Abstract
Curatively treated patients with early-stage head and neck squamous cell carcinoma (HNSCC) are at high risks for second primary tumor (SPT) and recurrence. The regulator of G-protein signaling (RGS) is important in essential signaling transduction and cellular activities. We hypothesize that genetic variations of RGS may modulate the risk of SPT/recurrence in patients with early-stage HNSCC. In a nested case–control study, we evaluated 98 single-nucleotide polymorphisms (SNPs) in 17 RGS genes for the risk of SPT/recurrence among 450 HNSCC patients. Eight SNPs showed significant associations with the risk of SPT/recurrence, with the most significant one of rs2179653, which is located in the 5′-flanking region of RGS2 gene. Under a recessive genetic model, the homozygous variant genotype of this SNP was associated with 2.95-fold [95% confidence interval (CI): 1.52–5.74] increased risk of SPT/recurrence. This association remained significant after the adjustment for multiple comparisons. Cumulative effects analysis revealed that the risk increased significantly with the increasing numbers of unfavorable genotypes. Compared with subjects carrying 0–2 unfavorable genotypes, the hazard ratios (95% CIs) for those carrying 3 or 4+ were 1.73 (1.10–2.70) and 3.05 (1.92–4.83), respectively. Furthermore, survival tree analysis revealed potential higher order gene–gene interactions and indicated different outcomes based on distinct genotype profiles. Genetic variations of RGS genes may modulate the susceptibility to SPT/recurrence in early-stage HNSCC patients individually and cumulatively. Our results stressed the importance of taking a polygenic approach to evaluate the cumulative and interaction effects of genetic variations in the prediction of cancer risk and prognosis.
Introduction
Surgery and radiotherapy are highly effective for patients with early-stage (I or II) head and neck squamous cell carcinoma (HNSCC) (1); however, up to 25% of these patients will develop second primary tumor (SPT) or local recurrence after 5 years of initial diagnosis (1,2), which has been a competing cause of posttreatment morbidity and mortality (3). Therefore, identification of clinically applicable biomarkers for the prediction of SPT/recurrence is important in the achievement of targeted interventions and long-term survival of early-stage HNSCC patients.
G-proteins are a family of proteins involved in cellular signal transduction (4). They are expressed in all cells of human body and function as ‘molecular switches’ by turning on intracellular signaling cascades in response to the activation of G-protein-coupled receptors (GPCRs) (5,6). GPCRs, with >800 members, comprise one of the largest families of cell-surface molecules (7). It plays a pivotal role in integrating the stimulatory signals and inhibitory signals by interplaying with G-proteins (8). The G-protein-coupled biological process is important for the development of increasing number of human diseases and requires fine-tuning through accessory molecules such as the regulator of G-protein signaling (RGS) (9). RGS are a family of cellular proteins with conserved domains of ∼120 amino acid residues (10). Aberration of RGS proteins has been implicated in the pathogenesis of many common human disorders and drug addiction (11–14). Nonetheless, the roles of RGS genes in tumorigenesis have remained largely unexplored.
There are multiple RGS subfamilies consisting of >20 different RGS proteins, ranging from small ones comprised solely of an RGS domain to multidomain proteins with functions in various signaling pathways (10,15). These multiple domains of RGS protein mediate interactions with other signaling pathways, allowing RGS proteins to serve as signaling scaffolds (15). Although the functions of various RGS genes are diverse, they all operate under similar mechanism (i.e. they all serve as guanosine triphosphatase-activating protein to accelerate guanosine triphosphate hydrolysis) to regulate various signaling pathways involved in growth and development. Genetic variations of these RGS subfamily genes have already shown to be associated with various common human diseases such as hypertension (16) and schizophrenia (17). Recent reports also have linked RGS domain containing genes to cancers (18–20). For instance, genetic fine mapping of chromosome 6p23–25 region revealed the potential link of single-nucleotide polymorphisms (SNPs) in RGS17 with familial lung cancer etiology (18). In addition, our group also found that potential functional SNPs in RGS2 and RGS6 gene might modulate the risk of bladder and lung cancer (19,20). Experimental studies have demonstrated that aberrant expression of RGS gene is associated with abnormal cell growth, which contributes to the carcinogenesis of thyroid and prostate cancer (21,22). Moreover, RGS expression has also been established as playing a pivotal role in vascular maturation and vessel remodeling during carcinogenesis (9). Angiogenesis from an existing vasculature is widely recognized as a necessary requirement for most tumor growth, which confers to the occurrence and progression of many cancers (23).
Hence, we hypothesize that genetic variations of RGS genes might modulate the risk of SPT/recurrence in curatively treated early-stage HNSCC patients. To test this hypothesis, we conducted a nested case–control study built upon a HNSCC prospective chemoprevention clinical trial to evaluate the effects of 98 SNPs in 17 RGS genes. To our knowledge, this is the first study to explore a comprehensive panel of genetic polymorphisms of RGS genes in the risk of SPT/recurrence.
Materials and methods
Study population
In this study, we enrolled patients from a randomized placebo-controlled Retinoid Head and Neck Second Primary Trial launched in 1991 and closed to new patient registration in 1999 (24). In this trial, patients with stage I or II head and neck cancer of the larynx, oral cavity or pharynx were randomized to receive either 13-cis-retinoic acid at daily low dose or placebo for 3 years followed by 4 years of observation. The stratification criteria for randomization included the primary tumor site, tumor stage and smoking status. A total of 1384 patients were registered and 1191 were eligible to be randomized into the study. Patients were followed up at 3, 6, 9, 12, 16, 20, 24, 28, 32 and 36 months after randomization, then semiannually followed up for 4 more years. Double-blind strategy was implemented to ensure that neither the patients nor the physicians/investigators were aware of which study agent was being taken. During the follow-up period, 354 patients developed SPT/recurrence. The definitions of SPT and recurrence following the Warren and Gates criteria were provided previously (24). Based on this prospective clinical trial, we conducted a nested case–control study including 150 HNSCC patients with SPT/recurrence (cases) that were frequently matched with 300 SPT/recurrence-free survivors (controls) by age (±5 years), sex and ethnicity. To further explore the potential selection bias in this study, we compared the distribution of key characteristics between these 150 cases and others with SPT/recurrence not being involved in the study. No significant difference was found on the distribution of age, gender, smoking status and clinical characteristics (P > 0.05).
Data collection
The study was approved by Institutional Review Board of The University of Texas M. D. Anderson Cancer Center. Written informed consent was obtained from all participants. Before randomization, patients were given a structured questionnaire that elicited information on socio-demographic factors, clinical information, tobacco exposure and alcohol consumption. Blood samples were collected and delivered to M. D. Anderson Cancer Center to be used for molecular analyses. Smoking status was assessed at the entry and during the study. The definitions of never-, former- and current-smoker were described previously (25). Never-smokers were individuals who had smoked <100 total cigarettes during their lifetime. Former-smokers were individuals who had stopped smoking for at least 1 year at the time of enrollment.
SNPs selection and genotyping
Seventeen genes in RGS family were selected from a customized cancer gene panel. The detailed procedure to compile this panel was described previously (25). The complete set of selected SNPs was sent to Illumina technical support for the Infinium II chemistry designability and bead type analyses using a program developed by Illumina (San Diego, CA). Supplementary Table 1 (available at Carcinogenesis Online) lists the complete set of genes and SNPs evaluated in this study. Genomic DNA was extracted from peripheral blood lymphocytes. Genotyping was carried out according to the standard protocol provided by Illumina.
Statistical analysis
Statistical analyses were performed using STATA 10.0 (StataCorp LP, College Station, TX) and the R software. The χ2 test (for categorical variables) or Student's t-test (for continuous variables) were used to compare characteristics between groups with and without SPT/recurrence. Multivariate Cox proportional hazard model was applied to calculate hazard ratios (HRs) and 95% confidence intervals (CIs) associated with the effect of SNPs on the development of SPT/recurrence while adjusting for age, sex, tobacco smoking, ethnicity, tumor stage, primary tumor site and treatment. Time to SPT/recurrence development was defined as the time between randomization and SPT/recurrence diagnosis. All SPT/recurrence-free patients were censored at the date of last follow-up. Time for controls without SPT/recurrence was defined as the time between randomization and the date of last follow-up or censor. We tested three different genetic models, including dominant model, recessive model and additive model. The model with the smallest P value was deemed to be the best-fitting model. To adjust for multiple comparisons, q-value was calculated using the R software. The q-value of a test measures the proportion of false positives incurred (false discovery rate) when that particular test of SNP is called significant in the main effect analysis (26,27). Higher order gene–gene interactions were explored with a survival tree-based data analysis, which was performed by using the software ‘Stree' (http://c2s2.yale.edu/software/stree/). Stree is a recursive partitioning technique, which allows identifying effect modifications between variables that are less visible by traditional regression model. Cumulative effects of SNPs in RGS genes were assessed using the unfavorable genotype analysis in which significant SNPs (P for the best-fitting model <0.05) identified from the single SNP analysis were combined by counting the number of unfavorable genotypes for each subject. Unfavorable genotypes were defined by referring the HRs of genotypes showing a significant association in single SNP analysis. We classified each subject into three risk categories based on the tertile distribution of the numbers of unfavorable genotypes in the SPT/recurrence-free group. We stratified the cumulative effects by selected host and environmental factors. Assessment of statistical interaction was conducted by adding a multiplicative term of genetic polymorphisms and environmental variables in the multivariate Cox regression model and the significance of the interaction term was tested by using likelihood ratio test. The differences between distinct risk categories were compared using the Kaplan–Meier curves and logrank test. All tests were two sided with a significant level of P <0.05 based.
Results
Patient characteristics
The host and clinical characteristics of the study subjects have been described elsewhere (25) (supplementary Table 2 is available at Carcinogenesis Online). After excluding subjects with >5% missing genotypes, 440 HNSCC patients (147 cases and 293 controls) were included in the analyses. The median time of follow-up was 2.3 years among cases (event-free time) and 5.0 years among controls. The majority of the patients were males (79.55%) and Caucasians (96.14%). The average age of all patients (mean ± SD) was 61.15 ± 10.25 years. The SPT/recurrence group had more patients with pharyngeal cancer (21.09 versus 8.53%) and fewer patients with laryngeal cancer (47.62 versus 62.82%) than the non-SPT/recurrence group (P < 0.001). No significance was found in the distribution of smoking status (P = 0.167), tumor stage (P = 0.169), surgery (P = 0.258), radiotherapy (P = 0.901) or 13-cis-retinoic acid treatment (P = 0.399).
Association between RGS SNPs and SPT/recurrence risk
After excluding SNPs with >5% missing calls, 95 of 98 SNPs in 17 RGS genes were analyzed in 440 study subjects (supplementary Table 2 is available at Carcinogenesis Online). Among them, eight SNPs (rs2179653 of RGS2, rs3795617 of RGS13, rs6670735 of RGS8, rs739999 of RGS11, rs11586945 of RGS5, rs3747813 of RGS3, rs6689169 and rs6700378 of RGS7) were significantly associated with an altered risk of SPT/recurrence (Table I). As rs6689169 and rs6700378 were completely linked (r2 = 1.0), only rs6689169 was kept in the following analyses. Among eight SNPs with significant main effects, two SNPs (rs2179653 and rs3795617) remained significant after adjustment for multiple comparisons at q-value <0.05 (q-values were 0.010 for rs2179653 and 0.015 for rs3795617, respectively). For rs2179653, homozygous variant genotype AA conferred to a 2.95-fold increased risk for SPT/recurrence, whereas AA genotype of rs3795617 was associated with a reduced risk (HR = 0.52, 95% CI: 0.34–0.81) (Table I).
Table I.
Associations between RGS genes genetic polymorphisms and SPT/recurrence in HNSCC
| Gene | SNP | Position | Region | Genotype | SPT/recurrence |
Best fitting model |
||||||
| Yes, n (%) | No, n (%) | HR (95% CI)a | P value | Modelb | HR (95% CI)a | P | q | |||||
| RGS2 | rs2179653 | chr1:191037685 | 5′ flanking | GG | 103 (70.07) | 208 (70.99) | 1 | |||||
| GA | 34 (23.13) | 81 (27.65) | 0.97 (0.65–1.44) | 0.877 | Rec | 2.95 (1.52–5.74) | 0.001 | 0.010 | ||||
| AA | 10 (6.80) | 4 (1.37) | 2.93 (1.50–5.73) | 0.002 | ||||||||
| RGS13 | rs3795617 | chr1:190870313 | Near 5′ | GG | 42 (28.57) | 87 (29.69) | 1 | |||||
| GA | 80 (54.42) | 119 (40.61) | 1.18 (0.81–1.72) | 0.395 | Rec | 0.52 (0.34–0.81) | 0.003 | 0.015 | ||||
| AA | 25 (17.01) | 87 (29.69) | 0.58 (0.35–0.96) | 0.035 | ||||||||
| RGS8 | rs6670735 | chr1:180909480 | Near 5′ | AA | 68 (46.26) | 111 (38.01) | 1 | |||||
| AG | 58 (39.46) | 147 (50.34) | 0.64 (0.45–0.92) | 0.015 | Dom | 0.69 (0.49–0.96) | 0.028 | 0.053 | ||||
| GG | 21 (14.29) | 34 (11.64) | 0.86 (0.52–1.42) | 0.553 | ||||||||
| RGS11 | rs739999 | chr16:259512 | Exon | AA | 113 (77.40) | 242 (82.59) | 1 | |||||
| AG | 28 (19.18) | 47 (16.04) | 1.16 (0.76–1.77) | 0.492 | Rec | 2.88 (1.06–7.78) | 0.037 | 0.053 | ||||
| GG | 5 (3.42) | 4 (1.37) | 2.99 (1.10–8.13) | 0.031 | ||||||||
| RGS5 | rs11586945 | chr1:161420559 | Intron | GG | 96 (65.31) | 206 (70.31) | 1 | |||||
| GC | 41 (27.89) | 76 (25.94) | 1.11 (0.77–1.62) | 0.568 | Rec | 2.00 (1.04–3.85) | 0.038 | 0.053 | ||||
| CC | 10 (6.80) | 11 (3.75) | 2.06 (1.06–3.99) | 0.033 | ||||||||
| RGS3 | rs3747813 | chr9:115367281 | 5′ UTR | GG | 136 (92.52) | 258 (88.05) | 1 | |||||
| GA | 11 (7.48) | 34 (11.60) | 0.54 (0.29–1.02) | 0.056 | Dom | 0.53 (0.28–0.98) | 0.043 | 0.053 | ||||
| AA | 0 | 1 (0.34) | — | |||||||||
| RGS7 | rs6689169c | chr1:239005040 | Near 3′ | AA | 120 (81.63) | 211 (72.01) | 1 | |||||
| AG | 24 (16.33) | 79 (26.96) | 0.60 (0.38–0.94) | 0.026 | Dom | 0.65 (0.42–0.99) | 0.047 | 0.053 | ||||
| GG | 3 (2.04) | 3 (1.02) | 1.65 (0.52–5.24) | 0.397 | ||||||||
UTR, untranslated region.
Adjusting for age, sex, smoking, ethnicity, tumor stage, primary tumor site and treatment.
Dom, dominant model; Rec, recessive model.
rs6700378 is completely linked with rs6689169 and is not shown in the table and involved in the analysis. Bold numbers represent P values that are statistically significant at P < 0.05.
Cumulative effects of unfavorable genotypes of RGS genes
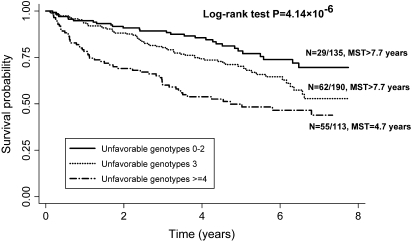
To further assess the cumulative effects of the SNPs in RGS genes on SPT/recurrence, we conducted an unfavorable genotype analysis by combining seven SNPs identified as significant in the main effect analysis (Table II). Compared with patients in the low-risk group with <3 unfavorable genotypes, the HRs (95% CIs) for medium-risk group (three unfavorable genotype) or high-risk group (≥4 unfavorable genotypes) were 1.73 (1.10–2.70) and 3.05 (1.92–4.83), respectively. In addition, we also observed a significant gene–dose effect with an increasing number of unfavorable genotypes (P for trend = 1.28 × 10−6) (Table II). The Kaplan–Meier survival curves for different risk groups were shown in Figure 1. The median survival times (event-free times) were 4.7 years for patients in the high-risk group but were longer than 7.7 years for patients in the low-risk and medium-risk groups, respectively (logrank test: P = 4.14 × 10−6). The effects of cumulative unfavorable genotypes were further stratified by selected variables including smoking status, tumor stage, primary tumor site and 13-cis-retinoic acid treatment (Table III). The gene dose remained evident in each stratum except for never-smokers (P for trend = 0.636). No significant interactions were found between cumulative effects of unfavorable genotypes and the stratified variables.
Table II.
The cumulative effects of unfavorable genotypes of RGS genes on SPT/recurrence in HNSCC
| Number of unfavorable genotypesa | SPT/recurrence | HR (95% CI)b | P | |
| Yes, n (%) | No, n (%) | |||
| 0–2 | 29 (19.86) | 106 (36.30) | 1 | |
| 3 | 62 (42.47) | 128 (43.84) | 1.73 (1.10–2.70) | 0.017 |
| ≥4 | 55 (37.67) | 58 (19.86) | 3.05 (1.92–-4.83) | 2.04 × 10−6 |
| P for trend | 1.28 × 10−6 | |||
Unfavorable genotypes: rs2179653 (AA), rs3795617 (GG + GA), rs6670735 (AA), rs739999 (GG), rs11586945 (CC), rs3747813 (GG) and rs6689169 (AA).
Adjusting for age, sex, smoking, ethnicity, tumor stage, primary tumor site and treatment. Bold numbers represent P values that are statistically significant at P < 0.05.
Fig. 1.
Kaplan–Meier survival estimates for patients carrying unfavorable genotypes of RGS gene in the risk of SPT/recurrence. MST, median survival time (event-free time).
Table III.
The cumulative effects of unfavorable genotypes of RGS genes stratified by selected factors
| Characteristics | Number of unfavorable genotypes | SPT/recurrence |
HR (95% CI)a | P | P for trend | |
| Yes, n (%) | No, n (%) | |||||
| Smoking | ||||||
| Never | 0–2 | 5 (26.32) | 18 (36.00) | 1 | ||
| 3 | 9 (47.37) | 16(32.00) | 1.67 (0.53–5.25) | 0.384 | ||
| ≥4 | 5 (26.32) | 16 (32.00) | 1.35 (0.37–4.84) | 0.649 | 0.636 | |
| Former | 0–2 | 14 (21.88) | 57 (39.58) | 1 | ||
| 3 | 29 (45.31) | 63(43.75) | 1.70 (0.87–3.32) | 0.118 | ||
| ≥4 | 21 (32.81) | 24(16.67) | 2.73 (1.33–5.60) | 0.006 | 0.006 | |
| Current | 0–2 | 10 (15.87) | 31 (31.63) | 1 | ||
| 3 | 24 (38.10) | 49 (50.00) | 1.53 (0.72–3.28) | 0.270 | ||
| ≥4 | 29 (46.03) | 18 (18.37) | 3.96 (1.89–8.28) | <0.001 | <0.001 | |
| Tumor stage | ||||||
| I | 0–2 | 15 (17.44) | 75 (38.86) | 1 | ||
| 3 | 40 (46.51) | 74 (38.34) | 2.42 (1.32–4.43) | 0.004 | ||
| ≥4 | 31 (36.05) | 44 (22.80) | 3.17 (1.69–5.95) | <0.001 | <0.001 | |
| II | 0–2 | 14 (23.33) | 31 (31.31) | 1 | ||
| 3 | 22 (36.67) | 54 (54.55) | 1.08 (0.55–2.13) | 0.826 | ||
| ≥4 | 24 (40.00) | 14 (14.14) | 3.20 (1.60–6.43) | 0.001 | 0.001 | |
| Primary tumor site | ||||||
| Larynx | 0–2 | 15 (21.43) | 61 (32.62) | 1 | ||
| 3 | 29 (41.43) | 88 (47.06) | 1.28 (0.68–2.42) | 0.445 | ||
| ≥4 | 26 (37.14) | 38 (20.32) | 2.64 (1.36–5.14) | 0.004 | 0.004 | |
| Oral cavity | 0–2 | 8 (17.78) | 31 (38.75) | 1 | ||
| 3 | 22 (48.89) | 30 (37.50) | 2.66 (1.16–6.10) | 0.021 | ||
| ≥4 | 15 (33.33) | 19 (23.75) | 2.64 (1.11–6.29) | 0.029 | 0.032 | |
| Pharynx | 0–2 | 6 (19.35) | 14 (56.00) | 1 | ||
| 3 | 11 (35.48) | 10 (40.00) | 1.92 (0.69–5.35) | 0.210 | ||
| ≥4 | 14 (45.16) | 1 (4.00) | 5.67 (1.91–16.87) | 0.002 | 0.002 | |
| 13 cis retinoic acid treatment | ||||||
| No | 0–2 | 15 (20.00) | 53 (38.13) | 1 | ||
| 3 | 34 (45.33) | 60 (43.17) | 1.84 (1.00–3.39) | 0.051 | ||
| ≥4 | 26 (34.67) | 26 (18.71) | 2.67 (1.41–5.07) | 0.003 | 0.002 | |
| Yes | 0–2 | 14 (19.72) | 53 (34.64) | 1 | ||
| 3 | 28 (39.44) | 68 (44.44) | 1.58 (0.81–3.08) | 0.175 | ||
| ≥4 | 29 (40.85) | 32 (20.92) | 3.43 (1.76–6.71) | <0.001 | <0.001 | |
Adjusting for age, sex, smoking, ethnicity, tumor stage, primary tumor site and treatment where appropriate. Bold numbers represent P values that are statistically significant at P < 0.05.
Multivariate analysis by tree-based method
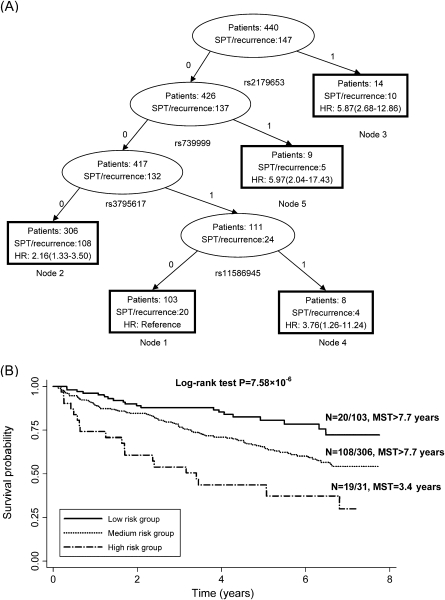
Survival tree model partitioning of 440 patients was performed and displayed in Figure 2. By using significant SNPs (P for the best-fitting model <0.05) identified from the single SNP analysis as attributes for tree construction, the resulting tree with five terminal nodes was first split by rs2179653 of RGS2, following by rs739999 of RGS11, rs3795617 of RGS13 and rs11586945 of RGS5 (Figure 2A). Furthermore, a multivariate proportional hazard model revealed the ability of predicting SPT/recurrence among early-stage HNSCC patients. Node 1 comprised individuals exhibiting the lowest risk (median survival time > 7.75 years) was defined as the reference node. Node 5 conferred to the highest risk for SPT/recurrence with HR (95% CI) of 5.97 (2.04–17.43). The terminal nodes were then categorized into three groups: low-risk (node 1, reference), medium-risk (node 2, HR > 1 but <3) and high-risk (node 3, 4 and 5, HR ≥ 3). Based on the risk classification from the survival tree model, Kaplan–Meier curves were plotted for groups 1–3 (Figure 2B). The risk for SPT/recurrence development was significantly different among these 3 groups (Log-rank test, P = 7.58 × 10−6). Compared with the low-risk group, the medium-risk and high-risk group conferred 2.16-fold (HR: 2.16, 95% CI: 1.33–3.50) and 5.24-fold (HR: 5.24, 95% CI: 2.71–10.11) increased SPT/recurrence risk, respectively (P trend <0.001) (Table IV).
Fig. 2.
Survival tree analysis on the association between RGS genetic polymorphisms and SPT/recurrence among HNSCC patients. (A) Survival tree model partitioning of 440 patients was performed. The resulting tree with five terminal nodes was split by rs2179653, rs739999, rs3795617 and rs11586945. For each SNP, ‘0’ represents common homozygous genotype; ‘1’ represents heterozygous or homozygous variant genotype. (B) Kaplan–Meier curves based on survival tree analysis in RGS genes for selected nodes. MST, median survival time (event-free time).
Table IV.
Cox proportional hazard model in HNSCC patients based on the survival tree analysis
| Group | Observed | SPT/recurrence (%) | HR (95% CI)a | P |
| Low-risk (node 1) | 103 | 20 (19.42) | 1 | |
| Medium-risk (node 2) | 306 | 108 (35.29) | 2.16 (1.33–3.50) | 0.002 |
| High-risk (nodes 3–5) | 31 | 19 (61.29) | 5.24 (2.71–10.11) | <0.001 |
Adjusting for age, sex, smoking, ethnicity, tumor stage, tumor site and treatment. Bold numbers represent P values that are statistically significant at P < 0.05.
Discussion
In this study, we investigated the effects of a comprehensive panel of 98 SNPs in 17 RGS genes on the risk of developing SPT/recurrence among curatively treated early-stage HNSCC patients. We found that genetic variations in RGS genes may modulate SPT/recurrence development individually, interactively and cumulatively.
RGS proteins directly control cellular homeostasis mediated by G-proteins and GPCRs through binding to active G subunits, activating guanosine triphosphatase and accelerating the kinetics and termination of G-protein-mediated signaling transduction (20,28,29). In our study, there are two SNPs remaining significant after multiple comparison adjustment. The most significant SNP is rs2179653 that is located in the 5′-flanking region of RGS2 gene. RGS2 was initially identified to be a kind of upregulated gene in the early response to activated T cells, and RGS2-deficient mice were found to have impaired T cell responses and emotive behaviors such as increased anxiety responses and decreased male aggressiveness (30). There are circumstantial evidences suggesting that RGS2 protein might play a critical role in various common human disorders including cardiovascular disease, hypertension and cancers of breast, prostate and ovary, whereas no study has been reported to date on the role of RGS2 in HNSCC (22,30–32). Another SNP remaining significant after the adjustment for multiple comparisons was rs3795617 located in the 5′ near region of RGS13 gene. As the smallest RGS protein in mammals, RGS13 is prominently expressed in immune tissues including tonsil, thymus, lymph node and spleen, which indicates its potential function in human immunity (33). Consistently, the expression of RGS13 has been implicated in the pathogenesis of hematopoietic malignancies such as leukemia and lymphomas (34–36). In the current study, these two significant SNPs are located either in the 5′ flanking region or near 5′ region of their host genes. These SNPs are potential functional variations that could modulate individual's cancer risk through regulating the promoter activity of their host genes. However, this hypothesis needs to be further confirmed in functional assays.
Five additional SNPs in RGS3, RGS5, RGS7, RGS8 and RGS11 also exhibited a significant association with an altered SPT/recurrence risk in the main effect analysis of individual SNP (Table I). Although none of these SNPs have been reported before, their host genes have all been associated with the etiology and prognosis of cancers. For instance, RGS3 has been reported to modulate glioma cell mobility and its expression has been associated with the prognosis of sarcoma and breast cancer (37–39). RGS5, a potential tumor angiogenesis factor, is dynamically regulated in various biological processes (9). Mice with RGS5 deficiency show substantial tumor development with poor survival, which may be caused by the normalized vasculature in the absence of RGS5 (9). RGS7 had overlapping distribution profiles with RGS17 and were both noticeably expressed in the cerebellum (40). In vivo, RGS7 can be rapidly upregulated after exposure to tumor necrosis factor-α (41). Tumor necrosis factor is a major inflammation cytokine and has been demonstrated to link with many human cancers (42). RGS8 is a brain-specific RGS protein of 180 amino acids (43). In situ hybridization analysis revealed that RGS8 are expressed widely but differentially in the central nervous system. RGS8 protein regulates the G protein-gated K (+) channels activities and RGS8 gene was also implicated in the region associated with the development of hereditary prostate cancer (43,44). Although RGS11 is a member of the R7 family of GGL (G protein gamma-like) domain-containing RGS proteins with its function largely unevaluated, the expression of RGS11 was reported to be significantly associated with the resistance to platinum therapy in colorectal cancer (45,46).
Though the role of RGS genes in cellular signaling transduction has been widely explored, the genetic etiology of RGS in most cancers remains largely unclear. It seems most plausible that more common genetic variants with low penetrance on disease susceptibility cause the bulk of this unexplained risk (47). Previous studies using a single candidate gene approach have not only identified a few cancer susceptibility loci but also produced a large number of false positive results (48). The application of polygenic approaches to genetic marker data is a viable alternative strategy and may represent a useful addition to single genetic variant analysis, where striking evidence of pathway enrichment often emerged in the absence of obvious single gene effects (49). For example, in our current study, when the cumulative effects of genetic variations were assessed by using the unfavorable genotype analysis, it was shown that the risk increased significantly with an increasing number of unfavorable genotypes. Compared with subjects carrying <3 unfavorable genotypes, the HR (95% CI) for those carrying 4+ unfavorable genotypes was 3.05-fold increased. These findings highlighted the importance of using a multigenic approach to identify signatures of genetic variations as predictors of cancer risk.
In addition to traditional Cox regression approach, a tree-based method was also applied in the multivariate analysis in order to gain insights into the prognostic effects of joint genetic variants of RGS genes. Survival tree analysis is an explorative, nonparametric approach that has been demonstrated in several studies in exploring high-order gene–gene and gene–environment interactions (50,51). The risk for SPT/recurrence development in each node with distinct genotype profiles differed significantly, suggesting a good discriminative ability of the survival tree analysis.
Nonetheless, in spite of the cancer relevance of RGS genes, most of the significant SNPs associated with SPT/recurrence development are located in non-coding regions. The detected associations might be either directly with causal variants or indirectly with markers linked with other causal variants. Further fine-mapping and functional studies are warranted to pinpoint the linked causative loci as well as their biological mechanisms.
Our results should be interpreted in the context of several caveats. First, sample size of this study was relatively small, which may have limited us from detecting effects that would have attained statistical significance in a larger sample. Second, replication of the current work in another independent study population is essential to confirm the findings. Third, although we adjusted for smoking, ethnicity, tumor stage, primary tumor site and treatment, additional confounders such as nutrient intake, socioeconomic status and other medications and their potential effects on the SPT/recurrence development remained to be determined.
Despite these limitations, our findings support the benefit of using a polygenic approach to evaluate the cumulative effects of genetic variations in the prediction of cancer risk and prognosis. In addition, our findings provide convergent evidence of RGS gene family in SPT/recurrence development and support further investigation of the RGS for potential biomarkers for prognosis among HNSCC patients.
Supplementary material
Supplementary Tables 1 and 2 can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health (CA52051, CA97007).
Supplementary Material
Acknowledgments
Dr W.K.Hong is an American Cancer Society Clinical Research Professor.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CI
confidence interval
- GPCR
G-protein-coupled receptor
- HR
hazard ratio
- HNSCC
head and neck squamous cell carcinoma
- RGS
regulator of G-protein signaling
- SPT
second primary tumor
- SNP
single-nucleotide polymorphism
References
- 1.Lefebvre JL. Current clinical outcomes demand new treatment options for SCCHN. Ann. Oncol. 2005;16(suppl. 6):vi7–vi12. doi: 10.1093/annonc/mdi452. [DOI] [PubMed] [Google Scholar]
- 2.Wu X, et al. Joint effect of mutagen sensitivity and insulin-like growth factors in predicting the risk of developing secondary primary tumors and tumor recurrence in patients with head and neck cancer. Clin. Cancer Res. 2006;12:7194–7201. doi: 10.1158/1078-0432.CCR-06-0671. [DOI] [PubMed] [Google Scholar]
- 3.Argiris A, et al. Competing causes of death and second primary tumors in patients with locoregionally advanced head and neck cancer treated with chemoradiotherapy. Clin. Cancer Res. 2004;10:1956–1962. doi: 10.1158/1078-0432.ccr-03-1077. [DOI] [PubMed] [Google Scholar]
- 4.Bastiani C, et al. Heterotrimeric G proteins in C. elegans. WormBook. 2006;13:1–25. doi: 10.1895/wormbook.1.75.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oldham WM, et al. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 2008;9:60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- 6.Siffert W. G protein polymorphisms in hypertension, atherosclerosis, and diabetes. Annu. Rev. Med. 2005;56:17–28. doi: 10.1146/annurev.med.56.082103.104625. [DOI] [PubMed] [Google Scholar]
- 7.Dorsam RT, et al. G-protein-coupled receptors and cancer. Nat. Rev. Cancer. 2007;7:79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 8.Ladds G, et al. Regulators of G protein signalling proteins in the human myometrium. Eur. J. Pharmacol. 2009;610:23–28. doi: 10.1016/j.ejphar.2009.03.042. [DOI] [PubMed] [Google Scholar]
- 9.Manzur M, et al. Modulation of g protein signaling normalizes tumor vessels. Cancer Res. 2009;69:396–399. doi: 10.1158/0008-5472.CAN-08-2842. [DOI] [PubMed] [Google Scholar]
- 10.Xie GX, et al. How regulators of G protein signaling achieve selective regulation. J. Mol. Biol. 2007;366:349–365. doi: 10.1016/j.jmb.2006.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu S, et al. RGS proteins: identifying new GAPs in the understanding of blood pressure regulation and cardiovascular function. Clin. Sci. (Lond.) 2009;116:391–399. doi: 10.1042/CS20080272. [DOI] [PubMed] [Google Scholar]
- 12.Hendriks-Balk MC, et al. Regulation of G protein-coupled receptor signalling: focus on the cardiovascular system and regulator of G protein signalling proteins. Eur. J. Pharmacol. 2008;585:278–291. doi: 10.1016/j.ejphar.2008.02.088. [DOI] [PubMed] [Google Scholar]
- 13.Hooks SB, et al. A role of RGS proteins in drug addiction. Biochem. Pharmacol. 2008;75:76–84. doi: 10.1016/j.bcp.2007.07.045. [DOI] [PubMed] [Google Scholar]
- 14.Riddle EL, et al. Multi-tasking RGS proteins in the heart: the next therapeutic target? Circ. Res. 2005;96:401–411. doi: 10.1161/01.RES.0000158287.49872.4e. [DOI] [PubMed] [Google Scholar]
- 15.Hurst JH, et al. Regulator of G-protein signaling (RGS) proteins in cancer biology. Biochem. Pharmacol. 2009;78:1289–1297. doi: 10.1016/j.bcp.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 16.Hahntow IN, et al. Are RGS2 gene polymorphisms associated with high blood pressure in an ethnicity- and gender-specific manner? Am. J. Hypertens. 2009;22:80–86. doi: 10.1038/ajh.2008.310. [DOI] [PubMed] [Google Scholar]
- 17.Liou YJ, et al. Analysis of genetic variations in the RGS9 gene and antipsychotic-induced tardive dyskinesia in schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2009;150B:239–242. doi: 10.1002/ajmg.b.30796. [DOI] [PubMed] [Google Scholar]
- 18.You M, et al. Fine mapping of chromosome 6q23-25 region in familial lung cancer families reveals RGS17 as a likely candidate gene. Clin. Cancer Res. 2009;15:2666–2674. doi: 10.1158/1078-0432.CCR-08-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu J, et al. A nonsynonymous single-nucleotide polymorphism in the PDZ-Rho guanine nucleotide exchange factor (Ser1416Gly) modulates the risk of lung cancer in Mexican Americans. Cancer. 2006;106:2716–2724. doi: 10.1002/cncr.21944. [DOI] [PubMed] [Google Scholar]
- 20.Berman DM, et al. A functional polymorphism in RGS6 modulates the risk of bladder cancer. Cancer Res. 2004;64:6820–6826. doi: 10.1158/0008-5472.CAN-04-1916. [DOI] [PubMed] [Google Scholar]
- 21.Nikolova DN, et al. Genome-wide gene expression profiles of thyroid carcinoma: identification of molecular targets for treatment of thyroid carcinoma. Oncol. Rep. 2008;20:105–121. [PubMed] [Google Scholar]
- 22.Cao X, et al. Regulator of G-protein signaling 2 (RGS2) inhibits androgen-independent activation of androgen receptor in prostate cancer cells. Oncogene. 2006;25:3719–3734. doi: 10.1038/sj.onc.1209408. [DOI] [PubMed] [Google Scholar]
- 23.O'Byrne KJ, et al. Tumour angiogenesis: a novel therapeutic target in patients with malignant disease. Expert Opin. Emerg. Drugs. 2001;6:155–174. doi: 10.1517/14728214.6.1.155. [DOI] [PubMed] [Google Scholar]
- 24.Khuri FR, et al. The impact of smoking status, disease stage, and index tumor site on second primary tumor incidence and tumor recurrence in the head and neck retinoid chemoprevention trial. Cancer Epidemiol. Biomarkers Prev. 2001;10:823–829. [PubMed] [Google Scholar]
- 25.Wu X, et al. Novel susceptibility loci for second primary tumors/recurrence in head and neck cancer patients: large-scale evaluation of genetic variants. Cancer Prev. Res. (Phila. Pa) 2009;2:617–624. doi: 10.1158/1940-6207.CAPR-09-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim KI, et al. Effects of dependence in high-dimensional multiple testing problems. BMC Bioinformatics. 2008;9:114. doi: 10.1186/1471-2105-9-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Storey JD, et al. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langer I, et al. Evidence for a direct and functional interaction between the regulators of G protein signaling-2 and phosphorylated C terminus of cholecystokinin-2 receptor. Mol. Pharmacol. 2009;75:502–513. doi: 10.1124/mol.108.051607. [DOI] [PubMed] [Google Scholar]
- 29.Hao J, et al. Regulation of cardiomyocyte signaling by RGS proteins: differential selectivity towards G proteins and susceptibility to regulation. J. Mol. Cell. Cardiol. 2006;41:51–61. doi: 10.1016/j.yjmcc.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 30.Le TH, et al. RGS2: a “turn-off” in hypertension. J. Clin. Invest. 2003;111:441–443. doi: 10.1172/JCI17836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smalley MJ, et al. Regulator of G-protein signalling 2 mRNA is differentially expressed in mammary epithelial subpopulations and over-expressed in the majority of breast cancers. Breast Cancer Res. 2007;9:R85. doi: 10.1186/bcr1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurst JH, et al. Regulator of G-protein signalling expression and function in ovarian cancer cell lines. Cell. Mol. Biol. Lett. 2009;14:153–174. doi: 10.2478/s11658-008-0040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson EN, et al. Functional characterization of the G protein regulator RGS13. J. Biol. Chem. 2002;277:16768–16774. doi: 10.1074/jbc.M200751200. [DOI] [PubMed] [Google Scholar]
- 34.Pise-Masison CA, et al. Gene expression profiling of ATL patients: compilation of disease-related genes and evidence for TCF4 involvement in BIRC5 gene expression and cell viability. Blood. 2009;113:4016–4026. doi: 10.1182/blood-2008-08-175901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chng WJ, et al. Gene expression profiling of pulmonary mucosa-associated lymphoid tissue lymphoma identifies new biologic insights with potential diagnostic and therapeutic applications. Blood. 2009;113:635–645. doi: 10.1182/blood-2008-02-140996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Islam TC, et al. High level of cannabinoid receptor 1, absence of regulator of G protein signalling 13 and differential expression of Cyclin D1 in mantle cell lymphoma. Leukemia. 2003;17:1880–1890. doi: 10.1038/sj.leu.2403057. [DOI] [PubMed] [Google Scholar]
- 37.Ooe A, et al. Possible involvement of CCT5, RGS3, and YKT6 genes up-regulated in p53-mutated tumors in resistance to docetaxel in human breast cancers. Breast Cancer Res. Treat. 2007;101:305–315. doi: 10.1007/s10549-006-9293-x. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi H, et al. Cancer diagnosis marker extraction for soft tissue sarcomas based on gene expression profiling data by using projective adaptive resonance theory (PART) filtering method. BMC Bioinformatics. 2006;7:399. doi: 10.1186/1471-2105-7-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tatenhorst L, et al. Regulators of G-protein signaling 3 and 4 (RGS3, RGS4) are associated with glioma cell motility. J. Neuropathol. Exp. Neurol. 2004;63:210–222. doi: 10.1093/jnen/63.3.210. [DOI] [PubMed] [Google Scholar]
- 40.Larminie C, et al. Selective expression of regulators of G-protein signaling (RGS) in the human central nervous system. Brain Res. Mol. Brain Res. 2004;122:24–34. doi: 10.1016/j.molbrainres.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 41.Benzing T, et al. Upregulation of RGS7 may contribute to tumor necrosis factor-induced changes in central nervous function. Nat. Med. 1999;5:913–918. doi: 10.1038/11354. [DOI] [PubMed] [Google Scholar]
- 42.Balkwill F. Tumour necrosis factor and cancer. Nat. Rev. Cancer. 2009;9:361–371. doi: 10.1038/nrc2628. [DOI] [PubMed] [Google Scholar]
- 43.Saitoh O, et al. Alternative splicing of RGS8 gene determines inhibitory function of receptor type-specific Gq signaling. Proc. Natl Acad. Sci. USA. 2002;99:10138–10143. doi: 10.1073/pnas.152085999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sood R, et al. Cloning and characterization of 13 novel transcripts and the human RGS8 gene from the 1q25 region encompassing the hereditary prostate cancer (HPC1) locus. Genomics. 2001;73:211–222. doi: 10.1006/geno.2001.6500. [DOI] [PubMed] [Google Scholar]
- 45.Saleem Y, et al. RGS11 interacts preferentially with R7BP over Galpha(oa)–characterization of Gbeta5-free RGS11. Biochem. Biophys. Res. Commun. 2009;386:65–70. doi: 10.1016/j.bbrc.2009.05.128. [DOI] [PubMed] [Google Scholar]
- 46.Martinez-Cardus A, et al. Pharmacogenomic approach for the identification of novel determinants of acquired resistance to oxaliplatin in colorectal cancer. Mol. Cancer Ther. 2009;8:194–202. doi: 10.1158/1535-7163.MCT-08-0659. [DOI] [PubMed] [Google Scholar]
- 47.Milne RL, et al. Current strategies in the search for low penetrance genes in cancer. Histol. Histopathol. 2008;23:507–514. doi: 10.14670/HH-23.507. [DOI] [PubMed] [Google Scholar]
- 48.Horikawa Y, et al. Genetic susceptibility to bladder cancer with an emphasis on gene-gene and gene-environmental interactions. Curr. Opin. Urol. 2008;18:493–498. doi: 10.1097/MOU.0b013e32830b88ff. [DOI] [PubMed] [Google Scholar]
- 49.Hong MG, et al. Strategies and issues in the detection of pathway enrichment in genome-wide association studies. Hum. Genet. 2009;126:289–301. doi: 10.1007/s00439-009-0676-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu X, et al. Germline genetic variations in drug action pathways predict clinical outcomes in advanced lung cancer treated with platinum-based chemotherapy. Pharmacogenet. Genomics. 2008;18:955–965. doi: 10.1097/FPC.0b013e32830efdd4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hildebrandt MA, et al. Genetic variations in the PI3K/PTEN/AKT/mTOR pathway are associated with clinical outcomes in esophageal cancer patients treated with chemoradiotherapy. J. Clin. Oncol. 2009;27:857–871. doi: 10.1200/JCO.2008.17.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.