Abstract
The radiation-induced bystander effect (RIBE) is a phenomenon whereby unexposed cells exhibit molecular symptoms of stress exposure when adjacent or nearby cells are traversed by ionizing radiation (IR). Recent data suggest that RIBE may be epigenetically mediated by microRNAs (miRNAs), which are small regulatory molecules that target messenger RNA transcripts for translational inhibition. Here, we analyzed microRNAome changes in bystander tissues after α-particle microbeam irradiation of three-dimensional artificial human tissues using miRNA microarrays. Our results indicate that IR leads to a deregulation of miRNA expression in bystander tissues. We report that major bystander end points, including apoptosis, cell cycle deregulation and DNA hypomethylation, may be mediated by altered expression of miRNAs. Specifically, c-MYC-mediated upregulation of the miR-17 family was associated with decreased levels of E2F1 and RB1, suggesting a switch to a proliferative state in bystander tissues, while priming these cells for impending death signals. Upregulation of the miR-29 family resulted in decreased levels of its targets DNMT3a and MCL1, consequently affecting DNA methylation and apoptosis. Altered expression of miR-16 led to changes in expression of BCL2, suggesting modulation of apoptosis. Thus, our data clearly show that miRNAs play a profound role in the manifestation of late RIBE end points. In summary, this study creates a roadmap for understanding the role of microRNAome in RIBE and for developing novel RIBE biomarkers.
Introduction
Bystander effects are non-targeted effects of radiation whereby unexposed cells exhibit the molecular symptoms of stress exposure when adjacent or nearby cells are traversed by ionizing radiation (IR). To date, a variety of radiation-induced bystander effect (RIBE) studies have been performed using cell culture models (1–4). However, these experiments utilized cell cultures in a single monolayer, making extrapolation to human exposure somewhat difficult. Recent work in tissue explants (5,6), spheroids (7), three-dimensional (3-D) cell cultures (8) and 3-D artificially reconstructed human tissues (9,10) has suggested that the cell–cell bystander effect operates in human 3-D systems. Thus, the RIBE remains an important and relevant consideration in the study of radiobiology.
RIBE encompass a wide range of genetic alterations, including gross genome rearrangements, chromosome aberrations, sister chromatid exchanges, deletions, duplications, mutations and amplifications (reviewed in refs 11–15). These effects also influence gene expression, cellular proliferation, cell cycle regulation, senescence and cell death (10,11) and are believed to be linked to IR-induced genome instability (13). However, although a great deal of data confirms the existence and manifestation of RIBE in cultured cells and 3-D tissues, the mechanisms are yet to be discovered.
The high frequency of induction and occurrence, as well as persistence of RIBE, has led some to suggest that epigenetic regulation may play an important role in bystander cells and tissues (10,16,17). Epigenetic changes are alterations in gene expression induced by DNA methylation, histone modifications and RNA-associated silencing (18). MicroRNAs (miRNAs) are important components of the RNA-associated silencing machinery. miRNAs are small regulatory molecules known to target messenger RNA transcripts for translational inhibition or, rarely, degradation in humans (19). Since their discovery, miRNAs have been found to play essential roles in regulating processes such as terminal differentiation (20), cell cycle (21), apoptosis (22) and DNA methylation (23). Further, genotoxic stress exposure deregulates cellular miRNA expression (24–26). Logically, deregulation of these miRNAs has been associated with a number of diseases, including cancer.
Based on the importance of miRNAs in the modulation of various cellular processes and the fact that they have been shown to be regulated in in vivo RIBE (24,27), we decided to analyze microRNAome changes in bystander tissues after α-particle microbeam irradiation of 3-D artificial human tissues using miRNA microarrays. Microarray data analysis has indicated that some of the molecular end points previously found in this bystander model may be the result of changes in miRNA expression. Our data suggest that miRNA regulation acting in concert with c-Myc activation in bystander tissues may sensitize bystander cells to apoptosis.
Materials and methods
Tissue systems and culture
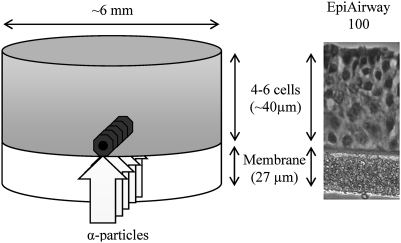
These experiments utilized a human 3-D tissue culture system (MatTek Corp, Ashland, MA). These artificial tissues reconstruct the normal tissue microarchitecture and preserve the in vivo differentiation patterns. They are mitotically and metabolically active, capable of releasing relevant cytokines, and contain gap junctions (28). They are stable and allow a high degree of experimental reproducibility (9,10). Specifically, we employed the EpiAirway™ (Air-112) tissue model (Figure 1). The tissues were cultured according to the manufacturer’s protocol, using an air–liquid interface tissue culture technique.
Fig. 1.
Tissue dimensions and irradiation setup. A narrow α-particle beam irradiated the EpiAirway tissue through the membrane to deliver approximately five α-particles to the nucleus of each cell in a single plane across the tissue base. Bystander tissues were harvested on either side of the irradiated plane for the microarray experiments.
Microbeam irradiation and delivered dose calculations
The Columbia University RARAF Singletron accelerator was used to produce a broad beam of 4He ions with an initial energy of 8.6 MeV. The ions passed through a Mylar scattering foil and a Havar metal vacuum window (together ∼20 μm thick) and 8 μm of air before entering the membranes on which tissue samples were grown.
Each tissue culture insert was placed on a plastic disc that had a hole in the center. Two 50 μm stainless steel half-discs (sufficient to stop 4He ions) were glued over each hole with a 25 μm gap between them. This allowed a single plane of tissue to be irradiated from below through the membrane that forms the base of the culture insert. The discs were placed on an irradiation wheel and rotated across the beam. The slits through which the tissue samples were irradiated were aligned with the direction of rotation so that there was almost no variation in the dose across the sample.
Assuming that the pores of the Teflon sponge membrane were filled with tissue medium, 4He ions had the energy of ∼4.5 MeV, linear energy transfer values of ∼110 keV/μm and the range of ∼30 μm at the membrane–tissue interface. The tissue is thicker than the range of 4He ions. As the ions penetrate the tissue, their linear energy transfer increases to ∼230 keV/μm just before they stop. Doses were measured using an ionization chamber with a Mylar window filled with methane-based tissue-equivalent gas. Based on a nuclear diameter of 5 μm, cell nuclei were irradiated with ∼5 4He ions per nucleus. Calculations of 4He ion energy, linear energy transfer and range were made for the irradiation conditions using tables of ranges and stopping powers for 4He ions generated by the computer program SRIM-2008 (29). The estimated dose for the first layer of cells is 5.4 Gy.
After irradiation, each tissue sample was returned to a multiwell dish filled with fresh medium and incubated at 37°C in a humidified atmosphere of 5% CO2. Tissues were removed and frozen on dry ice at 8 h and 1, 2, 3, 4, 5 and 7 days post-IR (d.p.i.). Time-matched mock-treated controls were available at 8 h, 3 days and 7 days post-mock IR. Mock-treated tissue frozen at the time of IR established the base-line expression level. The tissues were stored at −80°C before microdissection and RNA and protein extraction.
Tissue microdissection and RNA extraction
The line of irradiation was marked on the insert. Frozen tissues were placed on a liquid N2-chilled glass plates. Scalpels were used to cut ∼1 to 1.5 mm slabs from the center of the tissues along the irradiated plane to secure the bystander remaining tissues (∼2 mm thick on either side of the slabs) (Figure 1). The center slabs of the tissues represented mixed irradiated and bystander cell layers. The soley bystander tissues from either side of the slabs were pooled to a single microfuge tube.
Targeted and bystander tissues can be distinguished based on differences in kinetics of IR-induced and bystander double-strand break (DSB) formation and repair marked by the γ-H2AX antibody (30). Earlier, we reported that in marked contrast to DNA DSB dynamics in directly irradiated cells, in which maximal DSB formation was seen 30 min post-IR, the incidence of DSBs in bystander tissues reached a maximum much later, by 12–48 h post-IR (h.p.i.) (10). Thus, the central regions of the tissues (the 1.5 mm slabs containing irradiated planes of the tissues as well as unirradiated cells) show biphasic γ-H2AX focal kinetics, whereas bystander tissues exhibit the bystander-like delayed γ-H2AX focal kinetics providing additional verification of bystander tissue identity (supplementary Figure 1 is available at Carcinogenesis Online).
miRNA expression profiling
Three biological replicates were used for each of the experimental time points and treatment conditions. The bystander and mock-treated tissues were immersed in TRIzol (500 μl), and the samples were vortexed for 5–10 s to remove the tissues from the membrane. RNA extraction was carried out as per the manufacturer’s protocol (TRIzol; Invitrogen, Carlsbad, CA). The miRNA microarray analysis was performed by LC Sciences (Houston, TX) as a paid service as previously described (24,26).
In brief, total RNA (10 μg) was size fractionated (<200 nucleotides) using a mirVana kit (Applied Biosystems/Ambion, Austin, TX) and labeled with Cy3 and Cy5 fluorescent dyes. Dye switching was performed to eliminate any dye bias. Pairs of labeled samples were hybridized to dual-channel microarrays. Microarray assays were carried out on a ParaFlo microfluidics chip. Each of the detection probes on the chip contained a nucleotide sequence of coding segment complementary to a specific miRNA sequence and a long non-nucleotide molecule spacer that extended the detection probe away from the substrate (31). The miRNA detection signal threshold was defined as twice the maximum background signal. The maximum signal level of background probes was 180. To remove any system-related variations, normalization was performed using a cyclic LOWESS (locally weighted regression) method (32). Data adjustments included data filtering, log2 transformation and gene centering and normalization. The t-test analysis and analysis of variance (ANOVA) were conducted between the sample groups, samples and miRNA with P values <0.05 were selected for cluster analysis. The clustering analysis was done using a hierarchical method, average linkage and Euclidean distance metrics (31,33).
Western blotting
Protein extraction and western blotting was conducted as described (26,27). Membranes were incubated with antibodies against MYC, E2F1, RB1, DNMT3a, BCL2 (1:500; Santa Cruz Biotechnology, Santa Cruz, CA), MCL1 (1:1000; Cell Signalling, Bedford, MA) and actin (1:2000; Santa Cruz Biotechnology). Antibody binding was revealed by incubation with horseradish peroxidase-conjugated secondary antibodies and the ECL Plus TM immunoblotting detection system (GE Healthcare, Piscataway, NJ). Chemiluminescence was detected by Biomax MR film (Eastman Kodak, New Haven, CT) and scanned. Samples were loaded in a random manner to ensure lack of bias. For presentation purposes, after scanning pictures were arranged in the following order: mock 8 h, bystander 8 h, mock 3 days, bystander 3 days and bystander 5 days.
Immunohistochemistry
Frozen tissues were sectioned into 5–10 μm thick slices, which were perpendicular to the plane of the irradiated cells. Frozen sections were dried, fixed in 2% paraformaldehyde, permeabilized with 1% Triton X-100 and processed for immunostaining with a rabbit polyclonal anti-cleaved caspase-3 antibody (Trevigen, Gaithersburg, MD) and mouse monoclonal anti-γ-H2AX antibody (Abcam, Cambridge, MA) as previously described (34). Frequencies of cleaved caspase-3-positive cells were recorded for at least 1000 cells of side section areas. γ-H2AX foci were recorded by collapsing 0.5 μm optical Z-sections through the nuclei in a single plane and counting by eye in a blinded fashion in 100–200 randomly chosen cells of central and side areas of sections.
Statistical analysis
Statistical analysis was conducted using Student’s t-test. P-values <0.05 were considered significant.
Results
miRNAome deregulation in bystander tissues
We employed the EpiAirway™ (Air-112) tissue model, which consists of normal human-derived tracheal/bronchial epithelial cells that have been cultured to form a highly differentiated model that closely resembles the epithelial tissue of the respiratory tract. Figure 1 displays tissue dimensions and the location of irradiated and bystander tissues. In this model, the targeted and bystander tissues can be distinguished based on the different kinetics of IR-induced and bystander DSB formation and repair marked by the γ-H2AX antibody (10).
To analyze the role of the microRNAome in RIBE, miRNA expression profiles of bystander tissues were compared with time-matched mock controls at 8 h, 3 days and 7 d.p.i. miRNAs were considered for comparison only if their average expression levels were >1000 arbitrary units of fluorescence (AUF). The reason for this was 3-fold. Firstly, miRNAs with very low expression levels, <1000 AUF, are analyzed with less sensitivity by conventional microarray methods. Secondly, downstream studies involving quantitative reverse transcription–polymerase chain reaction methods are limited to miRNAs with expression levels of >1000 AUF. Finally, fold induction of miRNAs with low expression levels is less biologically significant than of those with higher expression levels.
At 8 h.p.i., four miRNAs (Student’s t-test, P < 0.05) changed significantly in bystander tissue over the time-matched controls. These were miR-22, -141 and -16 that were 2.0-, 1.8- and 1.2-fold upregulated and miR-183 that was 1.3-fold downregulated (Table I). At 3 d.p.i., six miRNAs were found to be significantly different from the mock control; miR-29a, -29c, -30a-5p and -20a were 2.45-, 1.62-, 1.47- and 1.25-fold upregulated, respectively, whereas miR-146a and -125b appeared to be 1.26- and 1.18-fold downregulated, respectively. The final time point, 7 d.p.i., yielded only two significantly regulated miRNAs, miR-181a and -181b, which were 1.5- and 1.2-fold downregulated, respectively (Table I). These fold changes in miRNA expression over the time course may initially seem small, but it is important to keep in mind that only a very small area of the tissue was actually irradiated. The signaling capacity of RIBE in terms of distance has not been precisely defined in this model. Thus, as all non-irradiated tissue used in this study was harvested as bystander tissue, some of the fold inductions may be diluted. Unfortunately, these tissue models are too scarce to be used in in situ miRNA analysis of selected miRNAs.
Table I.
Significantly regulated miRNAs in bystander tissue when compared with time-matched controls
| Days post irradiation | miRNA | Fold induction |
| 8 h | hsa-miR-22 | 1.95 |
| hsa-miR-141 | 1.79 | |
| hsa-miR-16 | 1.20 | |
| hsa-miR-183 | −1.31 | |
| 3 days | hsa-miR-29c | 2.45 |
| hsa-miR-29a | 1.62 | |
| hsa-miR-30a-5p | 1.47 | |
| hsa-miR-20a | 1.25 | |
| hsa-miR-146a | −1.26 | |
| hsa-miR-125b | −1.18 | |
| 7 days | hsa-miR-181a | −1.50 |
| hsa-miR-181b | −1.14 |
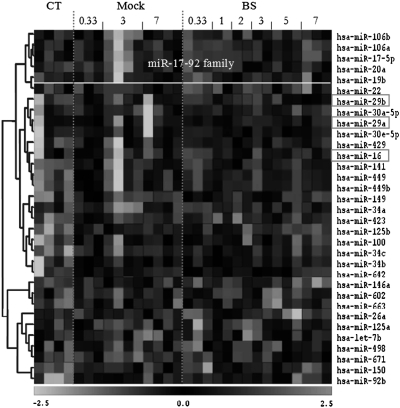
We examined expression levels of selected miRNAs at all time points after exposure and at available time-matched controls. This allowed us to establish an expression trend over the experimental period. Next, we analyzed patterns of miRNA expression at all time points based on ANOVA. The ANOVA-based expression patterns from all treatment groups can be seen in the heat map (P < 0.05; Figure 2). These miRNAs were clustered according to expression patterns. Viewing the array in this manner has revealed very intriguing groups of interrelated miRNAs. Among those, the miR-17-92 and miR-29 families exhibited the most interesting expression patterns.
Fig. 2.
Heat map of ANOVA (P < 0.05) expression analysis for all treatments. Vertical columns denote days after irradiation for the mock treatment and the bystander (BS) tissues. Control (CT)-unirradiated sample. The white box includes co-regulated members of the miR-17-92 family. Red boxes highlight important miRNAs discussed in the article. Each line represents an independent biological replicate.
Altered expression of the miR-17-92 family and deregulation of RB1 and E2F levels in bystander tissues
We identified a group of significantly changed miRNAs, including miR-106a, -106b, -17-5p, -20a and -19b that all belong to the same miR-17-92 family (Figure 2). Interestingly, this family has members that reside in three separate polycistrons on different chromosomes. The miR-17 cluster is located on chromosome 13 [13: 90800860-90801646 (+)] and consists of miR-17, -18a, -19a, -20a, 19b-1 and 92-1. The 106a cluster on chromosme X [X:133131148-133131894 (−)] is composed of miR-106a, -20b, -19b-2, -92-2 and -363. Finally, the mir-106b cluster composed of 106b, 93 and 25 is located on chromosome 7 [7:99529202-99528552 (−)] and contains miR-106b, 93 and 25. Both clusters 17 and 106a are intergenic, whereas the 106b cluster is located in intron 11 of the MCM7 gene. The regulation of this miRNA group in bystander tissues consisted of an initial increase at 8 h.p.i. that persisted before returning to control levels at 7 d.p.i. (Figure 2; supplementary Figure 2, available at Carcinogenesis Online).
Due to the similar regulation of these miRNAs located on different chromosomes, it is possible that they have similar regulatory elements. Both the miR-17 and -106b promoters have been predicted to be regulated by the c-MYC transcription factor (35,36).
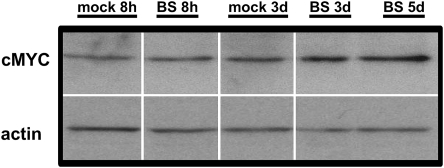
To see if c-MYC was deregulated in bystander tissues, we performed western blot analysis to assess protein levels. We observed an increased level of c-MYC in bystander tissues, suggesting that c-MYC upregulation mediates upregulation of the miR-17-92 family in bystander tissues (Figure 3). Further, the miR-17-92 family members are known to be oncogenic miRNAs, and they have found to be deregulated in a number of cancers (37). The miR-17-92 family is also known to regulate E2F transcription factors via miR-20a and miR-17-5p and possibly other miRNAs in these clusters. Interestingly, the E2F transcription factors are also induced by c-MYC and, conversely, c-MYC is induced by E2Fs (38). Importantly, in this study, we observed a notable upregulation of MYC and the miR-17-92 cluster and downregulation of E2F1 in bystander tissues (Figure 3 and Figure 4A).
Fig. 3.
Upregulation of cMYC levels in bystander tissues. Samples were loaded in a random manner to ensure lack of bias. For presentation purposes, after scanning pictures were arranged in the following order: mock 8h, bystander 8h, mock 3d, bystander 3d and bystander 5d. BS—bystander, 8h—8 h after irradiation, 3d—3 days after irradiation and 5d—5 days after irradiation.
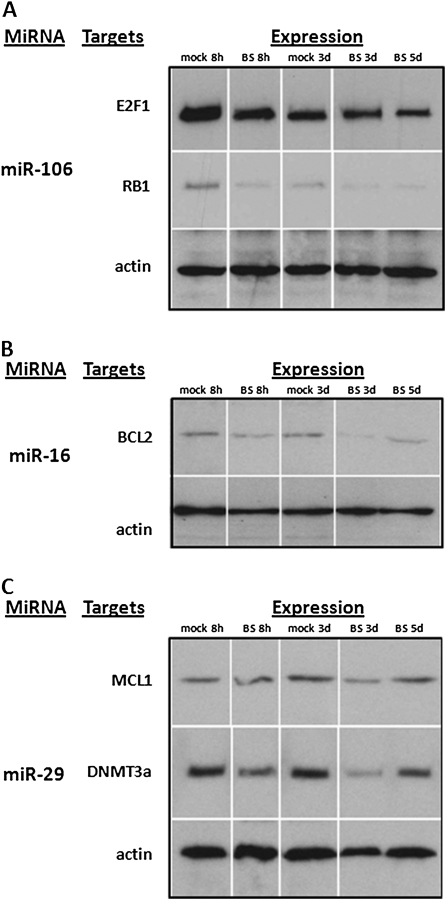
Fig. 4.
Altered expression of miRNA targets in bystander cells. (A) Targets of miR-106. (B) Targets of miR-16. (C) Targets of miR-29. Samples were loaded in a random manner to ensure lack of bias. For presentation purposes, after scanning pictures were arranged in the following order: mock 8h, bystander 8h, mock 3d, bystander 3d and bystander 5d. BS, bystander, 8h—8 h after irradiation, 3d—3 days after irradiation and 5d—5 days after irradiation.
Further, the miR-106 miRNAs have been shown to target retinoblastoma protein (RB1) (39), a well-known cell cycle regulator protein (40). Interestingly, the upregulation of miR-106 suggests a downregulation of RB1. Analyzing the levels of RB1 in bystander tissue, we found that the upregulation of miR-106 was correlated with downregulation of RB1 (Figure 4A).
MiR-16-mediated regulation of BCL2 in bystander tissues
MiR-16 was upregulated at 8 h.p.i. in bystander tissues (Table I); however, miR-16 expression showed a strong trend of upregulation for the duration of the experiment until day 7 where it returns to control levels (Figure 2).
It has been shown that miR-16 targets BCL2, a well-known anti-apoptotic protein, and it is the overexpression of BCL2 that largely contributes to the malignant phenotype in chronic lymphocytic leukemia (41). Here, we show that miR-16 is upregulated, suggesting cellular BCL2 levels would be lower, which indeed is the case (Figure 4B). Despite the absence of increased rates of apoptosis in the previous experiments at this time point, the elevated levels of BCL2 could induce a proapoptotic state in these cells, increasing their susceptibility to signaled cell death.
The miR-29 family affects MCL1 and DNMT3a and influences apoptosis and DNA hypomethylation in bystander tissues
The miR-29 family members are situated at two separate intergenic loci, miR-29a and -29b-1 on chromosome 7 [7:130212046-130212838 (−)] and miR-29c and -29b-2 on chromosome 1 [1:206041820-206042491 (−)]. MiR-29a and -29c are significantly upregulated in bystander tissues at 3 d.p.i. Mir-29b is also upregulated but with less confidence (P < 0.10). Thus, the entire family follows the same overall expression trends in bystander tissues (Figure 2).
MiR-29a was the most highly affected miRNA in the array. Though its fold induction was not the highest observed, it had the largest absolute change in expression, from ∼12 000 AUF in control cells to ∼20 000 AUF in bystander cells. Considering the competition for the miRNA machinery in the cell, it is probably that a change of this magnitude is more biologically significant than higher fold induction in less-expressed miRNAs, for example a 5-fold change of 50 AUF to 250 AUF.
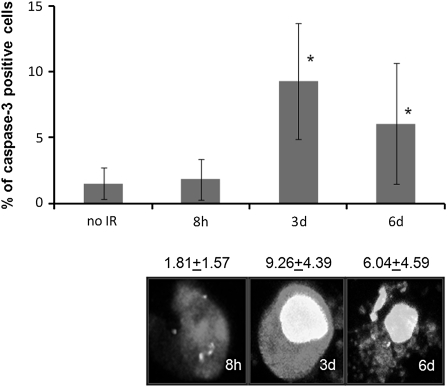
The miR-29 family has been well characterized in humans and has some very interesting and relevant targets. The miR-29 family targets MCL1, a tightly controlled BCL2 family member that is important in regulating tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis (42). In this study, upregulated levels of miR-29 members were paralleled by downregulation of MCL1 in bystander tissues (Figure 4B). Therefore, we suggest that significant upregulation of the miR-29 family in bystander tissues may be sensitizing or priming them for apoptosis. In agreement with our previous data based on this bystander model (10), apoptotic levels reached a maximum 3 and 4 d.p.i. before returning to basal levels at 7 d.p.i. These increases in apoptosis over the experimental period closely mirror the expression level of the miR-29 family over the same experimental period, further suggesting an important role of miR-29s and MCL1 in the regulation of apoptotis. As the next step of our study, we analyzed the levels of apoptosis by scoring the number of cells positive for cleaved caspase 3. Analysis revealed significantly (P < 0.05) elevated levels of apoptosis in bystander cells 3 d.p.i. (Figure 5). The miR-29 family also influences de novo DNA methyltransferases DNMT3a and DNMT3b. Fabbri et al. (23) have shown that the miR-29 family targets these methyltransferases and leads to aberrant methylation patterns. These data suggest that miR-29 expression can directly affect methylation patterns, and the possibility of this regulation in bystander tissues is a truly novel finding. We have previously reported that IR leads to hypomethylation in 3-D tissue models of RIBE (10). Here, we show that this hypomethylation corresponds to decreased levels of DNMT3a and increased levels of the miR-29 family in bystander tissues (Figure 4C).
Fig. 5.
Increase of apoptotic cells in bystander tissues following microbeam irradiation. IR-induced apoptotic cell death was scored in side areas of the sections containing unirradiated bystander cells; *significantly different from the control, P < 0.05, Student’s t-test. Cells positive for cleaved caspase-3 were recorded at various times post-IR in Air-100 epithelium. Representative images of apoptotic cells in Air-100 bystander epithelium positive for both cleaved caspase-3 (red) and γ-H2AX (green), 10 μm frozen sections, 40×, oil immersion. Cleaved caspase-3 is stained with Alexa 546 (red) and γ-H2AX is stained with Alexa 488 (green).
Interestingly, in our previous experiments, DNA hypomethylation levels in bystander tissues peaked at 3 d.p.i. and declined at 7 d.p.i. (10), a trend that is also mimicked by miR-29 expression observed in the current experiment. These data show that miR-29 family expression is very well correlated with DNMT3a protein levels. These factors may be responsible for hypomethylation previously seen in this bystander model.
Discussion
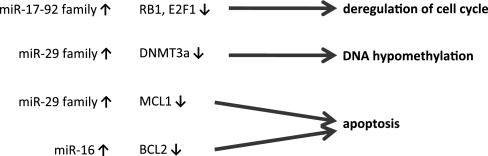
In this study, we analyzed microRNAome changes in bystander tissues in an ‘in vivo’ EpiAirway human tissue model. Bystander tissues in this model display profound cellular and molecular changes (9,10). Specifically, previous research showed an increase in apoptosis, DNA DSBs, micronucleus formation and cellular senescence as well as a decrease in global genome methylation. Additionally, RIBE is associated with deregulation of cell cycle control. Given that miRNAs are known to regulate some of these processes, we investigated the roles that miRNAs may play in the bystander response. Importantly, we found that major RIBE end points—apoptosis, cell cycle deregulation and global genome hypomethylation may be mediated by altered expression of miRNAs (Figure 6).
Fig. 6.
miRNAs that are involved in regulation of important bystander effect end points by targeting crucial regulator proteins.
Furthermore, we also show that altered miRNA expression in bystander tissues may be due to the bystander-induced expression of c-MYC. c-MYC is a widely studied gene involved in the control of cell size, cell cycle, proliferation and apoptosis (43). It has been shown that the proto-oncogene c-MYC transcription factor binds the miR-17-92 cluster’s promoter region, inducing these miRNAs to target E2F transcripts (35). The current paradigm is that the miR-17-92 cluster works as a regulator of a positive feedback loop between c-MYC and E2F transcription factors, preventing a reciprocal positive feedback from ‘runaway’ regulation (44). The upregulation of the miR-17-92 cluster via c-MYC to cause elevated expression levels of miR-17 and -20a is thought to indicate environmental signals to switch the cell to a proliferative state (44). Our study shows that upregulation of MYC and the miR-17-92 cluster and downregulation of E2F1 in bystander tissues, support the current paradigm. Furthermore, we found that the upregulation of miR-106 was correlated with downregulation of RB1.
It is interesting to speculate on the function of c-MYC activity in bystander tissues. Increased levels of c-MYC in the cell are most often associated with deregulated cell cycle and increased cellular proliferation, and the regulation of many miRNAs in the miR-17 family in bystander tissues suggests that this may be the case. Unfortunately, due to the scarcity of tissues, an increase in cellular proliferation in this bystander model has not been assayed, although it has been found in some bystander models (12–14). Future experiments in this model should assay whether cell proliferation actually occurs in bystander cells or whether the cell is just ‘poised’ to proliferate after further extracellular signals.
We can also look at these changes in cellular alignment governed by c-MYC and the miR-17 family in a different way that is more commonly associated with bystander effects. It is thought that c-MYC may prime the cell for apoptosis (45,46). Taken in this context, upregulation of c-Myc in these tissues may sensitize bystander cells for an impending death signal. Indeed, we have noted a significant increase in the level of apoptotic cells in bystander issues.
Further, c-MYC and BCL2 have long been shown to associate with cancer cells, whereby it is usually the overexpression of both that leads to the cancer phenotype. The result of this co-operation is that BCL2 suppresses c-MYC-driven apoptosis (45–47). However, our miRNA and protein expression patterns lead us to believe that BCL2 is downregulated via the action of miR-16. MiR-16 is significantly upregulated at 8 h.p.i., with a strong trend toward upregulation at 3 d.p.i. (Figure 2). It is possible that this decrease in cellular BCL2 is functioning to accent c-MYC-directed priming of apoptosis.
Moreover, apoptosis in bystander cells is also regulated via the miR-29 family, which mediates apoptosis through the regulation of MCL1. MCL1 is a tightly controlled BCL2 family member, which is important in regulating TRAIL-mediated apoptosis (42). MCL1 functions as a prosurvival protein by binding proapoptotic BH3-only BCL2 family members, such as BIM, BID, BIK, NOXA and PUMA (48). Although the exact mechanisms by which the BCL2 family members mediate apoptosis is unknown, it has been shown that MCL1 binding of BID and BIM proteins protects against TRAIL-induced cell death (42). Interestingly, it has been shown that TRAIL mediates cell–cell apoptotic RIBE (49,50). This study and our previous analysis show that bystander cells exhibit increased levels of apoptosis. Furthermore, the data of this study may suggest that tumor necrosis factor-mediated cytokine signaling in RIBE may be associated with miR-29 family.
Overall, the miR-29 family plays a dual role in bystander tissues promoting both global hypomethylation through the regulation of de novo methyltransferases DNMT3A and preparing the cell for TRAIL-mediated apoptosis through repression of anti-apoptotic MCL1. We also show that miRNA regulation in bystander tissues has been dramatically changed at 3 d.p.i. Interestingly, this corresponds to the apoptotic levels observed in this study and with the maximum apoptotic and hypomethylation levels previously seen in EpiAirway tissues under similar conditions (10). Given that there are significant changes in miRNA expression in bystander tissues, further investigation into the role of the miR-29 family in RIBE may come to discover that this family can acts as a good biomarker for two commonly observed bystander responses.
In previous experiments (10), there were no large-scale changes in molecular events at the 8 h.p.i. time point. This is logical, as it takes time for the bystander signal to propagate through cells and for global cellular changes to manifest. However, finding changes in miRNA expression at early time points, before the manifestation of bystander symptoms, could suggest that miRNA regulation is an upstream event of some bystander responses. Given that miRNAs may travel through gap junctions and gap junctions are necessary for bystander effects in some models (51), one can easily envisage a future model where miRNAs from irradiated cells are quickly transported through gap junctions to prepare bystander cells for future secreted signals.
Though miRNAs mediate crucial bystander effect end points and exhibit their effects as early as 8 h.p.i., they may not necessarily be primary bystander signals. Further studies are needed to dissect the potential role of miRNAs as bystander signals and to gain further mechanistic insight into the roles of miRNAs in irradiated and bystander tissues and the potential of miRNAs to enhance or suppress the RIBE in vivo. This challenging task can be achieved through transient transformation of irradiated and bystander cells and tissues using miRNAs or their inhibitors (antago-miRs or anti-miRs) (52,53). Following the transient transformation, the impact of miRNAs or anti-miRNAs on preventing or enhancing changes in the functional RIBE readouts can be established.
Interestingly, RIBE may be mediated by reactive oxygen species (ROS) (54). A recent study has confirmed that IR-induced oxidative stress significantly alters microRNAome of the exposed cells (55). Furthermore, the results clearly demonstrated a common miRNA expression signature in response to radiation, hydrogen peroxide and etoposide exposure (55). Though we have not seen a major overlap between our dataset and the reported IR- and ROS-induced miRNAs changes, some interesting parallels can be drawn. Specifically, miR-15b was upregulated by IR and ROS, whereas in our study, miR-16 was significantly upregulated in bystander tissues. These miRNAs belong to the same miRNA family and play roles in regulation of cell cycle and apoptosis (56–58). Their roles in ROS-induced bystander effects and their differential regulation in responses to direct IR and RIBE need to be further analyzed.
Overall, in the future, the current study may serve as a roadmap for further understanding the mechanistic roles of miRNAs in bystander effects and for dissecting a hierarchy and cross talk between epigenetic parameters (microRNAome and DNA methylation) and well-known RIBE manifestations.
Supplementary material
Supplementary Figures 1 and 2 can be found at http://carcin.oxfordjournals.org/
Funding
National Science and Engineering Research Council of Canada to O.K., F.J.Z., J.F., A.A.; The National Institute of Biomedical Imaging and Bioengineering to G.J.-B., S.A.M., D.J.B. (NIBIB 5 P41 EB002033-12); Intramural Research Program of the National Cancer Institute, Center for Cancer Research, National Institutes of Health (NIH) and the NIAID Radiation/Nuclear Countermeasures Program; the Intramural Research Program of the National Cancer Institute, Center for Cancer Research, NIH to J.S.D., W.M.B., O.A.S.
Supplementary Material
Acknowledgments
We appreciate the help of Dr Valentina Titova in proofreading this manuscript.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- ANOVA
analysis of variance
- AUF
arbitrary units of fluorescence
- 3-D
three-dimensional
- d.p.i.
days post-IR
- DSB
double-strand break
- h.p.i.
h post-IR
- IR
ionizing radiation
- miRNA
microRNA
- ROS
reactive oxygen species
- RIBE
radiation-induced bystander effect
- TRAIL
tumor necrosis factor-related apoptosis-inducing ligand
References
- 1.Maguire P, et al. Modulation of radiation responses by pre-exposure to irradiated cell conditioned medium. Radiat. Res. 2007;167:485–492. doi: 10.1667/RR0159.1. [DOI] [PubMed] [Google Scholar]
- 2.Sokolov MV, et al. Ionizing radiation induces DNA double-strand breaks in bystander primary human fibroblasts. Oncogene. 2005;24:7257–7265. doi: 10.1038/sj.onc.1208886. [DOI] [PubMed] [Google Scholar]
- 3.Lyng FM, et al. The involvement of calcium and MAP kinase signaling pathways in the production of radiation-induced bystander effects. Radiat. Res. 2006;165:400–409. doi: 10.1667/rr3527.1. [DOI] [PubMed] [Google Scholar]
- 4.Gaugler MH, et al. Intestinal epithelial cell dysfunction is mediated by an endothelial-specific radiation-induced bystander effect. Radiat. Res. 2007;167:185–193. doi: 10.1667/rr0702.1. [DOI] [PubMed] [Google Scholar]
- 5.Vines AM, et al. Bystander effect induced changes in apoptosis related proteins and terminal differentiation in in vitro murine bladder cultures. Int. J. Radiat. Biol. 2009;85:48–56. doi: 10.1080/09553000802635047. [DOI] [PubMed] [Google Scholar]
- 6.Belyakov OV, et al. Bystander-induced differentiation: a major response to targeted irradiation of a urothelial explant model. Mutat. Res. 2006;597:43–49. doi: 10.1016/j.mrfmmm.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 7.Persaud R, et al. Assessment of low linear energy transfer radiation-induced bystander mutagenesis in a three-dimensional culture model. Cancer Res. 2005;65:9876–9882. doi: 10.1158/0008-5472.CAN-04-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pinto M, et al. Bystander responses in three-dimensional cultures containing radiolabelled and unlabelled human cells. Radiat. Prot Dosimetry. 2006;122:252–255. doi: 10.1093/rpd/ncl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belyakov OV, et al. Biological effects in unirradiated human tissue induced by radiation damage up to 1 mm away. Proc. Natl Acad. Sci. USA. 2005;102:14203–14208. doi: 10.1073/pnas.0505020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sedelnikova OA, et al. DNA double-strand breaks form in bystander cells after microbeam irradiation of three-dimensional human tissue models. Cancer Res. 2007;67:4295–4302. doi: 10.1158/0008-5472.CAN-06-4442. [DOI] [PubMed] [Google Scholar]
- 11.Morgan WF, et al. Non-targeted bystander effects induced by ionizing radiation. Mutat. Res. 2007;616:159–164. doi: 10.1016/j.mrfmmm.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 12.Morgan WF. Non-targeted and delayed effects of exposure to ionizing radiation: I. Radiation-induced genomic instability and bystander effects in vitro. Radiat Res. 2003;159:567–580. doi: 10.1667/0033-7587(2003)159[0567:nadeoe]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 13.Morgan WF. Is there a common mechanism underlying genomic instability, bystander effects and other nontargeted effects of exposure to ionizing radiation? Oncogene. 2003;22:7094–7099. doi: 10.1038/sj.onc.1206992. [DOI] [PubMed] [Google Scholar]
- 14.Morgan WF. Non-targeted and delayed effects of exposure to ionizing radiation: II. Radiation-induced genomic instability and bystander effects in vivo, clastogenic factors and transgenerational effects. Radiat. Res. 2003;159:581–596. doi: 10.1667/0033-7587(2003)159[0581:nadeoe]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 15.Mothersill C, et al. Radiation-induced bystander effects—implications for cancer. Nat. Rev. Cancer. 2004;4:158–164. doi: 10.1038/nrc1277. [DOI] [PubMed] [Google Scholar]
- 16.Kaup S, et al. Radiation-induced genomic instability is associated with DNA methylation changes in cultured human keratinocytes. Mutat. Res. 2006;597:87–97. doi: 10.1016/j.mrfmmm.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 17.Wright EG, et al. Untargeted effects of ionizing radiation: implications for radiation pathology. Mutat. Res. 2006;597:119–132. doi: 10.1016/j.mrfmmm.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 18.Jaenisch R, et al. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33(suppl.):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 19.Eulalio A, et al. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 20.Foshay KM, et al. Small RNAs, big potential: the role of microRNAs in stem cell function. Curr. Stem Cell Res. Ther. 2007;2:264–271. doi: 10.2174/157488807782793781. [DOI] [PubMed] [Google Scholar]
- 21.Carleton M, et al. MicroRNAs and cell cycle regulation. Cell Cycle. 2007;6:2127–2132. doi: 10.4161/cc.6.17.4641. [DOI] [PubMed] [Google Scholar]
- 22.Jovanovic M, et al. miRNAs and apoptosis: RNAs to die for. Oncogene. 2006;25:6176–6187. doi: 10.1038/sj.onc.1209912. [DOI] [PubMed] [Google Scholar]
- 23.Fabbri M, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl Acad. Sci. USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koturbash I, et al. Sex-specific microRNAome deregulation in the shielded bystander spleen of cranially exposed mice. Cell Cycle. 2008;7:1658–1667. doi: 10.4161/cc.7.11.5981. [DOI] [PubMed] [Google Scholar]
- 25.Tamminga J, et al. DNA damage-induced upregulation of miR-709 in the germline downregulates BORIS to counteract aberrant DNA hypomethylation. Cell Cycle. 2008;7:3731–3736. doi: 10.4161/cc.7.23.7186. [DOI] [PubMed] [Google Scholar]
- 26.Ilnytskyy Y, et al. Altered microRNA expression patterns in irradiated hematopoietic tissues suggest a sex-specific protective mechanism. Biochem. Biophys. Res. Commun. 2008;377:41–45. doi: 10.1016/j.bbrc.2008.09.080. [DOI] [PubMed] [Google Scholar]
- 27.Koturbash I, et al. Role of epigenetic effectors in maintenance of the long-term persistent bystander effect in spleen in vivo. Carcinogenesis. 2007;28:1831–1838. doi: 10.1093/carcin/bgm053. [DOI] [PubMed] [Google Scholar]
- 28.Boelsma E, et al. Characterization and comparison of reconstructed skin models: morphological and immunohistochemical evaluation. Acta Derm. Venereol. 2000;80:82–88. [PubMed] [Google Scholar]
- 29.Ziegler A, et al. Aberration-corrected microscopy for structural biology applications. J. Microsc. 2009;233:170–177. doi: 10.1111/j.1365-2818.2008.03106.x. [DOI] [PubMed] [Google Scholar]
- 30.Bonner WM, et al. GammaH2AX and cancer. Nat. Rev. Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kovalchuk O, et al. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol. Cancer Ther. 2008;7:2152–2159. doi: 10.1158/1535-7163.MCT-08-0021. [DOI] [PubMed] [Google Scholar]
- 32.Bolstad BM, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 33.Eisen MB, et al. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogakou EP, et al. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 35.O'Donnell KA, et al. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 36.Petrocca F, et al. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68:8191–8194. doi: 10.1158/0008-5472.CAN-08-1768. [DOI] [PubMed] [Google Scholar]
- 37.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsumura I, et al. E2F1 and c-Myc in cell growth and death. Cell Cycle. 2003;2:333–338. [PubMed] [Google Scholar]
- 39.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leiderman YI, et al. Molecular genetics of RB1—the retinoblastoma gene. Semin. Ophthalmol. 2007;22:247–254. doi: 10.1080/08820530701745165. [DOI] [PubMed] [Google Scholar]
- 41.Calin GA, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mott JL, et al. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knoepfler PS. Myc goes global: new tricks for an old oncogene. Cancer Res. 2007;67:5061–5063. doi: 10.1158/0008-5472.CAN-07-0426. [DOI] [PubMed] [Google Scholar]
- 44.Coller HA, et al. "Myc'ed messages": myc induces transcription of E2F1 while inhibiting its translation via a microRNA polycistron. PLoS Genet. 2007;3:e146. doi: 10.1371/journal.pgen.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nieminen AI, et al. c-Myc primed mitochondria determine cellular sensitivity to TRAIL-induced apoptosis. EMBO J. 2007;26:1055–1067. doi: 10.1038/sj.emboj.7601551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nieminen AI, et al. c-Myc blazing a trail of death: coupling of the mitochondrial and death receptor apoptosis pathways by c-Myc. Cell Cycle. 2007;6:2464–2472. doi: 10.4161/cc.6.20.4917. [DOI] [PubMed] [Google Scholar]
- 47.Bissonnette RP, et al. Apoptotic cell death induced by c-myc is inhibited by bcl-2. Nature. 1992;359:552–554. doi: 10.1038/359552a0. [DOI] [PubMed] [Google Scholar]
- 48.Chen T, et al. Induction of apoptosis in mouse liver by microcystin-LR: a combined transcriptomic, proteomic, and simulation strategy. Mol. Cell. Proteomics. 2005;4:958–974. doi: 10.1074/mcp.M400185-MCP200. [DOI] [PubMed] [Google Scholar]
- 49.Huang X, et al. Cell to cell contact required for bystander effect of the TNF-related apoptosis-inducing ligand (TRAIL) gene. Int. J. Oncol. 2003;22:1241–1245. [PubMed] [Google Scholar]
- 50.Shareef MM, et al. Role of tumor necrosis factor-alpha and TRAIL in high-dose radiation-induced bystander signaling in lung adenocarcinoma. Cancer Res. 2007;67:11811–11820. doi: 10.1158/0008-5472.CAN-07-0722. [DOI] [PubMed] [Google Scholar]
- 51.Azzam EI, et al. Direct evidence for the participation of gap junction-mediated intercellular communication in the transmission of damage signals from alpha-particle irradiated to nonirradiated cells. Proc. Natl Acad. Sci. USA. 2001;98:473–478. doi: 10.1073/pnas.011417098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Rooij E, et al. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ. Res. 2008;103:919–928. doi: 10.1161/CIRCRESAHA.108.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elmen J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 54.Sedelnikova OA, et al. Role of oxidatively induced DNA lesions in human pathogenesis. Mutat. Res. 2010;704:152–159. doi: 10.1016/j.mrrev.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simone NL, et al. Ionizing radiation-induced oxidative stress alters miRNA expression. PLoS One. 2009;4 doi: 10.1371/journal.pone.0006377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo CJ, et al. miR-15b and miR-16 are implicated in activation of the rat hepatic stellate cell: an essential role for apoptosis. J. Hepatol. 2009;50:766–778. doi: 10.1016/j.jhep.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 57.Xia H, et al. MicroRNA-15b regulates cell cycle progression by targeting cyclins in glioma cells. Biochem. Biophys. Res. Commun. 2009;380:205–210. doi: 10.1016/j.bbrc.2008.12.169. [DOI] [PubMed] [Google Scholar]
- 58.Xia L, et al. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int. J. Cancer. 2008;123:372–379. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.