Abstract
Viral capsid assembly and stability in tailed, dsDNA phage and Herpesviridae are achieved by various means including chemical cross-links (unique to HK97), or auxiliary proteins (lambda, T4, ϕ29, and herpesviruses). All these viruses have coat proteins (CP) with a conserved, HK97-like core structure. We used a combination of trypsin digestion, gold-labeling, cryo-electron microscopy, 3D image reconstruction, and comparative modeling to derive two independent, pseudo-atomic models of bacteriophage P22 CP: before and after maturation. P22 capsid stabilization results from inter-subunit interactions among N-terminal helices and an extensive “P-loop”, which obviate the need for cross-links or auxiliary proteins. P22 CP also has a telokin-like, Ig domain that stabilizes the monomer fold so that assembly may proceed via individual subunit addition rather than via preformed capsomers as occurs in HK97. Hence, the P22 CP structure may be a paradigm for understanding how monomers assemble in viruses like ϕ29 and HSV-1.
Keywords: cryo-TEM, image reconstruction, coat protein, bacteriophage P22, virus assembly
Introduction
At least 1031 viruses are estimated to inhabit the biosphere and thus, they comprise the largest biomass on Earth (Bamford et al., 2005). Tailed dsDNA bacteriophage are the most abundant of all viruses and encapsulate their genomes in isometric or prolate heads composed primarily of many copies of a major coat protein (CP). The CPs of all tailed phage studied to date and herpesvirus share a common, HK97-like fold (Baker et al., 2005; Hendrix, 2002), which serves a critical role in phage assembly and is also believed to control key steps in the maturation of human pathogens like herpes. Genome packaging in phage and viruses with HK97-like CPs occurs via a process in which immature, metastable particles (procapsids) transform into more stable, mature capsids (King and Chiu, 1997). This transformation often proceeds through a sequence of several conformational changes in the CP involving, for example, unbending of α-helices, untwisting of β-sheets, and rigid body motions (Gertsman et al., 2009).
Bacteriophage P22 is one of the most studied systems in biology. However, a crystal structure of the 430 residue, ~47 kDa CP (gp5) has yet to be reported that would help integrate the extensive body of biochemical, molecular biological, and biophysical knowledge obtained from over more than three decades. Assembly of P22 proceeds through a nucleation-limited reaction in which 60–300 scaffolding proteins (gp8) co-polymerize 415 copies of CP, 12 copies of portal protein (gp1), and 12–20 copies each of three ejection proteins (gp7, gp16, and gp20) to form procapsids (King et al., 1976). The CP shell of P22 procapsids is an isometric structure comprised of the dodecameric portal complex and 71 capsomers, 60 of which are hexameric oligomers of CP (“hexons”) and eleven of which are pentameric oligomers of CP (“pentons”). Virus maturation continues when a complex of two additional proteins (gp2/3) packages the dsDNA genome and scaffolding protein is simultaneously released through large holes in the capsomers (Bazinet, 1988; Prasad et al., 1993). Final stages of assembly include ~10% expansion in volume of the head (Prasad et al., 1993) and addition of gp4, gp10, gp26, and the tail machinery (gp9) (Lander et al., 2009).
Recent advances in cryo-transmission electron microscopy (cryo-TEM) and three-dimensional (3D) image reconstruction have accelerated the means by which structures of large, macromolecular complexes can be determined at sub-nanometer resolutions (Zhou, 2008). These methods are therefore well suited to explore the mechanisms that drive capsid stabilization in viruses and we have used them to produce a nearly complete model for the P22 CP protein that helps identify how it transforms during capsid expansion. The modeling results were based on systematic investigation of several P22 specimens, including limited proteolysis and mass spectrometry of soluble monomers, and cryo-TEM of (1) wild-type (WT) empty procapsid shells (shells), (2) WT heat-expanded heads (ExH), (3) protease-digested ExH, and (4) three different, gold-labeled, cysteine variants of shells. We have modeled 91% of the P22 CP and have identified a separate, Ig-like domain with structural homology to telokin (Holden et al., 1992). This domain corresponds to the surface protrusions in P22 previously named “extra density” (ED) domains, although the function of this domain was not yet known (Jiang et al., 2003). ED domains have also been identified in related viruses, ϕ29 and HSV-1 (Baker et al., 2005; Morais et al., 2005). Furthermore, we show that the P22 CP undergoes a novel refolding that suggests a new mechanism for capsid stabilization, replacing the need to form covalent cross-links (“chain mail”) as uniquely occurs in HK97 (Gertsman et al., 2009; and references therein; Wikoff et al., 2000).
Though the atomic structure of HK97 (Gertsman et al., 2009; Wikoff et al., 2000) has emerged as the de facto standard for comparing lambda-like phage and Herpesviridae, our new model of the P22 CP highlights key differences that distinguish it from the HK97 CP and these differences answer as well as raise new questions about phage evolution (Table 1). Indeed, the vast majority of dsDNA phage/viruses may have evolved assembly mechanisms more like P22 and less like HK97 (Baker et al., 2005).
Table 1.
dsDNA phages/viruses with HK97 fold
| Virus | # CP a.a. § |
Cleavage † | Assembly | Capsid | Expansion†† | Capsid stabilization mechanism |
|---|---|---|---|---|---|---|
| HK97 | 282 | Yes | C | I | Yes | Chemical crosslinking |
| ε15 | 335 | N.A. | N.A. | I | N.A. | Auxiliary protein: gp10 |
| Lambda | 341 | No | N.A. | I | Yes | Auxiliary protein: gpD |
| T7 | 345 | No | N.A. | I | Yes | E-loop* |
| P22 | 430 | No | M | I | Yes | N-arm/P-loop interaction |
| ϕ29 | 448 | No | M | P | No | BIG2 ED bridges capsomers |
| T4** | 521 | Yes | C | P | Yes | Auxiliary protein: Soc |
| HSV-1 | 1374 | No | M | I | No | Auxiliary protein: triplex |
Table order based on CP length.
Abbreviations: N.A. = information not available; C= preformed capsomers; M = monomeric subunits; I = isometric; P = prolate;
proposed, but not confirmed experimentally (Agirrezabala et al., 2007)
T4 comparisons are done in terms of gp23, the major CP (forms hexons).
Is the CP cleaved during maturation?
Expansion ~10 % or greater upon dsDNA packaging?
Results
Structure of penton-less P22 capsids at sub-nanometer resolution
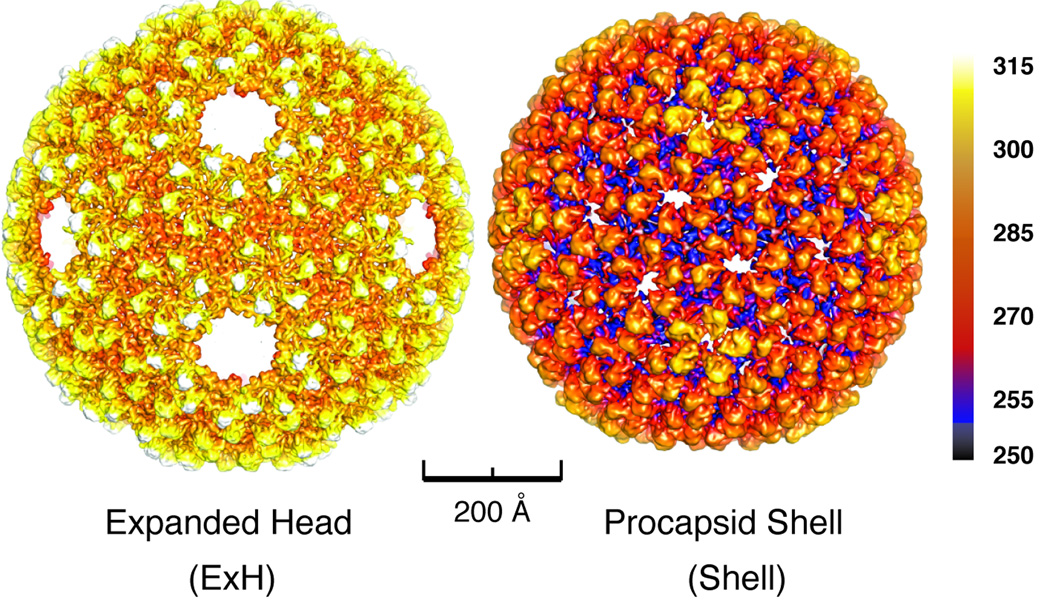
When heated in vitro, procapsids expand (~10% volume) and lose their pentons and portal, scaffold, and ejection proteins, resulting in so-called, “penton-less head” or ExH particles (Teschke et al., 2003). During this expansion, hexons change from a skewed, 2-fold symmetric structure into one that is nearly 6-fold symmetric (Teschke et al., 2003). Shells are produced when procapsids are treated with GuHCl and they only contain CP but have a full complement of pentons and hexons in the unexpanded, immature state (Experimental Procedures). The gross morphologies of P22 shells and ExH particles are revealed in icosahedral cryo-reconstructions, both obtained at sub-nanometer resolution (Figure 1 and Table 2).
Figure 1. 3D cryo-reconstructions of P22 expanded head and procapsid shell.
Radially color-cued surface representations of (left) ExH and (right) shell cryo-reconstructions, computed to 7.0- and 9.0-Å resolutions, respectively. Color bar indicates corresponding radii (in Å).
Table 2.
P22 image processing data
| CP type* | Caps id type**: | Images† | Defocus (µ.m) †† | Resolution (Å) *† |
|---|---|---|---|---|
| WT | ExH | 3,308 | 0.63 – 3.16 | 8.2 |
| WT-Try | ExH | 665 | 1.02 – 2.95 | 14.0 |
| WT | Shell | 11,997 | 0.41 – 3.65 | 9.1 |
| F170C-Au | Shell | 1,821 | 0.55 – 3.06 | 14.5 |
| A285C-Au | Shell | 4,679 | 0.71 – 3.75 | 11.3 |
| T379C-Au | Shell | 2,755 | 0.52 – 2.27 | 11.1 |
WT= Wild Type, Try = trypsin digested, Au= gold labeled
ExH = expanded heads, Shell = procapsid shell
Number of boxed particles used in image reconstruction
Range of objective lens underfocus settings for micrographs
Estimation based on FSC0.5criterion (van Heel and Schatz, 2005)
ExH particles are ideal for structural studies because they only contain CP and, the absence of pentons exposes a significant portion of one hexon subunit that otherwise forms close interactions with penton subunits in unexpanded procapsids. These properties of ExH particles and the sub-nanometer resolution achieved in the reconstruction helped us define the molecular envelopes of all individual hexon subunits, and this in turn was critical for building a near-complete, detailed model of the P22 CP structure. The absence of pentons in ExH also allowed us to detect conformational changes in CP that accompany different subunit:subunit interactions before and after expansion.
Location of N-arm revealed by limited proteolysis
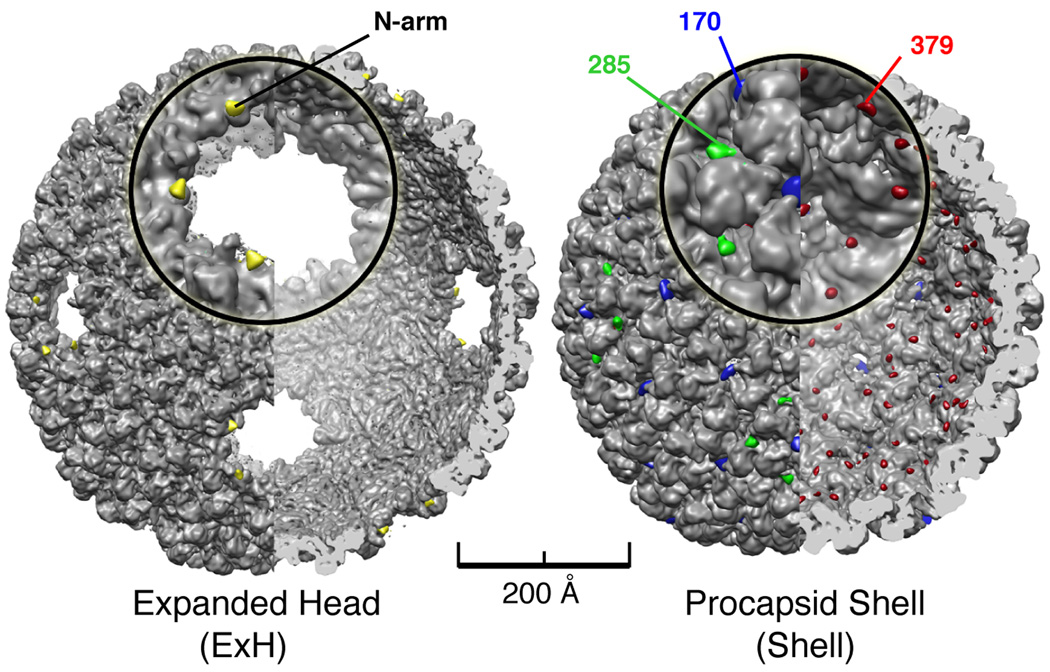
Trypsin digestion of ExH results in a single cleavage event that converts CP into products of ~43 and ~4 kDa (Capen and Teschke, 2000). Peptide masses were estimated from both migration distance in the SDS gel for the 43 kDa product and the predicted mass for a 42-residue peptide. Extensive trypsin digestion of ExH followed by MALDI-TOF-TOF MS/MS sequencing of the ~43 kDa product showed that it contains residues 43 to 430 (Supplemental Data). Hence, the ~4 kDa segment comprised the first 42 N-terminal residues. Cryo-TEM images of purified, digested ExH particles yielded a cryo-reconstruction at 14-Å resolution (Table 2). Subtraction of this reconstruction from that of un-digested ExH particles rendered at the same resolution produced a difference map showing the location of the N-arm in the subunit closest to the missing penton hole (Figure 2). The CP of mature, tail-less but genome-containing, heads is known to be trypsin-resistant (Lanman et al., 1999), so heat-induced release of pentons appears to expose a site that is normally inaccessible. Previous studies of the P22 CP structure were unable to determine the locations of any N- or C-termini (Jiang et al., 2003), and thus our identification of the N-arm in one ExH hexon subunit provided a key landmark for deriving a reliable CP model.
Figure 2. Identification of P22 N-arms in ExH and nano-gold labeled residues in shell mutants.
(Left) Cutaway view along two-fold direction of digested ExH reconstruction (computed to 7.0 Å, gray shaded surface) showing superimposed densities ascribed to the N-arms (yellow) obtained from an ExH minus digested ExH difference map. The ExH and digested ExH reconstructions were both computed at 14.0 Å resolution for difference map analysis. An enlarged view of one vertex appears in the insert. (Right) Same as left panel but showing that difference densities corresponding to nano-gold labels in reconstructions of three shell variants pinpoint the locations of residues 170 (blue), 285 (green), and 379 (red) in the WT procapsid shell (computed to 9.0Å, gray shaded-surface). Difference map analyses were all performed with gold-labeled and WT shell maps computed to 11.0 Å resolution.
CP residue locations identified in gold-labeled, procapsid mutants
As a further guide to accurate modeling of the CP structure, we used cryo-reconstruction methods to study shells that were engineered to include cysteine residues so electron-dense, 1.4 nm mono-maleimido nanogold could be added and specific residues located (Experimental Procedures). All mutant specimens behaved similarly to WT as monitored by standard phage assays that test for the ability of variant proteins to fold and assemble in vivo into infectious particles under physiological temperatures (data not shown). Candidates for nanogold labeling and cryo-TEM imaging were identified on the basis of a quantitative fluorescence assay, in which tetramethylrhodamine (TMRM) was used to determine the molar ratio of cysteines available for modification (Experimental Procedures, Table 3).
Table 3.
Tetramethylrhodamine-5-maleimide labeling
| CP type | Molar Ratio* |
|---|---|
| WT | 0.20 |
| F170C | 1.12 |
| A285C | 0.57 |
| T379C | 1.41 |
Molar ratio expressed as moles of TMRM per mole of CP; Typical error for these experiments is ± 0.12
Cryo-reconstructions of purified shells derived from three cysteine variants (F170C, A285C, and T379C) were compared via difference map analysis to an unlabeled, WT shell reconstruction (Figure 2, Table 2). None of the variants exhibited any obvious changes in CP secondary or tertiary structure compared to WT CP (data not shown). Neither cysteine substitution nor gold-labeling appear to cause structural perturbations, which raises the level of confidence that individual residue locations were identified and could be modeled accurately.
TMRM labeling of the A285C mutant showed the cysteines to be ~57% solvent-accessible (Table 3). The A285C reconstructed density map showed extra density was clearly present, but only on the penton surface (Figure 2). Though the TMRM labeling experiment indicated that not all hexon subunits receive label, those that do must occur randomly within the six, quasi-equivalent subunits in each asymmetric unit (Johnson, 1996). The averaging inherent in producing the icosahedral reconstruction would therefore reduce any signal attributed to labeled hexon subunits. In contrast to the A285C result, TMRM labeling indicated that 100% of the cysteine sites were solvent accessible in the F170C and T379C variants and gold label was observed in all subunits in the corresponding density maps (Figure 2). In F170C, label accumulated at the centers of the pentons and hexons, whereas in T379C, all subunits at the icosahedral and quasi three-fold axes of symmetry received label on the inner surface of the shell. WT P22 CP does contain a single cysteine residue (C405), but binds TMRM only marginally above background (Table 2). Consistent with this, a difference map computed between reconstructed maps of nano-gold treated and untreated (i.e. control) WT shells showed no evidence of label on particles (data not shown).
Stable, contiguous domain revealed by proteolysis of monomeric, C-terminal fragment
Modeling of the P22 CP subunit was further guided by the results of limited proteolysis of soluble CP. N-terminal (a.a 1–190) and C-terminal (a.a. 191–430) halves of the P22 CP form inclusion bodies when they are individually expressed in E. coli and only the C-terminal half can be refolded into soluble protein that exhibits high β content as revealed by circular dichroism (Kang and Prevelige, 2005). We used time-course trypsin digestion to produce proteolytic fragments from samples of the refolded, C-terminal half and analyzed these by SDS-PAGE and mass spectrometry (Supplemental Data). The presence of a well-defined, prominent band in the SDS gel suggested that a contiguous, folded domain existed within the CP C-terminal half (black arrow, Figure 3). MALDI-TOF-TOF and MS/MS mass spectrometry revealed this domain to be comprised of residues 223–349.
Figure 3. Residues 223–349 fold into a stable, contiguous domain.
SDS gel showing trypsin digestion of the C-terminal region (a.a. 191–430) of P22 CP. Lane 1 is the undigested C-terminal region, 2 is molecular weight markers, and 3 and 4 are of samples taken after 30 or 60 minutes of digestion at 20 °C, respectively. The black arrow points to the band corresponding to residues 223–349 (telokin domain). The white arrow points to a band with faster electrophoretic mobility that, when sequenced by MS/MS MALDI-TOF-TOF mass spectrometry, revealed multiple fragments with sequences all falling within in the 223–349 range.
Modeling of CP in mature (ExH) and immature (shell) states
We initiated comparative modeling of the P22 CP by employing the Fold and Function Assignment System (FFAS03), which provided an unbiased way to screen for candidate templates. This led to the crystal structures of HK97 Head II (PDB entry 1OHG) and Ig-like telokin (PDB entry 1FHG) as the best candidates, and these templates were used to model two, non-overlapping regions of the P22 CP sequence (Figure 4, Experimental Procedures). The presence of an Ig-like domain in the P22 CP is not surprising since Ig domains often occur in viruses and phage (Fraser et al., 2006). Structure-based sequence alignment provides a more accurate means to align sequences of structurally homologous proteins that exhibit low sequence similarity (Zhang and Skolnick, 2005). Therefore, we used the PROMALS3D server to align the entire P22 CP sequence against four structures homologous to the HK97 CP and two structures homologous to telokin (Figure 4, Experimental Procedures).
Figure 4. Sequence alignment and structure prediction of P22 coat protein.
The top two rows show secondary structure depictions of firstly, the final P22 CP model (yellow helices and cyan b-strands) as identified with the program Stride (Frishman and Argos, 1995) and secondly, as predicted (orange helices and violet b-strands) using the Jpred3 server (Cole et al., 2008). Next, the a.a. sequence of the P22 CP is listed above fifteen aligned sequences, which correspond to three different templates for HK97 gp5* (PDB ID: 1OHG), the CP of phage PfV virus-like particles (PDB ID: 2E0Z), bacteriophage T4 gp24 (PDB ID: 1YUE), hypothetical prophage proteins from B. bronchiseptica (PDB ID: 3BQW) and E. coli (PDB ID: 3BJQ), the encapsulin from T. maritima (PDB ID: 3DKT) (Pei et al., 2008), three templates of the D1 and one of the D2 domain of the human Fc receptor (PDB ID: 1E4J), telokin (PDB ID: 1FHG), and the D1 and D2 domains of human Ig Epsilon (PDB ID: 1F2Q). Alignments produced using the FFAS03 (Jaroszewski et al., 2005) and PROMALS3D (Pei et al., 2008) servers are labeled accordingly. Alignments for HK97 gp5* (Template), Ig Fc receptor D1 (Template), and telokin were adjusted manually using the described iterated procedure. Regions corresponding to the HK97 and telokin folds are shaded in green and pink, respectively. The area shaded in white corresponds to the unmodeled portion of the CP, and the region shaded in yellow corresponds to the 2-strand β-strand insert modeled from the hypothetical prophage protein. Sequence shading is based on the Jalview Blosum62 coloring. Briefly, the residue is colored dark blue (HK97) or dark purple (telokin) if it matches the consensus sequence (not shown) (Clamp et al., 2004).The residue is colored light blue (HK97) or light purple (telokin) if it does not match the consensus sequence, but has a positive Blosum62 score (Clamp et al., 2004). Colored circles beneath the aligned sequences identify residues 170 (blue), 285 (green), and 379 (red), where nanogold-labeling occurred in P22 shell variants studied by cryo-TEM. Regions of the CP identified by trypsin digestion are highlighted with yellow and pink bars, corresponding to the N-terminal arm and telokin domain, respectively.
Near-complete models for the CP structure (91 % of the sequence) in procapsid shells and in ExH were ultimately generated as follows. Each model consisted of two main components: an HK97 core (“core”) that accounted for the majority of the reconstructed density, and a telokin domain that accounted for the prominent P22 surface protrusion. Atomic models of the HK97 CP in Head II and Prohead II (Gertsman et al., 2009) (PDB entry 3E8K) particles served as templates to construct models of the P22 CP core in the ExH and shell reconstructions, respectively. The P22 protrusions in the ExH and shell reconstructions were both modeled using the same telokin template (Holden et al., 1992). The residues in P22 CP predicted by FFAS03 to adopt a telokin fold correspond to those that span the trypsin-resistant, C-terminal half described above, and highlight the notion that the ED is a stable, contiguous domain. The HK97 and telokin templates served as guides to manually segment out respective density volumes from the two reconstructed maps, corresponding to the core and ED portions of the P22 CP in ExH and shell particles. The X-ray templates were then fitted independently as rigid bodies into the segmented densities, and each was adjusted as needed to produce a backbone structure onto which the P22 sequence was threaded. This threading was greatly guided by knowledge gained from the gold-labeling (residue positions) and digestion (N-arm location) results described above.
The final two P22 CP models (Figure 5A) were derived through an exhaustive, iterative process involving several cycles of manual threading, energy minimization, molecular dynamics-driven flexible fitting, and density segmentation (Experimental Procedures). The reliability of each CP model was independently assessed and found to be consistent with the results of cross-linking (Kang et al., 2006), hydrogen/deuterium exchange (Kang and Prevelige, 2005), and proteolysis (Lanman et al., 1999) studies. Even though the models do not provide a full atomic description of the P22 CP, they do reliably represent the bulk of the CP structure before and after capsid maturation. Full asymmetric unit models for the ExH and shell particles were constructed by separately fitting and refining the respective CP models within the remaining unmodeled, reconstructed density for five (ExH) or six (shell) subunits (Figure 5B). These were then used to generate complete, icosahedral capsid models that accounted for and reliably represented all experimentally observed cryoTEM density (Figure 6).
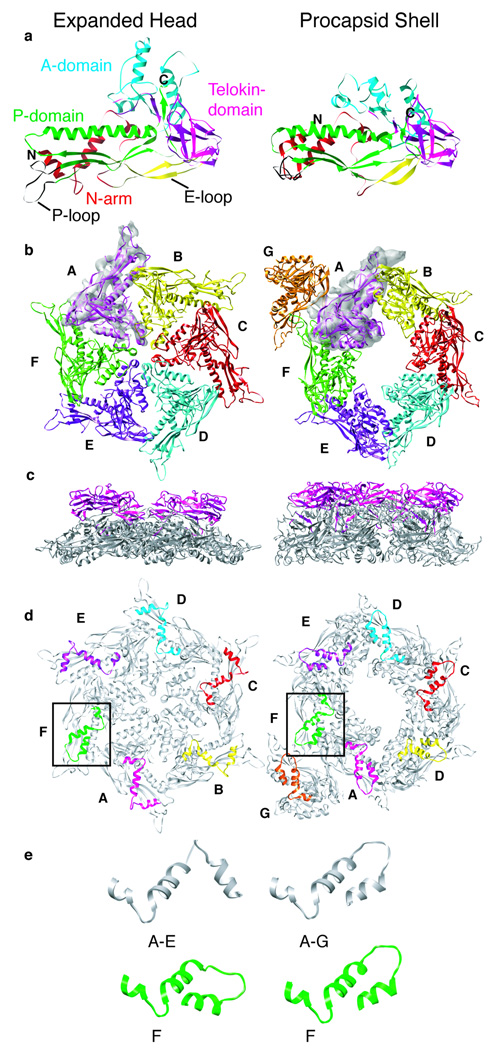
Figure 5. Models of P22 CP in ExH and shell particles.
(A) Final, refined models of the P22 CP in ExH (left) and shells (right), aligned with respect to their telokin domains (magenta). Structural landmarks previously identified in HK97 (Gertsman et al., 2009), including A- and P-domains, and the N-arm and E-loop, are highlighted in separate colors. Modeled N and C-termini are labeled “N” (residue 2) and “C” (residue 428). (B) Modeled asymmetric units of ExH (one hexon) and the shell (hexon plus one penton subunit), viewed along the hexon axis from outside the capsid. Segmented density for one subunit in each cryo-reconstruction is included to illustrate the quality of match between model and experimental data. Colors used to highlight the six (ExH) or seven (shell) quasi-equivalent subunits in each asymmetric unit (Subunit A-magenta, B-yellow, C-red, D-cyan, E-purple, F-green, G-orange). (C) Same as (B) but for side views (hexon axes vertical) with only telokin domains colored (D) Same as (B) but viewed from inside the capsid with only N-arms (a.a. 2–42) in color. Boxes draw attention to the N-arms of F-subunits, which lie closest to the missing penton. (E) Enlarged views of N-arms show that in five ExH subunits (A–E) they adopt one helix-loop-helix conformation whereas in all shell subunits (A–G) they adopt another, with the primary difference confined to the loop and N-terminal helix. Furthermore, this loop-helix motif in the ExH F-subunit is more similar to that observed in all shell subunits.
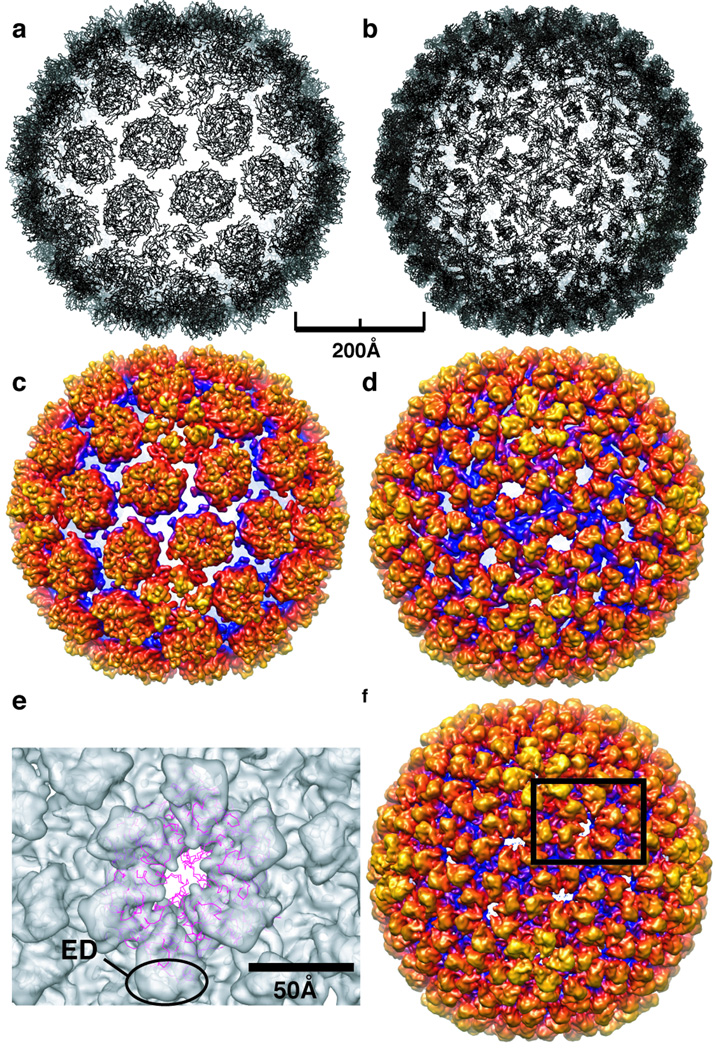
Figure 6. Comparison of HK97 Prohead II crystal structure and P22 shell model.
A) The crystal structure of HK97 Prohead II (PDB entry 3E8K), shown as a ribbon representation, docked as a rigid body into the P22 shell cryo-TEM density. Large gaps between capsomers are apparent. B). Final ribbon model of the P22 shell subunit, which shows no large gaps between capsomers. C & D) Density maps computed to 9.0-Å resolution from models in (A) and (B), both with a temperature factor of 250 Å2 applied and radially color-cued as in Figure 1. E) Magnified view of one asymmetric unit of the P22 shell cryo-TEM density map (shown in F) into which the HK97 Prohead II crystal structure is docked. Notably, the Prohead II model does not account for any of the shell density corresponding to the ED domain. F) Cryo-reconstruction of the P22 procapsid shell computed to 9.0-Å resolution (color cued as in Figure 1 and to which an inverse temperature factor of 1/250 Å−2 has been applied) shows excellent agreement with the P22 shell model (B,D).
An ExH hexon, viewed from the side, shows how the telokin-like ED domains project radially away from the surface of a relatively thin (~ 40 Å) capsid wall (Figure 5C). These domains make no quaternary contacts with one another either within or between hexons (Figure 1) and the closest points of contact occur at the icosahedral 2-folds where the EDs are separated by ~ 18 Å and ~9 Å gaps in ExH and shells, respectively (Figure 1). The apparent independence of the ED suggests it has no direct role in stabilizing the capsid in contrast to the putative role of the BIG2 ED in phage ϕ29 (Morais et al., 2005).
P22 CP structure reveals a novel mechanism of capsid stabilization
The P22 shell and ExH CP models suggest that two structural components primarily drive capsid stabilization during maturation. These include the N-arm and P-loop structures. The final P22 shell model showed that all seven, quasi-equivalent subunits have N-arms with nearly identical, helix-loop-helix structures (Figures 5D,E and 7). However, five of the six N-arms in ExH adopt a second, distinctly different helix-loop-helix structure. The notable exception occurs in the N-arm in the ExH subunit nearest the missing penton (Figure 5E, subunit “F”), which is more like the conformation seen in all seven procapsid shell subunits. Since heat treatment leads to penton release and formation of expanded particles, the penton:hexon interface likely represents the region of weakest intersubunit association in capsids. Loss of pentons removes the stabilizing interactions previously present between the penton and the N-arm of the nearby hexon subunit (Figure 7B,C) and this probably allows the N-arm of this one subunit to refold into the immature (procapsid) state. All these data are consistent with hydrogen/deuterium exchange experiments that show the N-arm becomes less solvent accessible during expansion (Kang and Prevelige, 2005). This therefore solidifies the notion that trimeric N-arm interactions contribute to capsid stability (Figure 7B,C).
Figure 7. N-arm, P-loop interactions stabilize capsids at 3 fold symmetry axes.
(A) An ExH CP model superimposed with corresponding segmented density from the ExH cryo-reconstruction with enlarged, stereo view of region (in box) containing N-arm shown at right. Residue 2 of the model is labeled “N” and the arrow points to the trypsin digestion site (R42:S43), which is rendered in “ball-and-stick” form. (B) ExH cryo-reconstruction with three hexon ribbon models docked into the density. The icosahedron line drawing shows the view direction is along a strict three-fold symmetry axis. The region of the model surrounded by a dashed envelope is shown in stereo at the right, which gives a closer view of stabilizing interactions formed by symmetry-related subunits. The P-loops are all in black, subunits are colored differently, and the N-arms corresponding to Figure 3D are shown in darker shades to highlight intersubunit interactions. Similar sets of interactions occur at the quasi three-fold axes (not shown). (C) Same as (B), but for the shell CP models and reconstruction.
The modeled P-loop in P22 is 17 residues longer than the P-loop in HK97 and forms extensive interactions with portions of adjacent three-fold- and local three-fold-related subunits, such as the N-arm (Figure 7B,C). The reconstructed densities corresponding to the P-loops in both maps are equally as strong as the remaining CP density, indicating that the P-loops are rigid components that contribute to the structural integrity of all subunits. However, P-loop/N-arm interactions alone are likely insufficient to confer complete stabilization of the entire phage structure. It is possible therefore, that additional particle stabilization could arise from dsDNA:protein interactions similar to those observed in ϕ29 (Tang et al., 2008).
Conclusions
The major CPs in the well-studied HK97 (Gertsman et al., 2009), T7 (Agirrezabala et al., 2007), T4 (Fokine et al., 2005), ϕ29 (Morais et al., 2005), ε15 (Jiang et al., 2008), lambda (Lander et al., 2008), and P22 (Jiang et al., 2003) bacteriophage, and in the human HSV-1 (Baker et al., 2005) all have structures consisting of two components. The first is a core of high α-helix content that forms the bulk of the capsid and adopts the classic HK97 fold. This common core provides the blueprint for building icosahedral capsids of all these viruses. Conservation of a CP core appears to be a recurring theme in icosahedral virus assembly (Bamford et al., 2005). Insertions and other additions to capsid cores provide adaptive functions such as occur in cell recognition and receptor binding (Kontou et al., 2005). However, this is likely not the primary determinant for host recognition in tailed dsDNA bacteriophage, since the highly specialized tail appendages are responsible for docking to cells and injecting the genomes. We propose that the ED protrusion of the P22 CP has a role in stabilizing the unassembled CP monomer.
Close scrutiny of the HK97-like CPs of viruses provides insights into the mechanisms that confer stability to different capsids (Table 1). Except for the simplest of icosahedral viruses (i.e. assembled from 60 identical CPs), capsid assembly and organization relies on subunit plasticity, which allows chemically identical CPs to adopt quasi-equivalent or different conformations (Johnson, 1996). Phage and viruses with isometric heads and triangulation numbers greater than T=3 generally have faceted heads composed of pentameric and hexameric or trimeric capsomers. Inter-capsomer interactions within a “flat face” are clearly distinct from those that occur across the more abrupt, sharply curved edges of the icosahedron. Capsid stabilization in dsDNA phage and viruses occurs via three basic mechanisms (Table 1), the most common of which involves the participation of auxiliary proteins. The bacteriophage T4 soc (Ishii et al., 1978), ε15 gp10 (Jiang et al., 2008), lambda gpD (Yang et al., 2000), and HSV-1 triplex proteins (Trus et al., 1996) are classic examples of virally-encoded proteins that bind to weak-link regions in capsids, most commonly at icosahedral two- or three-fold axes of symmetry (Zandi and Reguera, 2005). Along with soc, the N-termini of T4 CP may interact with each other to further stabilize the T4 capsid. Stabilization in HK97 has been extensively studied and is thus far unique among all viruses. HK97 capsids undergo a complex maturation event that involves protease cleavage of the precursor CP, 102-residue delta domain followed by covalent intersubunit cross-linking at a specific residue in an elongated E-loop, which leads to extensive, chain-mail interlocking of subunits (Gertsman et al., 2009). A comparative model derived for bacteriophage T7, where no cross-linking occurs, describes the homologous E-loop as a putative capsid stabilization element. (Agirrezabala et al., 2007). An alternative means of capsid stabilization has been proposed in ϕ29 (Morais et al., 2005), where the ED domain of the CP bridges neighboring capsomers as if to staple them together. P22 differs from T4, HK97, lambda, ε15, ϕ29, and HSV-1 because it has no auxiliary protein or covalent cross-links and its ED domain is an independent module. Hence, questions arise as to how stability is conferred to the mature P22 capsid and what role the ED domain plays.
In addition to lacking covalent crosslinks, P22 capsid maturation differs from HK97 in that large subunit rearrangements occur at the icoashedral and quasi three-fold axes and scaffolding protein is expressed separately. Indeed, the N-arms in P22 adopt a helix-loop-helix structure that reorganizes during maturation and creates stabilizing contacts with the P-loops at the inner surface of the capsid. In contrast, the N-arm of HK97 Head II assumes an extended conformation on the outer surface of the capsid and contributes an extra strand to the β structure of the E-loop (Gertsman et al., 2009). P-loops also move during P22 maturation, but they maintain a fixed conformation and act like molecular staples in HK97 (Gertsman et al., 2009). This striking difference may arise from alternate functions conferred by the scaffolding proteins in these two phage. Indeed, a property that P22, T7,ϕ29, and HSV-1 all share, which distinguishes them from other tailed, dsDNA phages, is that they all employ separately-expressed scaffold proteins. For example, the N-terminus in the immature HK97 CP includes the delta domain that is cleaved during maturation. In HK97 proheads, the delta domains cluster below the centers of all capsomers (Conway et al., 2007). In P22 and HSV-1, putative scaffolding binding sites occur at the strict and local three-fold axes (Thuman-Commike et al., 1999). Movement of P-loops and reorganization of N-arms during P22 maturation may alter the binding sites for scaffolding protein and facilitate its release during dsDNA packaging. The N-terminal 100 residues of the T7 CP may also bind scaffolding protein, but its proposed location is at the center of the capsomers, as in HK97 (Agirrezabala et al., 2005; Agirrezabala et al., 2007).
Assembly of all viruses listed in Table 1 proceeds via the formation of a precursor procapsid prior to genome packaging. In these species, procapsids are constructed in one of two ways. Procapsids of T4 (Stortelder et al., 2006) and HK97 are assembled from preformed capsomers (Conway et al., 1995; Duda et al., 1995; Stortelder et al., 2006), whereas those of P22 and ϕ29 are assembled by addition of CP monomers (Fu et al., 2007; Newcomb et al., 1996; Prevelige et al., 1993). HSV-1 assembly is also believed to occur by addition of CP monomers (Newcomb et al., 1999). Differential scanning calorimetry of preformed HK97 capsomers and P22 monomers yields a melting temperature difference of more than 20°C and demonstrates that capsomers are orders of magnitude more stable (Galisteo et al., 1995; Ross et al., 2006). This raises questions as to how and why certain viruses like P22, ϕ29, and HSV-1 utilize monomeric rather than preformed, oligomeric CPs for capsid assembly. Perhaps formation of pre-formed capsomers is dictated by the type of scaffolding protein-mediated assembly.
Bioinformatic searches clearly demonstrate that Ig-like folds commonly occur in phage CPs (Fraser et al., 2006) and suggest they may provide global, cell attachment interactions. Our results suggest an alternative role. In P22, the ED protrusions (a.a. 223–349) are Ig-like domains and residue variations in this region do not decrease viral infectivity as would be expected if the ED solely functioned in cell attachment (Gordon and King, 1994). Interestingly, eleven of eighteen CP variants that display dramatic protein folding defects are caused by a.a. substitutions found in this region (Gordon and King, 1993), which suggests that proper P22 CP folding is dictated by the telokin-like ED. The “towers” in HSV-1 and the ED domains in ϕ29 are also proposed to have Ig-like folds (Morais et al., 2005). Thus, the presence of an Ig fold may represent a prerequisite for phage and phage-like viruses that assemble from monomeric proteins. Previously, all tailed dsDNA phage and HSV-1 have been grouped together and characterized as being “HK97-like” or “lambda-like”. It seems more logical now to subdivide these viruses into “P22-like” and “HK97-like” groups, distinguished by whether assembly is driven by monomers or by capsomers.
Experimental Procedures
Purification of P22 ExH
Procapsid samples comprised of wild-type (WT) CP were isolated and purified as described previously and in the Supplemental Data. These were treated with 0.5 M GuHCl to produce shells (1 mg/mL) (Greene and King, 1994), then induced to form expanded, penton-less particles (ExH) by heat treatment at 71°C for 15 minutes (Teschke et al., 2003). ExH were concentrated to 20 mg/mL using a YM-30 Centricon (MILLIPORE), spun at 12k × g at 4°C and further purified by SEC using Sephacryl S1000 matrix (GE HEALTHCARE) in the presence of 7M urea. Dialysis until equilibrium was performed against 20 mM sodium phosphate buffer, pH 7.6 (Buffer A) at 4 °C and purified ExH were concentrated to ~10 mg/mL prior to vitrification for electron cryo-microscopy (cryo-TEM).
Trypsin digestion of ExH particles
ExH sample at ~3 mg/mL at 20°C was digested with free trypsin for 2 hours at a 1:300 molar ratio of trypsin:coat subunits and then incubated in 7M urea for 30 minutes at 20 °C. Digested ExH particles were isolated from the single resulting peptide by dialysis until equilibrium against Buffer A at 4°C using a 30 kDa molecular weight cutoff membrane (SPECTRAPOR) and were then concentrated to ~4 mg/mL via Centricon prior to sample vitrification for cryo-TEM.
Cryo-TEM
Small (3.5 µL) aliquots of purified P22 particles (~10 mg/mL) were vitrified and examined using standard procedures (Baker et al., 1999). Briefly, this involved applying samples to Quantifoil holey grids that had been glow-discharged for ~15 s in an Emitech K350 evaporation unit. Grids were then blotted with Whatman filter paper for ~5 s, plunged into liquid ethane, and transferred into a precooled, FEI Polara, multi-specimen holder, which maintained the specimen at liquid nitrogen temperature. Micrographs were recorded on Kodak KO-163 electron-image film at 200 keV in an FEI Polara microscope under minimal-dose conditions (~9 e/Å2) at a nominal magnification of 39,000. The range of objective lens defocus settings used to record each data set are listed in Table 2.
3D Image Reconstruction
Micrographs exhibiting minimal astigmatism and specimen drift were selected for further processing and digitized at 7 µm intervals (representing 1.795 Å pixels at the specimen) on a Zeiss SCAI scanner. The program RobEM (http://cryoEM.ucsd.edu/programs.shtm) was used to estimate micrograph defocus and astigmatism, extract individual particle images, and preprocess the images. For each specimen, 150 particle images were used as input to the random-model computation procedure to generate an initial 3D density map at ~25 Å resolution (Yan et al., 2007a). Each map was then used to initiate determination and refinement of particle orientations and origins for the complete set of images using the current version of AUTO3DEM (Yan et al., 2007b). Phases but not amplitudes of the particle structure factor data were corrected to compensate for the effects caused by the microscope contrast-transfer function (Bowman et al., 2002; Zhang et al., 2003). The Fourier Shell Correlation (FSC0.5) criterion was used to estimate the resolutions of all 3D reconstructions (Table 2) (van Heel and Schatz, 2005). To aid our analysis and interpretation of the reconstructed ExH data, we computed each final 3D map with an inverse temperature factor of 250 Å2 applied to enhance features out to a resolution limit of 7 Å (Havelka et al., 1995). Beyond this limit the inverse temperature factor was held constant and a Gaussian damping function was applied to attenuate the Fourier data smoothly to zero amplitude from 7.24 to 6.84 Å. Graphical representations were generated with the RobEM (http://cryoEM.ucsd.edu/programs.shtm) and Chimera (Goddard et al., 2007) visualization software packages.
Trypsin digestion of a C-terminal CP fragment
A pET-3a plasmid, containing the gene for a poly-histidine-tagged fragment of P22 CP (residues 191–430) was obtained from the Prevelige lab and the truncated protein was purified as described previously (Kang and Prevelige, 2005). The C-terminal fragment of CP at 1.5 mg/mL was mixed at 20 °C with varying concentrations of trypsin (0.1 – 16 µM) in Buffer B (50 mM Tris, 25 mM NaCl, 2 mM EDTA, pH 7.6); digestion was arrested by placing a portion of the reaction mixture in SDS sample buffer and heating for 5 minutes at 95°C. Samples were extracted at time points between 5 and 120 minutes, and the resulting peptides were analyzed by SDS-PAGE. Sequences of the resulting bands were determined by MALDI-TOF-TOF and MS/MS mass spectrometry (Supplemental Data).
Tetramethylrhodamine-5-maleimide (TMRM) labeling
Shells containing single cysteine mutations (Table 3) at 50 µM (subunits) in Buffer A were incubated with 9 mM TMRM in DMSO (Invitrogen) in the dark for 2 hours at 20°C. Free TMRM was separated from TMRM-labeled shells by SEC (Sepharose 4B, Buffer A). Labeled shells were disrupted with 6M GuHCl, and the molar ratio of TMRM per monomer was determined by absorbance measurements at both 280 nm and 555 nm. Contribution of signal at 280 nm from TMRM was compensated by subtracting the total A280 nm by 0.340*A555 nm according to Invitrogen protocol. Bound TMRM was then calculated as the molar ratio of TMRM to CP monomers. For the three cysteine variants, the TMRM:coat ratio is a combination of the accessibility of the WT C405 plus the engineered substitution.
Gold bead conjugation to WT, F170C, A285C, and 379C procapsid shells
P22 shells at 0.6 mg/mL final concentration were mixed with 4 M excess monomaleimido-nanogold (NANOPROBES) (Montesano-Roditis et al., 2001) in Buffer A and allowed to conjugate overnight at 4°C. Unbound gold was purified from goal-labeled shells by SEC using Sephadex G25 matrix (GE HEALTHCARE) in the presence of Buffer A. Labeled shells were concentrated to 4–8 mg/ml prior to vitrification.
CP modeling
The Fold and Function Assignment System (FFAS03) was used to initially identify the crystal structures of HK97 Head II (PDB entry 1OHG) and telokin (PDB entry 1FHG) as the best candidates for modeling the P22 CP structure (Figure 4). Alignment of the P22 CP “core” sequence (a.a. 1–222 and 350–430) to the HK97 Head II fold was obtained by using structure-based sequence alignment with the PROMALS3D server (Pei et al., 2008). Structures homologous to HK97 include: the bacteriophage T4 gp23 (PDB entry 1YUE), the virus-like particle from Pyrococcus furiosus (PDB entry 2E0Z), the phage related protein from B. branchiseptica (PDB entry 3BJQ), the putative capsid protein of E. coli prophage CFT073 (PDB entry 3BQW), and the T.maritimeencapsulin (PDB entry 3DKT). A similar procedure was used for the P22 CP (ED) protrusion (a.a. 223–349), where structures homologous to Telokin include two domains from each of the immunoglobulin gamma Fc and Epsilon receptors (PDB entries 1E4J and 1F2Q), and one domain from Telokin (PDB entry 1FHG).
These alignments were then manually refined using Swiss-PDB Viewer and SWISS-MODEL (Guex and Peitsch, 1997). This refinement was performed while paying particular attention to several relevant data. These included (i) the positions of the gold labels, (ii) results of the two partial-trypsin-digestion experiments, (iii) knowledge that regions of variations within the HK97-fold indicate a tolerance for insertions or deletions (Figure 4), (iv) the secondary structure prediction of the P22 CP, and (v) the sequence alignment of P22 with distant homologues identified by the Jpred 3 server (Cole et al., 2008). Jpred 3 alignment identifies insertions or deletions in region-specific sequences more similar to P22 than those provided by the original templates.
For each cycle of refinement, stretches of sequence predicted to form α-helix or β-strand were manually shifted towards the N- or C-terminus by three to six residues, one residue at a time, and the energy of each structure was calculated using the SWISS-MODEL server. This procedure was performed concurrently for both the ExH and shell forms of P22. The resulting two structures with lowest energies were then used as starting models for similar refinements that included the next secondary structure element in each form, and the whole process repeated until all elements of the core and ED portions of the P22 sequence had been included. The two final CP models were fitted into the remaining density in each asymmetric unit of the corresponding icosahedral cryo-reconstructions using UCSF Chimera (Pettersen et al., 2004). This led to six separate subunit fits for the ExH reconstruction and seven for the shell reconstruction. Density ascribed to each subunit model was segmented from the density maps with UCSF Chimera using a 6-Å mask. All models were then subjected to molecular-dynamics-driven, flexible fitting with the program MODELLER (Topf et al., 2005). Fitted models were visually assessed and reinterpreted to better fit the density as is common practice when constructing atomic models in X-ray crystallography. The program Coot (Emsley and Cowtan, 2004) was used to perform manual, rigid body movement and real space refinement of helices. MODELLER was then used to compute a second round of flexible fitting to help eliminate improper geometries introduced during manual modeling. The entire fitting process was iterated for 18 cycles, with the output of each cycle used as the input for the next cycle.
The final shell and ExH models deviated from the initial predictions made with the FFAS03 and PROMALS3D servers (Figure 4). This likely reflects the low level of sequence similarity between corresponding regions of P22 and HK97 (9.6%) and telokin (8.6%). The HK97 Prohead II and Head II crystal structures, when docked into the P22 shell and ExH image reconstructions respectively, leave large gaps between HK97 capsomers (Figure 6). This strongly suggested that P22 must include components, which are either not present or are refolded differently in HK97, that confer capsid stability in P22.
The HK97 and telokin crystal structures provided useful templates for generating all but about 19% of the P22 CP model. The HK97 Head II and Prohead II templates were modified or adjusted in four regions to better fit the P22 core densities in the two cryo-reconstructions. These included: (i) complete restructuring of the N-terminus (a.a. 2–40); (ii) substitution of an elongated loop (a.a. 191–218) in place of a shorter HK97 loop; (iii) substitution of a longer P-loop (a.a. 380–397): and (iv) shortening the E-loop (Figure 4). The HK97 N-termini in Head II and Prohead II have extended conformations that clearly did not correlate with either P22 density map. However, the N-terminus of the bacteriophage T4 gp23 (PDB entry 1YUE), which also adopts a helix-loop-helix structure, docked manually without requiring modification into the P22 ExH image reconstruction. Likewise, the elongated loop was modeled after the E. coli hypothetical prophage protein (PDB entry 3BQW) structure since it fit best into the ExH image reconstruction. Modification of the P-loop was not dictated by reference to an homologous structure, but instead was performed by simply modeling the longer P22 sequence to fit the shell and ExH density maps.
The P22 sequence for the ED portion was similarly threaded and refined into the telokin template. Interestingly, both the B. bronchiseptica and E. coli hypothetical prophage proteins have two-strand, β-sheet inserts in their HK97-like folds in the same loop where the P22 telokin fold occurs. This clearly demonstrated that the HK97-like fold tolerates an insertion within this loop (Figure 4). P22 residues 315 to 346 could not be modeled reliably since no template was identified for this region and it is too large to predict a loop structure with any accuracy. Small, finger-like protrusions in the ExH and shell density maps at the icosahedral and quasi 2-fold axes appear to correspond to at least some of the un-modeled residues and may represent a two-stranded β-sheet.
Supplementary Material
Acknowledgements
We thank the Prevelige lab (Univ. Alabama) for the gift of the pet3a plasmid, K. and J. Pogliano (Univ. California, San Diego) for use of lab space, R.S. Sinkovits for helpful discussions, and N.H. Olson for guidance in cryo-TEM. This work was supported in part by NIH grants R37 GM-033050 and 1S10 RR-020016 (TSB), GM-054076 (JEJ), and GM-076661 (CMT), by NIH fellowship F32A1078624 (KNP), by NIH fellowship F32A1065071 (RK) and support from UCSD and the Agouron Foundation (TSB).
Abbreviations
- CP
coat protein
- gp
gene product
- cryo-TEM
cryo-transmission electron microscopy
- 3D
three-dimensional
- WT
wild type
- ExH
WT heat-expanded (penton-less) heads
- shells
empty procapsid shells
- ED
extra density domain
- N-arm
N-terminal arm
- a.a
amino acids
- TMRM
tetramethylrhodamine-5-maleimide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Density maps of the ExH particles (EMD-4159) and procapsid shells (EMD-4150) are deposited at the Electron Microscopy Data Bank. C-alpha coordinates for the corresponding models were submitted to the Protein Data Bank under accession numbers 3IYI and 3IYH, respectively.
References
- Agirrezabala X, Martin-Benito J, Caston JR, Miranda R, Valpuesta JM, Carrascosa JL. Maturation of phage T7 involves structural modification of both shell and inner core components. EMBO J. 2005;24:3820–3829. doi: 10.1038/sj.emboj.7600840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirrezabala X, Velazquez-Muriel JA, Gomez-Puertas P, Scheres SH, Carazo JM, Carrascosa JL. Quasi-atomic model of bacteriophage t7 procapsid shell: insights into the structure and evolution of a basic fold. Structure. 2007;15:461–472. doi: 10.1016/j.str.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J. Virol. 2005;79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Olson NH, Fuller SD. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiology & Molecular Biology Reviews. 1999;63:862–922. doi: 10.1128/mmbr.63.4.862-922.1999. [erratum appears in Microbiol Mol Biol Rev 2000 Mar;64(1):237.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr Opin Struct Biol. 2005;15:655–663. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Bazinet C, Benbaset Julyet, King Jonathan, Carazo Jose Maria, Carrascosa Jose L. Purification and organization of the gene 1 portal protein required for phage P22 DNA packaging. Biochemistry. 1988;27:1849–1856. doi: 10.1021/bi00406a009. [DOI] [PubMed] [Google Scholar]
- Bowman VD, Chase ES, Franz AW, Chipman PR, Zhang X, Perry KL, Baker TS, Smith TJ. An antibody to the putative aphid recognition site on cucumber mosaic virus recognizes pentons but not hexons. J Virol. 2002;76:12250–12258. doi: 10.1128/JVI.76.23.12250-12258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capen CM, Teschke CM. Folding defects caused by single amino acid substitutions in a subunit are not alleviated by assembly. Biochemistry. 2000;39:1142–1151. doi: 10.1021/bi991956t. [DOI] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Research. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JF, Cheng N, Ross PD, Hendrix RW, Duda RL, Steven AC. A thermally induced phase transition in a viral capsid transforms the hexamers, leaving the pentamers unchanged. J Struct Biol. 2007;158:224–232. doi: 10.1016/j.jsb.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JF, Duda RL, Cheng N, Hendrix RW, Steven AC. Proteolytic and conformational control of a virus capsid maturation: the bacteriophage HK97 system. J Mol Biol. 1995;253:86–99. doi: 10.1006/jmbi.1995.0538. [DOI] [PubMed] [Google Scholar]
- Duda RL, Hempel J, Michel H, Shabanowitz J, Hunt D, Hendrix RW. Structural transitions during bacteriophage HK97 head assembly. J. Mol. Biol. 1995;247:618–635. doi: 10.1006/jmbi.1995.0168. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fokine A, Leiman PG, Shneider MM, Ahvazi B, Boeshans KM, Steven AC, Black LW, Mesyanzhinov VV, Rossmann MG. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc Natl Acad Sci U S A. 2005;102:7163–7168. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JS, Yu Z, Maxwell KL, Davidson AR. Ig-like domains on bacteriophages: a tale of promiscuity and deceit. J Mol Biol. 2006;359:496–507. doi: 10.1016/j.jmb.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Frishman D, Argos P. Knowledge-based secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- Fu CY, Morais MC, Battisti AJ, Rossmann MG, Prevelige PE., Jr Molecular dissection of o29 scaffolding protein function in an in vitro assembly system. J Mol Biol. 2007;366:1161–1173. doi: 10.1016/j.jmb.2006.11.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller MT, King J. Purification of the coat and scaffolding protein from procapsids of bacteriophage P22. Virology. 1981;112:529–547. doi: 10.1016/0042-6822(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Galisteo ML, Gordon CL, King J. Stability of wild-type and temperature-sensitive protein subunits of the phage P22 capsid. J. Biol. Chem. 1995;270:16595–16601. doi: 10.1074/jbc.270.28.16595. [DOI] [PubMed] [Google Scholar]
- Gertsman I, Gan L, Guttman M, Lee K, Speir JA, Duda RL, Hendrix RW, Komives EA, Johnson JE. An unexpected twist in viral capsid maturation. Nature. 2009;458:646–650. doi: 10.1038/nature07686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Gordon CL, King J. Temperature-sensitive mutations in the phage P22 coat protein which interfere with polypeptide chain folding. J. Biol. Chem. 1993;268:9358–9368. [PubMed] [Google Scholar]
- Gordon CL, King J. Genetic properties of temperature-sensitive folding mutants of the coat protein of phage P22. Genetics. 1994;136:427–438. doi: 10.1093/genetics/136.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene B, King J. Binding of scaffolding subunits within the P22 procapsid lattice. Virology. 1994;205:188–197. doi: 10.1006/viro.1994.1634. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environement for comparative modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Havelka WA, Henderson R, Oesterhelt D. Three-dimensional structure of halorhodopsin at 7 A resolution. J Mol Biol. 1995;247:726–738. doi: 10.1006/jmbi.1995.0176. [DOI] [PubMed] [Google Scholar]
- Hendrix RW. Bacteriophages: evolution of the majority. Theor Popul Biol. 2002;61:471–480. doi: 10.1006/tpbi.2002.1590. [DOI] [PubMed] [Google Scholar]
- Holden HM, Ito M, Hartshorne DJ, Rayment I. X-ray structure determination of telokin, the C-terminal domain of myosin light chain kinase, at 2.8Å resolution. J Mol Biol. 1992;227:840–851. doi: 10.1016/0022-2836(92)90226-a. [DOI] [PubMed] [Google Scholar]
- Ishii T, Yamaguchi Y, Yanagida M. Binding of the structural protein soc to the head shell of bacteriophage T4. J Mol Biol. 1978;120:533–544. doi: 10.1016/0022-2836(78)90352-2. [DOI] [PubMed] [Google Scholar]
- Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A. FFAS03: a server for profile-profile sequence alignments. Nucl. Acids Res. 2005;33 doi: 10.1093/nar/gki418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Baker ML, Jakana J, Weigele PR, King J, Chiu W. backbone structure of the infectious e15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1134. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- Jiang W, Li Z, Zhang Z, Baker ML, Prevelige PE, Chiu W. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat. Struct. Biol. 2003;10:131–135. doi: 10.1038/nsb891. [DOI] [PubMed] [Google Scholar]
- Johnson JE. Functional implications of protein-protein interactions in icosahedral viruses. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:27–33. doi: 10.1073/pnas.93.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Hawkridge AM, Johnson KL, Muddiman DC, Prevelige PJ. Identification of subunit-subunit interactions in bacteriophage P22 procapsids by chemical cross-linking and mass-spectrometry. J. Proteome Res. 2006;5:370–377. doi: 10.1021/pr050356f. [DOI] [PubMed] [Google Scholar]
- Kang S, Prevelige PE., Jr Domain study of bacteriophage p22 coat protein and characterization of the capsid lattice transformation by hydrogen/deuterium exchange. J Mol Biol. 2005;347:935–948. doi: 10.1016/j.jmb.2005.02.021. [DOI] [PubMed] [Google Scholar]
- King J, Botstein D, Casjens S, Earnshaw W, Harrison S, Lenk E. Structure and assembly of the capsid of bacteriophage P22. Philos. Trans. R. Soc. London B. 1976;276:37–49. doi: 10.1098/rstb.1976.0096. [DOI] [PubMed] [Google Scholar]
- King J, Chiu W. The procapsid-to-capsid transition in double-stranded DNA bacteriophages. In: Chiu W, Burnett R, Garcea R, editors. Structural Biology of Viruses. New York: Oxford University Press; 1997. pp. 288–311. [Google Scholar]
- Kontou M, Govindasamy L, Nam HJ, Bryant N, Llamas-Saiz AL, Foces-Foces C, Hernando E, Rubio MP, McKenna R, Almendral JM, Agbandje-McKenna M. Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice. J Virol. 2005;79:10931–10943. doi: 10.1128/JVI.79.17.10931-10943.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Evilevitch A, Jeemaeva M, Potter CS, Carragher B, Johnson JE. Bacteriophage lambda stabilization by auxiliary protein gpD: timing location, and mechanism of attachment determined by cryo-EM. Structure. 2008;16:1399–1406. doi: 10.1016/j.str.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Khayat R, Li R, Prevelige PJ, Potter CS, Carragher B, Johnson JE. The P22 tail machine at subnanometer resolution reveals the architechture of an infectious conduit. Structure. 2009;17:789–799. doi: 10.1016/j.str.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanman J, Tuma R, Prevelige PE., Jr Identification and characterization of the domain structure of bacteriophage P22 coat protein. Biochemistry. 1999;38:14614–14623. doi: 10.1021/bi9915420. [DOI] [PubMed] [Google Scholar]
- Montesano-Roditis L, Glitz DG, Traut RR, Stewart PL. Cryo-electron microscopic localization of protein L7/L12 within the Escherichia coli 70 S ribosome by difference mapping and Nanogold labeling. J Biol Chem. 2001;276:14117–14123. doi: 10.1074/jbc.M008430200. [DOI] [PubMed] [Google Scholar]
- Morais MC, Choi KH, Koti JS, Chipman PR, Anderson DL, Rossmann MG. Conservation of the capsid structure in the tailed dsDNA bacteriophages: the pseudoatomic structure of phi29. Mol Cell. 2005;18:149–159. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Newcomb W, Homa F, Thomsen D, Booy F, Trus B, Steven A, Spencer J, Brown J. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 1996;263:432–446. doi: 10.1006/jmbi.1996.0587. [DOI] [PubMed] [Google Scholar]
- Newcomb WW, Homa FL, Thomsen DR, Trus BL, Cheng N, Steven A, Booy F, Brown JC. Assembly of the herpes simplex virus procapsid from purified components and identification of small complexes containing the major capsid and scaffolding proteins. J Virol. 1999;73:4239–4250. doi: 10.1128/jvi.73.5.4239-4250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Kim B-H, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucl. Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Prasad BVV, Prevelige PE, Jr, Marieta E, Chen RO, Thomas D, King J, Chiu W. Three-dimensional transformation of capsids associated with genome packaging in a bacterial virus. J. Mol. Biol. 1993;231:65–74. doi: 10.1006/jmbi.1993.1257. [DOI] [PubMed] [Google Scholar]
- Prevelige PE, Jr, Thomas D, King J. Nucleation and growth phases in the polymerization of coat and scaffolding subunits into icosahedral procapsid shells. Biophys. J. 1993;64:824–835. doi: 10.1016/S0006-3495(93)81443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross PD, Conway JF, Cheng N, Dierkes L, Firek BA, Hendrix RW, Steven AC, Duda RL. A free energy cascade with locks drives assembly and maturation of bacteriophage HK97 capsid. J Mol Biol. 2006;364:512–525. doi: 10.1016/j.jmb.2006.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stortelder A, Hendriks J, Buijs JB, Bulthuis J, Gooijer C, van der Vies SM, van der Zwan G. Hexamerization of the bacteriophage T4 capsid protein gp23 and its W13V mutant studied by time-resolved tryptophan fluorescence. J Phys Chem B. 2006;110:25050–25058. doi: 10.1021/jp064881t. [DOI] [PubMed] [Google Scholar]
- Tang J, Olson N, Jardine PJ, Grimes S, Anderson DL, Baker TS. DNA poised for release in bacteriophage phi29. Structure. 2008;16:935–943. doi: 10.1016/j.str.2008.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschke CM, McGough A, Thuman-Commike PA. Penton release from P22 heat-expanded capsids suggests importance of stabilizing penton-hexon interactions during capsid maturation. Biophysical Journal. 2003;84:2585–2592. doi: 10.1016/S0006-3495(03)75063-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuman-Commike PA, Greene B, Malinski JA, Burbea M, McGough A, Chiu W, Prevelige PEJ. Mechanism of scaffolding-directed virus assembly suggested by comparison of scaffolding-containing and scaffolding-lacking P22 procapsids. Biophys. J. 1999;76:3267–3277. doi: 10.1016/S0006-3495(99)77479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topf M, Baker ML, John B, Chiu W, Sali A. Structural characterization of components of protein assemblies by comparative modeling and electron cryo-microscopy. J Struct Biol. 2005;149:191–203. doi: 10.1016/j.jsb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Trus BL, Booy FP, Newcomb WW, Brown JC, Homa FL, Thomsen DR, Steven AC. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- van Heel M, Schatz M. Fourier shell correlation threshold criteria. J Struct Biol. 2005;151:250–262. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- Yan X, Dryden KA, Tang J, Baker TS. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J Struct Biol. 2007a;157:211–225. doi: 10.1016/j.jsb.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Sinkovits RS, Baker TS. AUTO3DEM-an automated and high throughput program for image reconstruction of icosahedral particles. J Struct Biol. 2007b;157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Forrer P, Dauter Z, Conway JF, Cheng N, Cerritelli ME, Steven AC, Pluckthun A, Wlodawer A. Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD. Nature Struct. Biol. 2000;230:230–237. doi: 10.1038/73347. [DOI] [PubMed] [Google Scholar]
- Zandi R, Reguera D. Mechanical properties of viral capsids. Phys Rev E Stat Nonlin Soft Matter Phys. 2005;72 doi: 10.1103/PhysRevE.72.021917. 021917. [DOI] [PubMed] [Google Scholar]
- Zhang X, Walker SB, Chipman PR, Nibert ML, Baker TS. Reovirus polymerase lambda 3 localized by cryo-electron microscopy of virions at a resolution of 7.6 A. Nat. Struct. Biol. 2003;10:1011–1018. doi: 10.1038/nsb1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. The protein structure prediction problem could be solved using the current PDB library. Proc Natl Acad Sci U S A. 2005;102:1029–1034. doi: 10.1073/pnas.0407152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH. Towards atomic resolution structural determination by single-particle cryo-electron microscopy. Curr Opin Struct Biol. 2008;18:218–228. doi: 10.1016/j.sbi.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.