict of interest statement
No direct financial gain will be achieved by publication of our manuscript. No potential commercial partners have been involved in the study design, performance of work, analysis of data, preparation of the manuscript, or decision to submit.
References
- 1.Cornwall GA, Vreeburg JT, Holland MK, Orgebin-Crist MC. Interactions of labeled epididymal secretory proteins with spermatozoa after injection of 35S-methionine in the mouse. Biol Reprod. 1990;43:121–129. doi: 10.1095/biolreprod43.1.121. [DOI] [PubMed] [Google Scholar]
- 2.Frenette G, Girouard J, Sullivan R. Comparison Between Epididymosomes Collected in the Intraluminal Compartment of the Bovine Caput and Cauda Epididymidis. Biol Reprod. 2006;75:885–890. doi: 10.1095/biolreprod.106.054692. [DOI] [PubMed] [Google Scholar]
- 3.Chang MC. Fertilizing Capacity of Spermatozoa deposited into the Fallopian Tubes. Nature. 1951;168:697–698. doi: 10.1038/168697b0. [DOI] [PubMed] [Google Scholar]
- 4.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 5.Pike LJ. Lipid rafts: bringing order to chaos. J Lipid Res. 2003;44:655–667. doi: 10.1194/jlr.R200021-JLR200. [DOI] [PubMed] [Google Scholar]
- 6.Zheng YZ, Berg KB, Foster LJ. Mitochondria do not contain lipid rafts, and lipid rafts do not contain mitochondrial proteins. J Lipid Res. 2009;50:988–998. doi: 10.1194/jlr.M800658-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kusumi A, Suzuki K. Toward understanding the dynamics of membrane-raft-based molecular interactions. Biochim Biophys Acta. 2005;1746:234–251. doi: 10.1016/j.bbamcr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Hancock JF. Lipid rafts: contentious only from simplistic standpoints. Nat Rev Mol Cell Biol. 2006;7:456–462. doi: 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asano A, Selvaraj V, Buttke DE, Nelson JL, et al. Biochemical characterization of membrane fractions in murine sperm: identification of three distinct sub-types of membrane rafts. J Cell Physiol. 2009;218:537–548. doi: 10.1002/jcp.21623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bou Khalil M, Chakrabandhu K, Xu H, Weerachatyanukul W, et al. Sperm capacitation induces an increase in lipid rafts having zona pellucida binding ability and containing sulfogalactosylglycerolipid. Dev Biol. 2006;290:220–235. doi: 10.1016/j.ydbio.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 11.Nixon B, Bielanowicz A, McLaughlin EA, Tanphaichitr N, et al. Composition and significance of detergent resistant membranes in mouse spermatozoa. J Cell Physiol. 2008 doi: 10.1002/jcp.21575. [DOI] [PubMed] [Google Scholar]
- 12.Selvaraj V, Asano A, Buttke DE, Sengupta P, et al. Mechanisms underlying the micron-scale segregation of sterols and GM1 in live mammalian sperm. J Cell Physiol. 2009;218:522–536. doi: 10.1002/jcp.21624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selvaraj V, Asano A, Buttke DE, McElwee JL, et al. Segregation of micron-scale membrane sub-domains in live murine sperm. J Cell Physiol. 2006;206:636–646. doi: 10.1002/jcp.20504. [DOI] [PubMed] [Google Scholar]
- 14.Travis AJ, Merdiushev T, Vargas LA, Jones BH, et al. Expression and localization of caveolin-1, and the presence of membrane rafts, in mouse and Guinea pig spermatozoa. Dev Biol. 2001;240:599–610. doi: 10.1006/dbio.2001.0475. [DOI] [PubMed] [Google Scholar]
- 15.Toshimori K, Higashi R, Oura C. Filipin-sterol complexes in golden hamster sperm membranes with special reference to epididymal maturation. Cell Tissue Res. 1987;250:673–680. doi: 10.1007/BF00218962. [DOI] [PubMed] [Google Scholar]
- 16.Pelletier RM, Friend DS. Development of membrane differentiations in the guinea pig spermatid during spermiogenesis. Am J Anat. 1983;167:119–141. doi: 10.1002/aja.1001670110. [DOI] [PubMed] [Google Scholar]
- 17.Elias PM, Friend DS, Goerke J. Membrane sterol heterogeneity. Freeze-fracture detection with saponins and filipin. J Histochem Cytochem. 1979;27:1247–1260. doi: 10.1177/27.9.479568. [DOI] [PubMed] [Google Scholar]
- 18.Lin Y, Kan FW. Regionalization and redistribution of membrane phospholipids and cholesterol in mouse spermatozoa during in vitro capacitation. Biol Reprod. 1996;55:1133–1146. doi: 10.1095/biolreprod55.5.1133. [DOI] [PubMed] [Google Scholar]
- 19.Enders GC, Werb Z, Friend DS. Lectin binding to guinea-pig sperm zipper particles. J Cell Sci. 1983;60:303–329. doi: 10.1242/jcs.60.1.303. [DOI] [PubMed] [Google Scholar]
- 20.Friend DS. Sperm maturation: membrane domain boundaries. Ann N Y Acad Sci. 1989;567:208–221. doi: 10.1111/j.1749-6632.1989.tb16472.x. [DOI] [PubMed] [Google Scholar]
- 21.Selvaraj V, Buttke DE, Asano A, McElwee JL, et al. GM1 dynamics as a marker for membrane changes associated with the process of capacitation in murine and bovine spermatozoa. J Androl. 2007;28:588–599. doi: 10.2164/jandrol.106.002279. [DOI] [PubMed] [Google Scholar]
- 22.Bearer EL, Friend DS. Modifications of anionic-lipid domains preceding membrane fusion in guinea pig sperm. J Cell Biol. 1982;92:604–615. doi: 10.1083/jcb.92.3.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 24.Travis AJ, Jorgez CJ, Merdiushev T, Jones BH, et al. Functional relationships between capacitation-dependent cell signaling and compartmentalized metabolic pathways in murine spermatozoa. J Biol Chem. 2001;276:7630–7636. doi: 10.1074/jbc.M006217200. [DOI] [PubMed] [Google Scholar]
- 25.Travis AJ, Foster JA, Rosenbaum NA, Visconti PE, et al. Targeting of a germ cell-specific type 1 hexokinase lacking a porin-binding domain to the mitochondria as well as to the head and fibrous sheath of murine spermatozoa. Mol Biol Cell. 1998;9:263–276. doi: 10.1091/mbc.9.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas PD, Campbell MJ, Kejariwal A, Mi H, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35:D247–252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pike LJ. Rafts defined: a report on the Keystone symposium on lipid rafts and cell function. J Lipid Res. 2006;47:1597–1598. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 29.Shaikh SR, Edidin MA. Membranes are not just rafts. Chem phys lipids. 2006;144:1–3. doi: 10.1016/j.chemphyslip.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 30.Wu S, Wan P, Li J, Li D, et al. Multi-modality of pI distribution in whole proteome. Proteomics. 2006;6:449–455. doi: 10.1002/pmic.200500221. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz R, Ting CS, King J. Whole proteome pI values correlate with subcellular localizations of proteins for organisms within the three domains of life. Genome Res. 2001;11:703–709. doi: 10.1101/gr.gr-1587r. [DOI] [PubMed] [Google Scholar]
- 32.Ho E, Hayen A, Wilkins MR. Characterisation of organellar proteomes: a guide to subcellular proteomic fractionation and analysis. Proteomics. 2006;6:5746–5757. doi: 10.1002/pmic.200600241. [DOI] [PubMed] [Google Scholar]
- 33.Chan P, Lovric J, Warwicker J. Subcellular pH and predicted pH-dependent features of proteins. Proteomics. 2006;6:3494–3501. doi: 10.1002/pmic.200500534. [DOI] [PubMed] [Google Scholar]
- 34.Frenette G, Sullivan R. Prostasome-like particles are involved in the transfer of P25b from the bovine epididymal fluid to the sperm surface. Mol Reprod Dev. 2001;59:115–121. doi: 10.1002/mrd.1013. [DOI] [PubMed] [Google Scholar]
- 35.Ekstedt E, Holm L, Ridderstråle Y. Carbonic Anhydrase in Mouse Testis and Epididymis; Transfer of Isozyme IV to Spermatozoa During Passage. J Mol Histol. 2004;35:167–173. doi: 10.1023/b:hijo.0000023387.02793.af. [DOI] [PubMed] [Google Scholar]
- 36.Zhou CX, Zhang YL, Xiao L, Zheng M, et al. An epididymis-specific beta-defensin is important for the initiation of sperm maturation. Nat Cell Biol. 2004;6:458–464. doi: 10.1038/ncb1127. [DOI] [PubMed] [Google Scholar]
- 37.Kohane AC, Gonzalez Echeverria FM, Pineiro L, Blaquier JA. Interaction of proteins of epididymal origin with spermatozoa. Biol Reprod. 1980;23:737–742. doi: 10.1095/biolreprod23.4.737. [DOI] [PubMed] [Google Scholar]
- 38.Tubbs CE, Hall JC, Scott RO, Clark VP, et al. Binding of protein D/E to the surface of rat epididymal sperm before ejaculation and after deposition in the female reproductive tract. J Androl. 2002;23:512–521. [PubMed] [Google Scholar]
- 39.Boue F, Blais J, Sullivan R. Surface localization of P34H an epididymal protein, during maturation, capacitation, and acrosome reaction of human spermatozoa. Biol Reprod. 1996;54:1009–1017. doi: 10.1095/biolreprod54.5.1009. [DOI] [PubMed] [Google Scholar]
- 40.Olson LM, Zhou X, Schreiber JR. Immunolocalization of apolipoprotein E in the testis and epididymis of the rat. Biol Reprod. 1994;50:535–542. doi: 10.1095/biolreprod50.3.535. [DOI] [PubMed] [Google Scholar]
- 41.Fargeas CA, Joester A, Missol-Kolka E, Hellwig A, et al. Identification of novel Prominin-1/CD133 splice variants with alternative C-termini and their expression in epididymis and testis. J Cell Sci. 2004;117:4301–4311. doi: 10.1242/jcs.01315. [DOI] [PubMed] [Google Scholar]
- 42.Zhang N, Li L. Effects of common surfactants on protein digestion and matrix-assisted laser desorption/ionization mass spectrometric analysis of the digested peptides using two-layer sample preparation. Rapid Commun Mass Spectrom. 2004;18:889–896. doi: 10.1002/rcm.1423. [DOI] [PubMed] [Google Scholar]
- 43.Shadan S, James PS, Howes EA, Jones R. Cholesterol efflux alters lipid raft stability and distribution during capacitation of boar spermatozoa. Biol Reprod. 2004;71:253–265. doi: 10.1095/biolreprod.103.026435. [DOI] [PubMed] [Google Scholar]
- 44.Arnoult C, Zeng Y, Florman HM. ZP3-dependent activation of sperm cation channels regulates acrosomal secretion during mammalian fertilization. J Cell Biol. 1996;134:637–645. doi: 10.1083/jcb.134.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomes CN, Michaut M, De Blas G, Visconti P, et al. SNARE complex assembly is required for human sperm acrosome reaction. Dev Biol. 2002;243:326–338. doi: 10.1006/dbio.2002.0567. [DOI] [PubMed] [Google Scholar]
- 46.de Curtis I, Fumagalli G, Borgese N. Purification and characterization of two plasma membrane domains from ejaculated bull spermatozoa. J Cell Biol. 1986;102:1813–1825. doi: 10.1083/jcb.102.5.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soucek DA, Vary JC. Some properties of acid and alkaline phosphates from boar sperm plasma membranes. Biol Reprod. 1984;31:687–693. doi: 10.1095/biolreprod31.4.687. [DOI] [PubMed] [Google Scholar]
- 48.Dietrich C, Volovyk ZN, Levi M, Thompson NL, Jacobson K. Partitioning of Thy-1, GM1, and cross-linked phospholipid analogs into lipid rafts reconstituted in supported model membrane monolayers. Proc Natl Acad Sci U S A. 2001;98:10642–10647. doi: 10.1073/pnas.191168698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simons M, Friedrichson T, Schulz JB, Pitto M, et al. Exogenous administration of gangliosides displaces GPI-anchored proteins from lipid microdomains in living cells. Mol Biol Cell. 1999;10:3187–3196. doi: 10.1091/mbc.10.10.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mazzone A, Tietz P, Jefferson J, Pagano R, LaRusso NF. Isolation and characterization of lipid microdomains from apical and basolateral plasma membranes of rat hepatocytes. Hepatology. 2006;43:287–296. doi: 10.1002/hep.21039. [DOI] [PubMed] [Google Scholar]
- 51.Damek-Poprawa M, Golub E, Otis L, Harrison G, et al. Chondrocytes Utilize a Cholesterol-Dependent Lipid Translocator To Externalize Phosphatidylserine. Biochemistry. 2006;45:3325–3336. doi: 10.1021/bi0515927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mulgrew-Nesbitt A, Diraviyam K, Wang J, Singh S, et al. The role of electrostatics in protein-membrane interactions. Biochim Biophys Acta. 2006;1761:812–826. doi: 10.1016/j.bbalip.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 53.Lavon U, Boursnell JC. Characterization of boar seminal plasma, vesicular secretion and epididymal plasma proteins by gel disc electrophoresis and isoelectric focusing on polyacrylamide. J Reprod Fertil. 1971;27:227–232. doi: 10.1530/jrf.0.0270227. [DOI] [PubMed] [Google Scholar]
- 54.Moore HD, Hibbitt KG. The binding of labelled basic proteins by boar spermatozoa. J Reprod Fertil. 1976;46:71–76. doi: 10.1530/jrf.0.0460071. [DOI] [PubMed] [Google Scholar]
- 55.Kawano N, Yoshida K, Iwamoto T, Yoshida M. Ganglioside GM1 Mediates Decapacitation Effects of SVS2 on Murine Spermatozoa. Biol Reprod. 2008 doi: 10.1095/biolreprod.108.069054. [DOI] [PubMed] [Google Scholar]
- 56.Robert M, Gibbs BF, Jacobson E, Gagnon C. Characterization of prostate-specific antigen proteolytic activity on its major physiological substrate, the sperm motility inhibitor precursor/semenogelin I. Biochemistry. 1997;36:3811–3819. doi: 10.1021/bi9626158. [DOI] [PubMed] [Google Scholar]
- 57.Luo CW, Lin HJ, Chen YH. A novel heat-labile phospholipid-binding protein, SVS VII, in mouse seminal vesicle as a sperm motility enhancer. J Biol Chem. 2001;276:6913–6921. doi: 10.1074/jbc.M006954200. [DOI] [PubMed] [Google Scholar]
- 58.Robert M, Gagnon C. Purification and characterization of the active precursor of a human sperm motility inhibitor secreted by the seminal vesicles: identity with semenogelin. Biol Reprod. 1996;55:813–821. doi: 10.1095/biolreprod55.4.813. [DOI] [PubMed] [Google Scholar]
- 59.de Lamirande E, Yoshida K, Yoshiike TM, Iwamoto T, Gagnon C. Semenogelin, the main protein of semen coagulum, inhibits human sperm capacitation by interfering with the superoxide anion generated during this process. J Androl. 2001;22:672–679. [PubMed] [Google Scholar]
- 60.Rashid-Doubell F, Tannetta D, Redman CW, Sargent IL, et al. Caveolin-1 and lipid rafts in confluent BeWo trophoblasts: evidence for Rock-1 association with caveolin-1. Placenta. 2007;28:139–151. doi: 10.1016/j.placenta.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 61.Li WP, Liu P, Pilcher BK, Anderson RG. Cell-specific targeting of caveolin-1 to caveolae, secretory vesicles, cytoplasm or mitochondria. J Cell Sci. 2001;114:1397–1408. doi: 10.1242/jcs.114.7.1397. [DOI] [PubMed] [Google Scholar]
- 62.Nakamura M, Sakurai Y, Takeda Y, Toda T. Comparative proteomics of flotillin-rich Triton X-100-insoluble lipid raft fractions of mitochondria and synaptosome from mouse brain. J Electrophoresis. 2005;49:77–83. [Google Scholar]
- 63.Storey BT. Strategy of oxidative metabolism in bull spermatozoa. J Exp Zool. 1980;212:61–67. doi: 10.1002/jez.1402120109. [DOI] [PubMed] [Google Scholar]
- 64.Runnebaum B, Rabe T, Kiesel L. Different approaches in contraception. Contracept Deliv Syst. 1984;5:15. [PubMed] [Google Scholar]
- 65.Cosentino MJ, Matlin SA. Pharmacological developments in male contraception. Expert Opin Investig Drugs. 1997;6:635–653. doi: 10.1517/13543784.6.6.635. [DOI] [PubMed] [Google Scholar]
- 66.Cooper TG, Yeung CH. Approaches to post-testicular contraception. Asian J Androl. 1999;1:29–36. [PubMed] [Google Scholar]
- 67.Cooper TG, Yeung CH. Recent biochemical approaches to post-testicular, epididymal contraception. Hum Reprod Update. 1999;5:141–152. doi: 10.1093/humupd/5.2.141. [DOI] [PubMed] [Google Scholar]
- 68.Dorus S, Wasbrough ER, Busby J, Wilkins EC, Karr TL. Sperm Proteomics Reveals Intensified Selection on Mouse Sperm Membrane and Acrosome Genes. Mol Biol Evol. 2010 doi: 10.1093/molbev/msq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Findlay GD, Swanson WJ. Proteomics enhances evolutionary and functional analysis of reproductive proteins. Bioessays. 2010;32:26–36. doi: 10.1002/bies.200900127. [DOI] [PubMed] [Google Scholar]
- 70.Sullivan R, Frenette G, Girouard J. Epididymosomes are involved in the acquisition of new sperm proteins during epididymal transit. Asian J Androl. 2007;9:483–491. doi: 10.1111/j.1745-7262.2007.00281.x. [DOI] [PubMed] [Google Scholar]
- 71.Law GL, McGuinness MP, Linder CC, Griswold MD. Expression of apolipoprotein E mRNA in the epithelium and interstitium of the testis and the epididymis. J Androl. 1997;18:32–42. [PubMed] [Google Scholar]
- 72.Moura AA, Chapman DA, Koc H, Killian GJ. Proteins of the cauda epididymal fluid associated with fertility of mature dairy bulls. J Androl. 2006;27:534–541. doi: 10.2164/jandrol.05201. [DOI] [PubMed] [Google Scholar]
- 73.Jha KN, Shumilin IA, Digilio LC, Chertihin O, et al. Biochemical and structural characterization of apolipoprotein A-I binding protein, a novel phosphoprotein with a potential role in sperm capacitation. Endocrinology. 2008;149:2108–2120. doi: 10.1210/en.2007-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.