Abstract
The genetic component of colorectal cancer (CRC) predisposition has been only partially explained. We recently suggested that a subtle decrease in the expression of one allele of the TGFBR1 gene was a heritable quantitative trait predisposing to CRC. Here, we refined the measurements of allele-specific expression (ASE) of TGFBR1 in a population-based series of CRC patients and controls. Five single-nucleotide polymorphisms (SNPs) in the 3′-untranslated region of the gene were genotyped and used for ASE determination by pyrosequencing. After eliminating non-informative samples and samples with RNA of insufficient quality 109 cases and 125 controls were studied. Allelic ratios ranged between 0.74 and 1.69 without evidence of bimodality or cutoff points for ‘ASE’ versus ‘non-ASE’. Treating ASE as a continuous variable, cases had non-significantly different values than controls (P = 0.081 when comparing means by permutation test). However, cases had significantly higher ASE values when comparing medians by permutation test (P = 0.0027) and when using Wilcoxon test (P = 0.0094). We conclude that with the present-day technology, ASE differences between individuals and between cases and controls are too subtle to be used to assess CRC risk. More advanced technology is expected to resolve this issue as well as the low informativity caused by the limited heterozygosity of transcribed SNPs.
Introduction
Based on twin studies, colorectal cancer (CRC) displays ∼35% heritability (1). Case control studies suggest a first-degree family risk ratio ∼3 (2). Among all CRC patients, a positive family history of colon cancer in a first-or second-degree relative occurs in 20–30%. High-penetrance susceptibility genes account for <5% of all CRC (3). In a recent review, it was concluded that 59% of the population-attributable fraction of CRC predisposition could presently be accounted for when considering all or most genetic mechanisms detected or proposed so far (4). This assessment included the assumption that several recently discovered single-nucleotide polymorphism (SNP) associations with low or ultralow effect size detected by genome-wide association studies (GWAS) would turn out to be real and account for 52% of the total population-attributable fraction. This leaves ∼40% of the genetic predisposition unexplained. The putative mutations have been hypothesized to be relatively rare (allele frequency <1%; not readily detectable by GWAS) and/or of low but not ultralow effect size (odds ratio 2–5 and therefore not readily detectable by linkage) (5).
These data lead to the assumption that the remaining predisposing genes can be found neither by GWAS nor by linkage analysis. There is some hope that whole genome sequencing will eventually provide the answers; however, at present, the preferred way of finding such intermediate-effect size gene mutations is the candidate gene approach. The transforming growth factor-β pathway is heavily involved in CRC (6,7). Of the two receptors, the type 2 receptor has been conclusively implicated as a tumor suppressor in CRC (8), whereas the type 1 receptor has received less attention. Recently, when Tgfbr1 was knocked out in mice, homozygous loss was lethal but heterozygous loss conferred no specific phenotype. When heterozygous knockout mice were bred into mice heterozygous for the ApcMin mutation, the double mutants acquired an ∼2-fold increase in the number of intestinal adenomas in comparison with ApcMin mice, and importantly, the Tgfbr1+/−;ApcMin/+ mice acquired colonic carcinomas, suggesting that haploinsufficiency for Tgfbr1 predisposes to colon cancer (9).
Allele-specific expression (ASE) was recently mentioned as a mutational mechanism with phenotypic consequences (10). Contrary to the common concept that the two alleles of a locus are equally expressed, ASE means that one allele is expressed at a lower level than the other or conversely one is expressed at a higher level than the other. Driven by these findings, we used the SNaPshot technology to study the ASE of TGFBR1 in unaffected human tissue (blood) and found that moderately lowered expression from one allele was a measurable quantitative trait that was more common in CRC patients (∼10%) than in controls (∼1.5%) (11). This ASE of TGFBR1 could, if confirmed in larger populations, be interpreted to account for as much as 10% of the population-attributable fraction. To validate the findings, an alternative method based on pyrosequencing was applied to a smaller series of lymphoblastoid cells from familial CRC patients and controls from another cohort. In this experiment, the proportion of ASE in CRC patients was lower (2 of 50 cases) when the same cutoff (ASE ratio 1.5) was applied (12). This prompted us to undertake the present experiments to further explore ASE of TGFBR1.
Material and methods
CRC cases and controls
The patients belonged to a series of 1566 consecutively ascertained, unselected consenting CRC cases diagnosed in 1999–2004 in the six main hospitals in metropolitan Columbus, Ohio (13,14). These hospitals perform the vast majority of operations for CRC or suspected CRC in the Columbus, Ohio, metropolitan area (population, 1.5 million). The research protocol and consent form were approved by the institutional review board at each participating hospital, and all patients provided written informed consent. Cases of familial adenomatous polyposis or the rare polyposis syndromes were not accrued. Microsatellite instability-positive cases were excluded from the study. Totally 960 cases were considered for this study.
The control samples (n = 900) were provided by the Ohio State University Medical Center’s Human Genetics Sample Bank, which is a collection of control samples for use in human genetics research that includes both donors’ anonymized biological specimens and linked phenotypic data. The data and samples are collected under the protocol ‘Collection and Storage of Controls for Genetics Research Studies’, which is approved by the Biomedical Sciences Institutional Review Board at Ohio State University Medical Center. Recruitment takes place in Ohio State University Medical Center primary care and internal medicine clinics. If individuals agree to participate, they provide written informed consent, complete a questionnaire, which includes demographic, medical and family history information, and donate a blood sample. As a result, the controls were derived from the same Columbus-area population as the CRC patients. After removing cases and controls due to the lack of informative markers and/or insufficient RNA quantity or quality, the ASE study was performed on 109 cases and 125 controls. Cases and controls were frequency matched by age groups: ≤45, 46–55, 56–65, 66–75 and ≥76 with Chi-square P-value = 0.053. The median age of the informative cases was 61 years and of the controls 58 years with Wilcoxon rank sum test (P-value = 0.060). Permutation testing of the age showed variable significance between cases and controls when comparing means (P = 0.015) and medians (P = 0.208).
Age range for informative cases was 32–89 years, whereas for the controls, it was 18–94 years. Ethnicity in the cases was 95.4% Caucasian and 4.6% African–American, and in the controls, 89% Caucasian and 11% African–American (Chi-square test P-value = 0.108). There were 48% females among the cases and 50% among the controls (Chi-square test P-value = 0.47).
DNA–RNA extraction, RNA quality and complementary DNA synthesis
Extraction of genomic DNA from peripheral blood was performed by a standard phenol–chloroform procedure. For total RNA extraction, cells were processed with TRIzol reagent (Invitrogen, Carlsbad, CA). Samples for the RNA extraction were stored either as dry white blood cell pellets or as cells lysed in TRIzol. The quality of total RNA was checked using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). We determined the RNA Integrity Number (RIN) of each sample and it emerged that RNA extracted from frozen pellets was often highly degraded (RIN < 3.0), whereas the RNA obtained from TRIzol fractions was generally good (RIN > 8.0). For the purpose of the ASE study, we chose only samples with a RIN of ≥6.0 (15). Total RNA was treated with DNase I (DNAfree™; Ambion, Austin, TX) prior to reverse transcription with AMV Reverse Transcriptase (Roche, Indianapolis, IN) using a gene-specific primer annealing to the 3′-end of the most downstream amplicon (containing SNP rs1590) used in the ASE analysis.
Genotyping samples for informativity
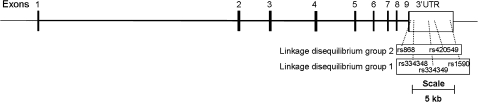
With present-day methods, ASE can only be readily determined in samples that are heterozygous for SNPs residing in the transcribed sequence of the gene. For large-scale ASE assessment, the markers must be reproducibly typeable and reasonably informative. In TGFBR1, there are only five such SNPs; their minor allele frequencies range from 9 to 38%. All five markers are in the 3′-untranslated region as shown in Figure 1. The markers comprise three that are in strong linkage disequilibrium (LD) with each other (LD group 1; R2 > 0.95) and two that are in equally strong LD with one another (LD group 2; R2 > 0.95) but only partial LD with group 1 (R2 = 0.68). We typed the markers in LD group 1 (rs334348, rs334349 and rs1590) and LD group 2 (rs868 and rs420549). For the genotyping, two polymerase chain reaction (PCR) amplicons were obtained from genomic DNA for each sample using GoldTaq polymerase (PE Applied Biosystems, Foster City, CA) in the presence of 5%dimethyl sulfoxide. The first amplicon contained SNPs rs868 and rs334348 (forward: 5′-CAGCTTTGCCTGAACTCTCC-3′ and reverse: 5′-TGCAAAAGCTTGATGTGAGAA-3′) and the second amplicon contained SNPs rs334349, rs420549 and rs1590 (forward: 5′-ACCTGCTCTCCTGCTTGCT-3′ and reverse: 5′-CTGTAGACAGGTCCATCATGC-3′). The PCR conditions included 36 cycles of 95°C for 15 s, 56°C for 15 s and 72°C for 60 s (first amplicon) or 120 s (second amplicon). Multiplexed SNaPshot reactions were performed on amplicon one and amplicon two using two or three extension primers, respectively. Extension primers used were: rs868, 5′-CTCTCAGTGAGGTAGAACAA-3′; rs334348, 5′-CTTGATGTGAGAATATTCAAACATGA-3′; rs420549, 5′-TTGTTGTGCACTCTAACGAT-3′; rs1590, 5′-GAGATCACCTGTAGACAGGTCCATCA-3′ and rs334349 5′-CCCTGACGCAGAGACC-3′. Roughly 29% of samples were informative for LD group 1 and 11% of samples were informative for all five SNP markers.
Fig. 1.
Diagram of the TGFBR1 genomic region. Shown are the locations of the five SNPs in the 3′-untranslated region used for ASE determination.
Pyrosequencing and allele quantitation
For the samples in which all five SNPs were informative, three SNP markers were used for pyrosequencing: rs868, rs334348 and rs334349. When samples were only informative for SNPs in LD group 1, all three SNP markers were used, whereas in the case of SNPs in LD group 2, both markers were used. For each SNP, the PCR reactions for DNA and RNA were performed in triplicates in a 30 μl reaction volume using AmpliTaq Gold polymerase (PE Applied Biosystems). Primers and PCR conditions were as described (12).
After PCR, the DNA and RNA amplification products were sequenced using the sequencing primers (12) on a PyroMark, MD pyrosequencing instrument (QIAGEN, Chatsworth, CA) as per the manufacturer’s instructions. The proportions of individual alleles for each SNP were obtained using the PyroMark MD software package (QIAGEN) and the ratio of allele 1 versus allele 2 in the DNA and RNA was then calculated. The results of triplicate measurements for each single SNP for DNA and RNA were averaged and the standard deviation (SD) was calculated. In only 8% of the measurements, the SD exceeded a value of 0.1. The final ASE ratio for each SNP of each sample measured was calculated using the formula: ASE ratio = RNA (allele 1 expression/allele 2 expression)/genomic DNA (allele 1 expression/allele 2 expression). For the five SNPs used, the (allele 1/allele 2) was as follows: rs868 (A/G), rs334348 (T/C), rs334349 (C/T), rs1590 (T/G) and rs420549 (C/G). For each sample, the final ASE value was calculated as the median of the ASE values for two or three SNPs typed (see supplementary Table S1, available at Carcinogenesis Online).
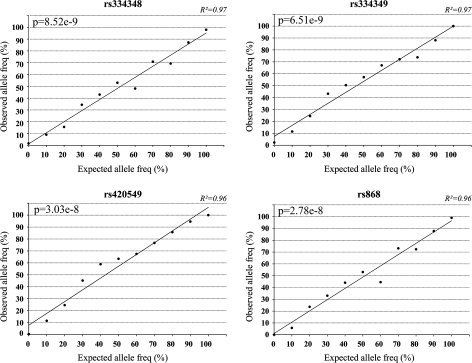
To validate the use of pyrosequencing for ASE measurements, mixing experiments were performed in which DNA homozygous for allele 1 was mixed in known proportions with DNA homozygous for allele 2. The purpose was to establish whether the peak strengths of the two alleles are comparable at different proportions of the two alleles. Results from four of the SNPs are shown graphically in Figure 2. A consistent rectilinear correlation between input of the two allelic variants and the resulting ratio is demonstrated.
Fig. 2.
Accuracy of pyrosequencing to quantify the two alleles of SNPs. Genomic DNA was extracted from cells homozygous for either allele of the SNP markers and quantified using NanoDrop. For each SNP, the two DNAs representing the alternative alleles were mixed at different ratios to simulate expected allele ratios. PCR was performed on each mixture, followed by pyrosequencing to determine the observed allelic ratios. Pearson’s correlation test is used to calculate the P-value.
Statistical analysis
The nonparametric Wilcoxon rank sum test was used to compare ASE values between cases and controls. Moreover, the means and the medians of the two groups were compared using a permutation test with 100 000 permutations. All tests were two sided. The ethnicity, gender and age group distributions between cases and controls were compared using Chi-squared test.
Results
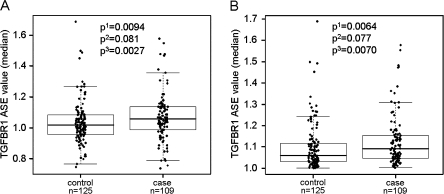
The ASE values of the cases and controls are shown in Figure 3A. The values range from 0.74 to 1.69, with two cases and two controls having values ≥1.5, the cutoff previously suggested to distinguish between ASE and non-ASE based on a receiver operating characteristic analysis (11). The visual impression of the scatter plot does not suggest any obvious cutoff point or bimodality in the data. The receiver operating characteristic analysis estimating the sensitivity, specificity and the Youden’s index defined as ‘sensitivity + specificity−1’ that measures the overall diagnostic accuracy of varied cutoff points was performed (Table I). The best cutoff was obtained by maximizing Youden’s index at ASE value of 1.1, which signifies a roughly 10% difference in expression between the two alleles.
Fig. 3.
TGFBR1 ASE distribution in 109 CRC patients and 125 controls studied by pyrosequencing. (A) ASE values obtained from pyrosequencing. (B) The same data shown after ASE values <1.0 have been transformed to their reciprocals. Superscript numerals with probability values refer to the following: 1, Paired Wilcoxon test; 2, permutation test comparing means; and 3, permutation test comparing medians.
Table I.
Sensitivity, specificity and Youden’s index for different ASE cutoff values
| ASE cutoff | Sensitivity | Specificity | Youden’s index |
| 1.00 | 1 | 0 | 0 |
| 1.05 | 0.706 | 0.432 | 0.138 |
| 1.10 | 0.468 | 0.672 | 0.140 |
| 1.15 | 0.257 | 0.808 | 0.065 |
| 1.20 | 0.165 | 0.840 | 0.005 |
| 1.25 | 0.128 | 0.912 | 0.040 |
| 1.30 | 0.101 | 0.952 | 0.053 |
| 1.35 | 0.073 | 0.968 | 0.041 |
| 1.40 | 0.046 | 0.968 | 0.014 |
| 1.45 | 0.046 | 0.968 | 0.014 |
| 1.50 | 0.018 | 0.992 | 0.010 |
| 1.55 | 0.018 | 0.992 | 0.010 |
| 1.60 | 0 | 0.992 | −0.008 |
| 1.65 | 0 | 0.992 | −0.008 |
| 1.70 | 0 | 1 | 0 |
The cutoff with the highest Youden’s index is bolded.
Comparison of high (ASE ≥ 1.1 and ASE ≤ 0.9) and low (0.9 < ASE < 1.1) ASE between the case and control groups was done by applying a univariate logistic regression analysis (odds ratio = 1.68, P-value = 0.055). This suggests that with the methods used here, classifying cases and controls as ASE versus non-ASE is not feasible. When ASE is treated as a continuous variable; however, the difference between cases and controls shows a significantly higher ASE value in cases than in controls (P = 0.0094 by Wilcoxon test), whereas the permutation testing is borderline (P = 0.081) when comparing means. Permutation testing comparing medians results in a significantly higher ASE value in cases than in controls (P = 0.0027).
Joint evaluation of ASE values > and <1 is problematic since a value of 1.1 corresponds to a value of 0.9 but the statistical analysis does not give the same weight to these two reciprocal values. To overcome this problem, values <1 were reciprocally converted as shown in Figure 3B. Analyzing the differences in this way, ASE values in cases are higher than in controls (P = 0.0064) by Wilcoxon test and borderline (P = 0.077) using permutation testing when comparing means. Permutation testing comparing medians showed a significantly higher ASE value in cases than in controls (P = 0.0070).
There was no difference in ASE values between older and younger CRC cases (median cut, Wilcoxon test P = 0.82 and Permutation test, based on medians P = 0.48).
Discussion
While conducting our final analysis, the results of two studies on ASE of TGFBR1 were published. In the first study, no evidence of an association between TGFBR1 polymorphisms and CRC risk was detected, and ASE was reported to be equally rare in cases as in controls (16). In the second study, ASE was found in ∼10% (10/98) of all Caucasian and 7% (1/14) of all African–American patients; however, there were no healthy controls studied for comparison (17). An association between ASE and three SNPs in TGFBR1 was noted. Whereas the first study apparently contradicts our findings, the second study agrees with our initial report (11) but differs from our most recent results. How can such contrasting results be explained?
There are major differences between our present study and the three published reports referred to above. Firstly, all three studies were conducted using the SNaPshot technology that we have found to give inconsistent results as evidenced by considerably larger SDs compared with the pyrosequencing method (SNaPshot: median SD = 0.13, average SD = 0.15; pyrosequencing: median SD = 0.076, average SD = 0.094). Secondly, in our previous study (11), we had included SNP rs7871490 that accounted for ∼35% informative cases and ∼28% informative controls. This SNP resides in a region of repetitive sequence. Our repeated attempts with SNaPshot yielded inconsistent results, possibly due to polymerase slippage. We therefore elected to remove SNP rs7871490 from the study. We note that a significant number of ASE-positive cases in reference (11) stem from results using this marker. Thirdly, using microsatellites such as the 9A/6A marker (16) to measure ASE has not proven to be reproducible in our hands. Fourthly, we have noticed that high-quality RNA is essential for reproducibility of ASE. Even just somewhat degraded RNA can produce inconsistent results with either method.
In our current study, 49 cases were the same as in our previous study and they consistently showed lower ASE ratios by pyrosequencing (supplementary Figure S1 is available at Carcinogenesis Online; paired Wilcoxon test P = 6.747e-06). However, the calculation of ASE differed between our two studies. In the present study, the ASE value for each sample was calculated as a median value of at least two or three SNPs (see Materials and Methods; supplementary Table S1 is available at Carcinogenesis Online). In Valle et al. (11), ASE was calculated as the average of measurements across multiple SNPs obtained for each sample. In this way, one exceptionally high measurement (‘outlier’) can bias the ASE ratio and we realized that this had pertained in particular to SNP rs7871490. However, even after removing rs7871490, we observe a bias using SNaPshot that evidently exaggerates the allelic differences. (supplementary Figure S1 is available at Carcinogenesis Online). Thus, the difference in ASE results between this study and those reported before (11,12,16,17) originates in different strategies and methods, possibly study populations and numbers of studied individuals. While such differences may explain some of the incongruity, we wish to raise here the possibility that none of the present methods are sensitive enough to measure subtle ASE on a case-by-case basis.
Our present findings support the notion that the two alleles in TGFBR1 are not always equally expressed, constituting a subtle quantitative trait that is weakly associated with the risk for CRC. However, our results suggest that the unequal expression of TGFBR1 from the two alleles in a given individual cannot be used to classify the individual as unequivocally ASE positive or ASE negative as we suggested previously (11). At the present time, this precludes ASE from being used as a predictive marker of CRC risk until a genomic cause for ASE can be determined. Nevertheless, to assess ASE, pyrosequencing has proven to be more robust than SNaPshot in direct comparison.
The fact that the ASE phenomenon is widespread not only in humans (10,18) and mice (19) but also in most organisms is becoming well known and widely publicized (20,21). Moreover, the differential expression of alleles in autosomal loci is often inherited and can be highly context-specific (22). We show that ASE of TGFBR1 is slightly more common in patients with CRC than in controls. ASE as a cause of disease predisposition has been documented before, for instance, regarding the APC gene in familial adenomatous polyposis (23) and the DAPK1 gene in chronic lymphocytic leukemia (24). Further examples are being proposed and reported increasingly, e.g. ASE of BRCA1 contributing to breast cancer (25). Notably, in most of the cases referred to, the putative genomic cause of the ASE phenomenon has so far been elusive.
The extreme subtlety of the expression differences that we describe should be kept in mind. A more definitive evaluation of the ASE phenomenon in TGFBR1 will probably only come with technological and conceptual advances that will allow greater precision and circumvent the need for naturally occurring transcribed SNPs as an obligatory tool. We note that with the present technology, it is possible to score an ASE value in only approximately one-third of all individuals at best, leaving open the possibility that ASE occurs or does not occur preferentially in those individuals who are uninformative for the relevant SNPs.
With the present limitations (lack of informativity; lack of a cutoff ratio defining ASE), it is challenging to try to clarify the underlying putative constitutional (‘germ line’, ‘genomic’) cause of ASE. We previously sequenced some 96 kb of genomic DNA comprising TGFBR1 and adjacent sequence in six CRC patients classified as having ASE. This led to the detection of some 200 sequence changes, mainly SNPs, of which approximately half were not listed in the databases. However, no particular sequence change stood out as being a probable candidate for an ASE-causing mutation (11). Similarly, exonic sequencing of 96 CRC patients by others failed to identify any probable mutations (16).
Hence, the cause of ASE of TGFBR1 remains unresolved. It is presently not clear if the cause is in cis or in trans. It is also totally unclear if an expression difference between the two alleles per se is relevant or if the sum of the expression level is important. Furthermore, it remains obscure how small allelic imbalances could cause a phenotype. However, there are now several precedents that subtle gene expression differences can have a significant phenotypic impact. GWAS have implicated SMAD7 as a locus for CRC risk (26). Functional analysis of a novel intronic SNP in the gene showed that a very small difference in expression levels of just 11% between the two alleles was detectable and associated with a 1.4-fold elevated CRC risk (27). This is also apparently true for a SNP (rs6983267) in chromosome band 8q24 that is associated with an increased risk for CRC (28,29,30). The underlying mechanism was suggested to be a different binding affinity of the two alleles for a transcription factor that affects the locus’ function as a transcriptional enhancer (31,32). Again, the experimental difference between the two alleles was of the order of only 30%. At this point, we do not know which if any of the variants around TGFBR1 is responsible for ASE in cis. Moreover, we have not seen any indication that ASE of TGFBR1 is age-related or tissue-type specific but more studies along these lines are expected.
Supplementary material
Supplementary Figure S1 and Table S1 can be found at http://carcin.oxfordjournals.org/
Funding
National Cancer Institute (CA16058 to The Ohio State University Comprehensive Cancer Center, CA67941 to A.dlC., CA130901 to S.D.M.).
Supplementary Material
Acknowledgments
We thank Laura Valle and Stephen Gruber for helpful discussions.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- ASE
allele-specific expression
- CRC
colorectal cancer
- GWAS
genome-wide association studies
- LD
linkage disequilibrium
- PCR
polymerase chain reaction
- RIN
RNA integrity number
- SD
standard deviation
- SNP
single-nucleotide polymorphism
References
- 1.Lichtenstein P, et al. Environmental and heritable factors in the causation of cancer-analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 2.Risch N. The genetic epidemiology of cancer: interpreting family and twin studies and their implications for molecular genetic approaches. Cancer Epidemiol. Biomarkers Prev. 2001;10:733–741. [PubMed] [Google Scholar]
- 3.Lynch HT, et al. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin. Genet. 2009;76:1–18. doi: 10.1111/j.1399-0004.2009.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hemminki K, et al. Surveying the genomic landscape of colorectal cancer. Am. J. Gastroenterol. 2009;104:789–790. doi: 10.1038/ajg.2008.100. [DOI] [PubMed] [Google Scholar]
- 5.Kemp Z, et al. An update on the genetics of colorectal cancer. Hum. Mol. Genet. 2004;13:R177–R185. doi: 10.1093/hmg/ddh247. [DOI] [PubMed] [Google Scholar]
- 6.Massague J, et al. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 7.Bierie B, et al. TGF-beta and cancer. Cytokine Growth Factor Rev. 2006;17:29–40. doi: 10.1016/j.cytogfr.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, et al. Demonstration that mutation of the type II transforming growth factor beta receptor inactivates its tumor suppressor activity in replication error-positive colon carcinoma cells. J. Biol. Chem. 1995;270:22044–22049. doi: 10.1074/jbc.270.37.22044. [DOI] [PubMed] [Google Scholar]
- 9.Zeng Q, et al. Tgfbr1 haploinsufficiency is a potent modifier of colorectal cancer development. Cancer Res. 2009;69:678–686. doi: 10.1158/0008-5472.CAN-08-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan H, et al. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- 11.Valle L, et al. Germline allele-specific expression of TGFBR1 confers an increased risk of colorectal cancer. Science. 2008;321:1361–1365. doi: 10.1126/science.1159397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guda K, et al. Infrequent detection of germline allele-specific expression of TGFBR1 in lymphoblasts and tissues of colon cancer patients. Cancer Res. 2009;69:4959–4961. doi: 10.1158/0008-5472.CAN-09-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hampel H, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N. Engl. J. Med. 2005;352:1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 14.Hampel H, et al. Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J. Clin. Oncol. 2008;26:5783–5788. doi: 10.1200/JCO.2008.17.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strand C, et al. RNA quality in frozen breast cancer samples and the influence on gene expression analysis - a comparison of three evaluation methods using microcapillary electrophoresis traces. BMC Mol. Biol. 2007;8:38. doi: 10.1186/1471-2199-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carvajal-Carmona LG, et al. Comprehensive assessment of variation at the transforming growth factor β type 1 receptor locus and colorectal cancer predisposition. Proc. Natl Acad. Sci. USA. 2010;107:7858–7862. doi: 10.1073/pnas.1002816107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pasche B, et al. Constitutively decreased TGFBR1 allelic expression is a common finding in colorectal cancer and is associated with three TGFBR1 SNPs. J. Exp. Clin. Cancer Res. 2010;29:57. doi: 10.1186/1756-9966-29-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lo HS, et al. Allelic variation in gene expression is common in the human genome. Genome Res. 2003;13:1855–1862. doi: 10.1101/gr.1006603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cowles CR, et al. Detection of regulatory variation in mouse genes. Nat. Genet. 2002;32:432–437. doi: 10.1038/ng992. [DOI] [PubMed] [Google Scholar]
- 20.Verlaan DJ, et al. Targeted screening of cis-regulatory variation in human haplotypes. Genome Res. 2009;19:118–127. doi: 10.1101/gr.084798.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ge B, et al. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat. Genet. 2009;41:1216–1222. doi: 10.1038/ng.473. [DOI] [PubMed] [Google Scholar]
- 22.Knight JC. Allele-specific gene expression uncovered. Trends Genet. 2004;20:113–116. doi: 10.1016/j.tig.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Yan H, et al. Small changes in expression affect predisposition to tumorigenesis. Nat. Genet. 2002;30:25–26. doi: 10.1038/ng799. [DOI] [PubMed] [Google Scholar]
- 24.Raval A, et al. Downregulation of death-associated protein kinase 1 (DAPK1) in chronic lymphocytic leukemia. Cell. 2007;129:879–890. doi: 10.1016/j.cell.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X, et al. Allelic imbalance in BRCA1 and BRCA2 gene expression is associated with an increased breast cancer risk. Hum. Mol. Genet. 2008;17:1336–1348. doi: 10.1093/hmg/ddn022. [DOI] [PubMed] [Google Scholar]
- 26.Broderick P, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat. Genet. 2007;39:1315–1317. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 27.Pittman AM, et al. The colorectal cancer risk at 18q21 is caused by a novel variant altering SMAD7 expression. Genome Res. 2009;19:987–993. doi: 10.1101/gr.092668.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomlinson I, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat. Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 29.Zanke BW, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat. Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 30.Haiman CA, et al. A common genetic risk factor for colorectal and prostate cancer. Nat. Genet. 2007;39:954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pomerantz MM, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat. Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuupanen S, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat. Genet. 2009;41:885–890. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.