Abstract
Secretory and membrane proteins that are destined for intracellular organelles in eukaryotes are first synthesized at the endoplasmic reticulum (ER) and are then delivered to their final destinations. The ER contains high concentrations of molecular chaperones and folding enzymes that assist substrates to acquire their native conformations. However, protein misfolding is an inevitable event especially when cells are exposed to stress or during development or aging. ER-associated degradation (ERAD) is a major mechanism to eliminate misfolded proteins from the secretory pathway. The importance of ERAD is underscored by the fact that mutations in secretory and membrane proteins or corruption of the ERAD machinery have been linked to human diseases. Many components involved in ERAD have been identified by a genetic analysis using the yeast Saccharomyces cerevisiae, and it now appears that most of these factors are conserved in higher eukaryotes. In this chapter, we describe a method to recapitulate the ubiquitination and extraction of misfolded polytopic membrane proteins in vitro using materials prepared from yeast. These techniques provide a powerful tool to further dissect the ERAD pathway into elementary steps.
Keywords: Endoplasmic reticulum, ER-associated degradation, ATP, proteasome, Ufd2, Cdc48/p97, microsomes, yeast
1. Introduction
Newly synthesized secretory and membrane proteins that fail to achieve their native conformations are retained in the endoplasmic reticulum (ER) and may be degraded. This process is referred to as ER-associated degradation (ERAD). From studies over the past 13 years, it is now clear that ERAD substrates are first recognized in the ER and are then retrotranslocated back to the cytoplasm where they are ubiquitinated and degraded by the proteasome (1–5). Earlier genetic studies using the yeast Saccharomyces cerevisiae have identified core components required for ERAD, including membrane-associated E2/E3 ubiquitination enzymes, cytoplasmic and luminal chaperones, and the proteasome. Although the detailed mechanism for substrate recognition and retrotranslocation is not yet clear, current evidence suggests that, depending on the location of the misfolded lesion, molecular chaperones and chaperone-like lectins either in the ER or in the cytoplasm help select ERAD substrate (6–9). To further dissect the ERAD reaction into elementary steps and to characterize the functions of known and novel components, it is vital to biochemically reconstitute ERAD.
Ste6p is a yeast a-factor mating pheromone transporter that is synthesized in the ER and is delivered to and functions at the plasma membrane. A mutant form of Ste6p, which is called Ste6p*, is retained in the ER and is degraded by the proteasome via ERAD (10). Ste6p* has 12 transmembrane domains and is structurally similar to the cystic fibrosis transmembrane conductance regulator (CFTR), which is also an ERAD substrate and which when mutated results in cystic fibrosis. Genetic analysis has shown that Ste6p* degradation is slowed when specific E2 ubiquitin-conjugating enzymes (Ubc6p and Ubc7p), E3 ubiquitin ligases (Doa10p and Hrd1p), cytoplasmic Hsp70 and Hsp40 chaperones (Ssa1p and Ydj1p/Hlj1p), and a AAA-ATPase Cdc48p are disabled (6, 11). Although the ERAD pathway for Ste6p* is relatively well-defined, until recently it was not clear how this substrate is selected for ubiquitination and whether it is degraded in the cytoplasm or at the ER membrane.
We recently reconstituted the ubiquitination and extraction of Ste6p* using materials prepared from yeast (12). This assay has proven that Ssa1p is essential for ubiquitination. Moreover, ubiquitinated Ste6p* is extracted from the ER membrane to the cytosol in an ATP- and Cdc48p-dependent manner. We also discovered that Ufd2p, an E4 polyubiquitin chain-extending enzyme, elongates ubiquitin chains. Theoretically, this assay can be applied to any misfolded membrane protein that can be expressed in yeast. This assay also has the potential to further dissect the pathway of these ERAD substrates using yeast genetic mutants.
2. Materials
2.1. Preparation of ER-Derived Microsomes
Plasmids that encode misfolded polytopic membrane substrates: Ste6p*-3HA is encoded by pSM1082 (2μ URA3 pste6ste6-166∷HA) or pSM1911 (2μ URA3 pPGKste6-166∷HA) and CFTR-3HA is encoded by pSM1152 (2μ URA3 pPGKCFTR∷HA) (11, 13). Yeast cells are transformed with one of these plasmids and grown in a selective medium using established methods (14). Filter-sterilized medium is used for the protocol in Section 3.1.2.
Lyticase: It was obtained commercially or has been produced using a heterologous expression system (15, 16).
Lyticase buffer: 0.7 M sorbitol, 0.75% (w/v) yeast extract, 1.5% (w/v) bacto peptone, 0.5% glucose, 10 mM tris(hydroxymethyl)aminomethane (Tris)–hydrochloric acid (HCl), pH 7.4
Lysis buffer: 0.1 M sorbitol, 50 mM potassium acetate (KOAc), 2 mM ethylenediaminetetraacetic acid (EDTA), 20 mM N-(2-hydroxyethyl)piperazine-N-'(2ethanesulfonic acid) (HEPES)–NaOH, pH 7.4. The following reagents were added immediately prior to use: 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 μg/mL leupeptin, and 0.5 μg/mL pepstatin A.
Cushion 1: 0.8 M sucrose, 1.5% Ficoll 400, 20 mM HEPES–NaOH, pH 7.4
Cushion2: 1.0 M sucrose, 50 mM KOAc, 20 mM HEPES–NaOH, pH 7.4. DTT (1 mM) is added immediately prior to use.
Buffer 88 (B88): HEPES–NaOH, pH 6.8, 150 mM KOAc, 5 mM magnesium acetate (MgOAc), 250 mM sorbitol in double-deionized water (ddH2O). The solution should be filter sterilized and stored at 4°C.
2.2. Preparation of Yeast Cytosol
Liquid nitrogen (~4 L)
Stainless-steel blender
B88 (see Section 2.1, item 7)
2.3. Ubiquitination of Ste6p*-3HA
B88 (see Section 2.1., item 7).
Microsomal membranes (see Section 3.1).
Yeast cytosol (see Section 3.2).
10× ATP-regenerating system: 10 mM ATP, 500 mM creatine phosphate, 2 mg/mL of creatine phosphokinase in B88. We typically store this solution in aliquots of approximately 100 μL at −80°C and use only once (i.e., do not re-freeze).
125I-labeled ubiquitin: Bovine ubiquitin (Sigma) is dissolved in phosphate-buffered saline at a concentration of 10 μg/μL and labeled with 125I (NEN Research, BioRad) using the ICl method (17, 18). The labeled ubiquitin is enriched with a D-salt Excellulose Desalting column (Pierce) and is stored at a final concentration of 0.2 μg/μL(~1.0 × 106 cpm/μL) (see Notes 1 and 7).
Apyrase (Sigma).
Methylated ubiquitin (Boston Biochem).
1.25% SDS stop solution: 50 mM Tris–Cl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1.25% sodium dodecyl sulfate (SDS). The following reagents are added immediately prior to use: 1 mM PMSF, 1 μg/mL leupeptin, 0.5 μg/mL pepstatin A, and 10 mM N-ethylmaleimide (NEM).
2 or 1% Triton X-100 solution: 50 mM Tris–Cl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 2 or 1% Triton X-100. The following reagents are added immediately prior to use: 1 mM PMSF, 1 μg/mL leupeptin, 0.5 μg/mL pepstatin A, and 10 mM NEM.
2× SDS–PAGE sample buffer: 4% β-mercaptoethanol, 4% SDS, 130 mM Tris–Cl, pH 6.8, 20% glycerol, 10 mg/mL bromophenol blue.
Trichloroacetic acid (TCA) sample buffer: 80 mM Tris–Cl, pH 8.0, 8 mM EDTA, 0.25 M DTT, 3.5% SDS, 15% glycerol, 0.08% Tris-base, 0.01% bromophenol blue.
Anti-HA antibody: 5 mg/mL (Roche).
Protein A-Sepharose: Sepharose 50% (v/v) (GE Health care) is equilibrated with a buffer (50 mM Tris–Cl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1 mM azide) and is stored at 4°C.
IP wash buffer: 50 mM Tris–Cl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.2% SDS. 10 mM NEM (1 M stock in dimethyl sulfoxide) is added immediately prior to use.
SDS–PAGE fixative: 25% (v/v) isopropanol, 10% (v/v) glacial acetic acid in ddH2O.
3. Methods
3.1. Preparation of ER-Derived Microsomes
In vitro ubiquitination of Ste6p* and CFTR depends on relevant ubiquitination enzymes (e.g., Ubc6p/7p, Hrd1p/Doa10p, and Ufd2p) and Hsp70 and Hsp40 molecular chaperones (e.g., Ssa1p and Ydj1p/Hlj1p). To assay the effects of these agents, yeast microsomes are prepared in one of three different ways from mutant cells and isogenic wild-type cells expressing Ste6p* or CFTR. When microsomes are prepared from deletion mutant cells (e.g., ubc6Δubc7Δ, hrd1Δ, doa10Δ, hrd1Δdoa10Δ, and ufd2Δ) and isogenic wild-type strains, the cell walls are first digested with lyticase at room temperature or at 30°C for <1 h before the preparation of cell homogenate (see Section 3.1.1). However, during this incubation at the permissive temperature, the temperature-sensitive defect may be lost. Therefore, when microsomes are instead prepared from temperature-sensitive mutants (e.g., ssa1–45, a mutant form of SSA1, and the ydj1–151/hlj1Δ strains) and isogenic wild-type strains, cells are grown at a permissive temperature of 26°C and then are shifted to a restrictive temperature of 37°C. Cells are then collected on ice and are physically disrupted with glass beads by keeping them on ice to strictly control the temperature (see Section 3.1.2 or 3.1.3)(see Note 4).
3.1.1. Preparation of Microsomes from Homogenates After Spheroplast Formation (Large Scale)
The following procedure, used routinely in our laboratory, is based on a protocol previously described (19–21).
Yeast microsomes are prepared from cells expressing the desired substrate (Ste6p* or CFTR) grown to log to late-log phase (optical density at 600 nm [OD600] of 2–3). Typically yeast cells are grown in 1–2 L of selective medium.
The cell walls are digested with lyticase, and the resulting spheroplasts are collected by centrifugation through Cushion 1. The plasma membrane is then broken with a Teflon-glass motor-driven homogenizer.
Lysates are layered onto Cushion 2 and centrifugation is used to obtain a crude microsomal fraction, which is then concentrated and washed with B88 by centrifugation at approximately 15,000 g for 10 min.
The concentration of microsomes is adjusted to approximately 10 mg protein/mL (OD280 = 40 in 2% SDS) with B88. Microsomes should be stored in single-use aliquots (~50 μL), which are stable indefinitely at −80°C and should be thawed on ice immediately before use.
3.1.2. Preparation of Microsomes From Homogenates After Glass Bead Disruption (Small Scale)
Cells are grown to log phase (OD600 = 0.7–1.5) at a permissive temperature (e.g., ssa1-45 and ydj1-151/hlj1Δ at room temperature) and are shifted to 37°C for approximately 1 h. A shaking water bath is used to strictly control the temperature.
Approximately 20–30 OD600 equivalents of cells are collected by centrifugation at 4,300 g for 5 min at 4°C and are washed once with 20–30 mL of ice-cold distilled water. The pelleted cells are resuspended in 1 mL of ice-cold water, transferred to a new microcentrifuge tube, and recentrifuged and the remaining water is removed. The cells are then frozen in liquid nitrogen and stored at −80°C.
To prepare a crude membrane fraction, add 250 μL of lysis buffer to the frozen cells and disperse the cells quickly by agitation. Transfer the cell suspension to 13 × 100 mm glass test tube (VWR International) and add acid-washed 0.5-mm glass beads (Scientific Industries) to the meniscus.
The cells are broken by vigorous agitation on a vortex mixer for 30 s 10 times, with 30-s intervals on ice between each treatment.
Add 500 μL of ice-cold B88 to the homogenate and agitate for approximately 1 s.
Transfer cell suspension to a pre-cooled microcentrifuge tube on ice.
Wash the glass beads with 500 μL of ice-cold B88 by brief agitation and pool the wash with the homogenate above (step 6).
Remove unbroken cells by two rounds of centrifugation at 830 g for 5 min at 4°C in a refrigerated microcentrifuge.
To obtain the subcellular membrane fraction, the resulting supernatant is centrifuged at 18,000 g for 20 min at 4°C.
Wash the membrane fraction with 1 mL of ice-cold B88 and re-collect the membranes as above (step 9).
Adjust the protein concentration and store at −80°Casin Section 3.1.1, step 4.
3.1.3. Preparation of Microsomes from Homogenate After Glass Beads Disruption (Medium Scale)
Cells are grown to log phase (OD600 = 0.7–1.5) at a permissive temperature and are shifted to 37°C for approximately 1 h (see Section 3.1.2, step 1).
Approximately 200 OD600 equivalents of cells are collected by centrifugation at 4,300 g for 5 min at 4°C and are washed and re-collected by centrifugation two times with 20–30 mL of ice-cold distilled water. Cells can be placed in a polycarbonate centrifugation tube and frozen in liquid nitrogen and stored at −80°C.
To prepare a crude membrane fraction, add 2 mL of lysis buffer to the frozen cells and glass beads to the meniscus.
The cells are broken as in Section 3.1.2, step 4.
Add 5 mL of ice-cold B88 to the homogenate and agitate for approximately 1 s.
Transfer the cell homogenate to a new pre-cooled tube on ice.
Wash the glass beads with 5 mL of ice-cold B88 by brief agitation and pool this wash with the homogenate above (step 6).
Overlay the cell homogenate (~15 mL) onto 15 mL of Cushion 2 in a polycarbonate centrifugation tube and centrifuge in a swinging-bucket HB-6 rotor at 7,000 g for 10 min at 4°C.
Transfer the upper layer to a new polycarbonate tube and centrifuge at 15,000 g for 10 min at 4°C.
Wash the pelleted membrane fraction with 20 mL of B88 and centrifuge again as above (step 9).
Adjust the concentration and store at −80°C as in Section 3.1.1, step 4.
3.2. Preparation of Yeast Cytosol
The following procedure, used routinely in our laboratory, is based on a protocol previously described (21).
Grow yeast cells in rich medium to log phase (OD600 = ~2.0) at 30°C. When the cdc48-3 mutant and isogenic wild-type cells are being used, the yeast are propagated at room temperature and shifted to a restrictive temperature (37°C) for 5 h (22).
Collect the cells and wash with distilled water.
Resuspend the cells in a minimal amount of B88 to form a thick yeast slurry (e.g., <6 mL of B88 per 6 L of initial yeast culture).
Freeze the cells by drop-wise addition to 500 mL liquid nitrogen in a plastic beaker. After the excess liquid nitrogen evaporates, store these “popcorn”-like particles at −80°C.
Add the particles to approx 500 mL of liquid nitrogen and blend at high speed for 8–10 min in a stainless-steel blender. Maintain the volume of liquid nitrogen above the rotating blades by periodic addition of liquid nitrogen during blending.
After the liquid nitrogen evaporates, transfer the powder containing broken cells to a 50 mL Falcon tube, which can be stored at −80°C.
Place the tube on ice and add a minimal amount of B88 (e.g., ~0.5 mL/40 mL of broken yeast slurry) containing 1 mM DTT. Then, thaw the cells in a room temperature water bath.
After thawing, centrifuge the lysate at 10,000 g for 10 min at 4°C. The supernatant is then collected and centrifuged at 300,000 g for 1 h at 4°C to remove membranes/aggregated protein.
The supernatant from this final spin is aliquoted (~100–200 μL), snap-frozen in liquid nitrogen, and stored at −80°C. The concentration of resultant cytosol is usually 20–30 mg/mL (see Note 2). Avoid repeated freeze and thaw cycles as the activity of the lysate diminishes upon each cycle.
3.3. Ubiquitination of Ste6p*
The in vitro ubiquitinated Ste6p* and the presence of unmodified Ste6p* can be detected by autoradiography and by western blotting, respectively. The following procedure results in sample volumes of approximately 28 μL, but 12-μL samples are sufficient for autoradiography or western blotting. The same protocol can be used to detect in vitro ubiquitinated CFTR.
Combine the reagents in the following order on ice: B88 (sufficient amount for an initial reaction volume of 18 μL), 2 μL of microsomes, 2 μL of 10× ATP-regenerating system, and the appropriate final concentration of cytosol (typically 1–4 mg/mL). As a negative control, microsomes prepared from the strain lacking the Ste6p* expression vector and B88 instead of cytosol can be used. Add reaction inhibitors such as apyrase (ATP control) or methylated ubiquitin (inhibitor of ubiquitin extension; see Fig. 21.1) at this point (see Note 5).
Pre-incubate the reaction at 23°C for 10 min.
Add 2 μL 125I-labeled ubiquitin (see Note 7).
Incubate up to 1 h at 23°C (see Note 3).
At the desired time point, add 80 μL of 1.25% SDS stop solution and briefly agitate (~2 s) on a vortex mixer at high speed.
Incubate at 37°C for 30 min.
Add 400 μL of 2% Triton X-100 solution or 900 μL of 1% Triton X-100 solution and place the tubes on ice.
Add 2 μL of anti-HA (10 μg) antibody and gently mix the solution by rotating overnight at 4°C.
Add 30 μL of 50% (v/v) Protein A-Sepharose and continue to rotate at 4°C for 2–3 h.
Harvest and then wash the immunoprecipitates with 800 μL of ice-cold IP wash buffer four times (collect the Sepharose beads by centrifugation at 2,100g for 10 s at room temperature using a mini-centrifuge). The samples are placed on ice between each step. After the final wash, remove as much buffer as possible from the Sepharose with a gel-loading micropipet tip.
Add 30 μL of either 2× SDS–PAGE sample buffer or TCA sample buffer and elute bound proteins by incubating at 37°C for 30 min.
Spin down the Sepharose by centrifugation at 7,000 g for 10 s at room temperature.
Transfer the supernatant (~28 μL) to a new Eppendorf tube and analyze via 6% SDS–PAGE (see Section 3.5).
Fig. 21.1.
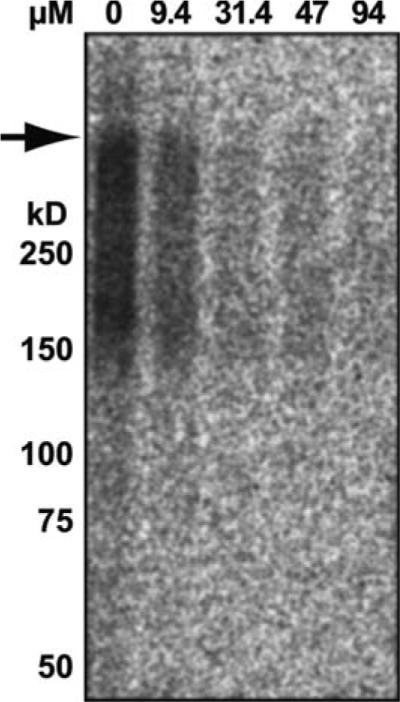
The in vitro ubiquitination assay. The assay was performed essentially as described in Section 3.3 using wild-type ER-derived microsomes (prepared as in Section 3.1.1) from yeast expressing Ste6p*-3HA. The microsomes were incubated with a final concentration of 6.5 mg/mL cytosol, the ATP-regenerating mix, 125I-labeled ubiquitin, and the indicated concentration of methylated ubiquitin at 30°C for 15 min. The arrow indicates the boundary between the stacking and running gel.
3.4. Membrane Extraction of Ubiquitinated Ste6p*
The membrane extraction assay is similar to the ubiquitination assay except that each sample is separated into membrane and cytosolic fractions by centrifugation after the ubiquitination reaction.
Set up 25-μL reactions using the procedure outlined in Section 3.3, steps 1–4 (we typically set up 25-μL reactions to obtain a 20-μL supernatant, which does not disturb the pellet). To observe Cdc48p/p97-dependent extraction of ubiquitinated Ste6p*,use cdc48-3 mutant cytosol and its isogenic wild-type cytosol prepared as in Section 3.2. Optimal Cdc48 dependence is observed when the membranes are incubated for 5 min on ice and washed 2 times with B88.
Following the incubation, pellet the microsomes in a refrigerated microcentrifuge at 18,000 g for 10 min at 4°C.
Return the reaction tube to ice and quickly transfer 20 μL of supernatant (containing extracted/ubiquitinated Ste6p*)to a new microcentrifuge tube on ice. Be careful not to disturb the pellet.
Remove the remaining supernatant completely and resuspend the pelleted microsomes in 25 μL of ice-cold B88. Transfer 20 μL of suspension to a new tube.
Add 80 μL of 1.25% SDS stop solution to the supernatant and resupspended microsomes and briefly agitate the mixture on a vortex mixer. Process the samples as in Section 3.3, steps 6–13.
3.5. Data Collection and Analysis
3.5.1. Autoradiography
Half of the final sample (~12 μL, see Section 3.3, step 13) is analyzed by SDS–PAGE. Typically, 6 cm 6% gels are used to resolve the ubiquitinated species and are run at 20 mA (constant current) until the bromophenol blue dye front is at the bottom of the gel (see Note 6). Unmodified Ste6p* will reside approximately in the center of the separating phase of the gel and the “smear” of ubiquitinated Ste6p* will reside in the upper half of the gel.
The gel is gently placed in SDS–PAGE fixative for 15 min to 2 h and gently shaken at room temperature. Gels are dried on filter paper for approximately 45 min on a vacuum drier with heating (~80°C) and are then cooled to room temperature before the vacuum is broken. Typically, the resulting autoradiograph requires 1 day of exposure on a phosphoimage screen although the use of old label may require significantly longer exposure times (e.g., ~1 month).
3.5.2. Western Blotting
The other half of the sample (~12 μL) is used to detect unmodified Ste6p* by western blotting in the same manner as described in Section 3.5.1, step 1.
Proteins are transferred from gels to a nitrocellulose membrane, which is then blotted with anti-HA antibody followed by decoration with horseradish peroxidase-conjugated secondary antiserum. The bound secondary antibody is detected with the SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) according to the manufacture's instructions.
Footnotes
We typically store 125I-labeled ubiquitin at −80°C in aliquots of 20 μL. Although repeated freeze and thaw cycles (~3 times) do not seem to be detrimental to activity, best results are seen when the reagent is used within 2 months (half-life of 125I is ~60 days) after preparation. Non-labeled ubiquitin is also stored at −80°C in aliquots of 20 μL.
Protein concentration of the cytosol is measured by the Bradford method with the protein assay kit (Bio-Rad). Bovine serum albumin (BSA) is used as the standard. No detectable loss of the activity was seen when cytosol was stored at −80°C for up to approximately 12 months.
The in vitro ubiquitination of Ste6p* requires physiological temperature and does not occur on ice. The optimal temperature is 23°C and the extent of ubiquitination becomes inefficient at higher temperatures (e.g., 37°C), possibly because the misfolded substrate protein aggregates. However, the phenotype of some temperature-sensitive mutant alleles (e.g., ssa1–45), which is most evident at 37°C in vivo, is exhibited at 23°C in the in vitro reaction.
Microsomes prepared from homogenates after spheroplast formation (see Section 3.1.1) or after glass beads disruption in a medium scale (see Section 3.1.3) are more E3 ligase enzyme-dependent than microsomes prepared from homogenates after a small-scale glass bead disruption (see Section 3.1.2).
The addition of an inhibitor for deubiquitination (ubiquitin aldehyde) or a proteasome inhibitor (MG132 “n-cbz-leu-leu-leu-al”) does not result in increased ubiquitin chain extension. In addition, higher concentrations of cytosol (>~8 mg/mL) decrease the signal intensity.
Cytosol at a final concentration of 1–2 mg/mL results in a low-molecular weight ubiquitinated species, but addition of more cytosol (at a final concentration of 4–6 mg/mL) “shifts” the ubiquitinated species to a higher molecular weight. The use of a 6% gel is critical to differentiate these two species, and this molecular weight shift is due to Ufd2p in the cytosol (12).
All reaction samples containing 125I-labeled ubiquitin should be shielded by an approximately 1-mm lead plate to prevent excess exposure. The aliquots of 125I-labeled ubiquitin should be stored in a substantially more shielded lead container. Radioactivity at each step of this protocol should be surveyed with a γ-detecting monitor, and all items that contact 125I should be properly disposed.
References
- 1.Tsai B, Ye Y, Rapoport TA. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell Biol. 2002;3:246–255. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- 2.Kostova Z, Wolf DH. For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J. 2003;22:2309–2317. doi: 10.1093/emboj/cdg227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat. Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 4.Römisch K. Endoplasmic reticulum-associated degradation. Annu. Rev. Cell Dev. Biol. 2005;21:435–456. doi: 10.1146/annurev.cellbio.21.012704.133250. [DOI] [PubMed] [Google Scholar]
- 5.Nakatsukasa K, Brodsky JL. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic. 2008;9:861–870. doi: 10.1111/j.1600-0854.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vashist S, Ng DT. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishikawa S, Brodsky JL, Nakatsukasa K. Roles of molecular chaperones in endoplasmic reticulum (ER) quality control and ER-associated degradation (ERAD) J. Biochem. 2005;137:551–555. doi: 10.1093/jb/mvi068. [DOI] [PubMed] [Google Scholar]
- 8.Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 9.Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 10.Loayza D, Tam A, Schmidt WK, Michaelis S. Ste6p mutants defective in exit from the endoplasmic reticulum (ER) reveal aspects of an ER quality control pathway in Saccharomyces cerevisiae. Mol. Biol. Cell. 1998;9:2767–2784. doi: 10.1091/mbc.9.10.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huyer G, Piluek WF, Fansler Z, Kreft SG, Hochstrasser M, Brodsky JL, Michaelis S. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J. Biol. Chem. 2004;279:38369–38378. doi: 10.1074/jbc.M402468200. [DOI] [PubMed] [Google Scholar]
- 12.Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 2008;132:101–112. doi: 10.1016/j.cell.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Nijbroek G, Sullivan ML, McCracken AA, Watkins SC, Michaelis S, Brodsky JL. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol. Biol. Cell. 2001;12:1303–1314. doi: 10.1091/mbc.12.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaiser CA, Michaelis S, Mitchell A. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1994. [Google Scholar]
- 15.Nossal NG, Heppel LA. The release of enzymes by osmotic shock from Escherichia coli in exponential phase. J. Biol. Chem. 1966;241:3055–3062. [PubMed] [Google Scholar]
- 16.Shen SH, Chrétien P, Bastien L, Slilaty SN. Primary sequence of the glucanase gene from Oerskovia xanthineolytica. Expression and purification of the enzyme from Escherichia coli. J. Biol. Chem. 1991;266:1058–1063. [PubMed] [Google Scholar]
- 17.McFarlane AS. Efficient trace-labelling of proteins with iodine. Nature. 1958;182:53. doi: 10.1038/182053a0. [DOI] [PubMed] [Google Scholar]
- 18.Helmkamp RW, Goodland RL, Bale WF, Spar IL, Mutschler LE. Cancer Res. 1960;20:1495–1500. [PubMed] [Google Scholar]
- 19.Rothblatt JA, Meyer DI. Secretion in yeast: reconstitution of the translocation and glycosylation of alpha-factor and invertase in a homologous cell-free system. Cell. 1986;44:619–628. doi: 10.1016/0092-8674(86)90271-0. [DOI] [PubMed] [Google Scholar]
- 20.Deshaies RJ, Schekman R. SEC62 encodes a putative membrane protein required for protein translocation into the yeast endoplasmic reticulum. J. Cell Biol. 1989;109:2653–2664. doi: 10.1083/jcb.109.6.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brodsky JL, Schekman R. A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J. Cell Biol. 1993;123:1355–1363. doi: 10.1083/jcb.123.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Latterich M, Fröhlich KU, Schekman R. Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER. Cell. 1995;82:885–893. doi: 10.1016/0092-8674(95)90268-6. [DOI] [PubMed] [Google Scholar]


