SUMMARY
The cranial neural crest (CNC) are multipotent embryonic cells that contribute to craniofacial structures and other cells and tissues of the vertebrate head. During embryogenesis, CNC is induced at the neural plate boundary through the interplay of several major signaling pathways. Here we report that the metalloproteinase activity of ADAM13 is required for early induction of CNC in Xenopus. In both cultured cells and X. tropicalis embryos, membrane-bound Ephrins (Efns) B1 and B2 were identified as substrates for ADAM13. ADAM13 upregulates canonical Wnt signaling and early expression of the transcription factor snail2, whereas EfnB1 inhibits the canonical Wnt pathway and snail2 expression. We propose that by cleaving class B Efns, ADAM13 promotes canonical Wnt signaling and early CNC induction.
INTRODUCTION
The cranial neural crest (CNC) is a highly migratory population of embryonic cells. During early vertebrate embryogenesis, CNC is induced at the anterior border of neural plate and epidermis. This induction/specification is controlled by several major signaling pathways, including Wnt, BMP, FGF and Notch signaling (Basch and Bronner-Fraser, 2006). The specified CNC cells then undergo epithelial-to-mesenchymal transformation and subsequently migrate to populate the face and pharyngeal arches, where they contribute to craniofacial structures such as cartilages, bones, and connective tissues, as well as the peripheral nervous system. A recent study in Xenopus shows that CNC induction involves two steps: an initial induction of the prospective neural crest and a subsequent “maintenance” of the neural crest territory. The initial induction of the prospective CNC occurs during gastrula stages and requires canonical Wnt signaling from the dorsolateral mesoderm (Steventon et al., 2009).
Protein ectodomain shedding by members of the ADAM family of metalloproteinases has emerged as a key regulatory event in cell signaling. ADAMs are type I transmembrane proteins with an extracellular metalloproteinase domain and disintegrin and cysteine-rich domains (Edwards et al., 2008; White et al., 2005). We previously showed that ADAM13, an ADAM metalloproteinase, is expressed in CNC cells and early mesoderm in Xenopus laevis, and that overexpression of a protease dead mutant of ADAM13 impairs CNC migration (Alfandari et al., 2001; Alfandari et al., 1997). To better understand the roles of ADAM13 in vertebrate development, we carried out loss-of-function studies in Xenopus tropicalis and observed a previously unreported early function of ADAM13 in CNC induction. ADAM13 is expressed in the mesoderm during gastrula stages and cleaves Efns B1 and B2 in vivo. This cleavage is essential for upregulation of canonical Wnt signaling and early expression of the neural crest marker snail2 (also known as slug). Our data provide evidence for a crosstalk between EfnB signaling and canonical Wnt signaling, and for an important role of ADAM13 protease activity in regulating this crosstalk and CNC induction.
RESULTS
ADAM13 Metalloproteinase Activity Is Required for Normal CNC Induction
We cloned a cDNA encoding X. tropicalis ADAM13 and used it to generate probes for in situ hybridization. As in X. laevis (Alfandari et al., 1997), endogenous adam13 mRNA was detected in the mesoderm of early gastrula stage X. tropicalis embryos (Figures S1A and S1A’). By the end of gastrulation (stage ~13), adam13 was strongly expressed in the anterolateral border of the neural plate, where the CNC is induced. Expression was maintained in the developing CNC throughout neurula stages, and was present in migrating CNC cells (Figures S1B and S1C). To characterize the function of ADAM13 in X. tropicalis embryos, antisense morpholinos (MOs 13-1 and 13-3) were designed to block ADAM13 translation (Figure 1A). We first assessed MO efficacy and specificity by determining the translational inhibition of co-injected mRNAs encoding different myc-tagged X. tropicalis ADAMs. As shown in Figure 1B, MO 13-1 knocked down expression of exogenous ADAM13 while having no effect on the closely related ADAM12. A slight decrease in the expression of another paralogue, ADAM19, was detected in 13-1 morphants, likely due to a complex cross-regulation between the ADAM13 and 19 proteins but not MO specificity (Wei et al., unpublished results). MO knockdown (KD) of endogenous ADAM13 protein was also confirmed by Western blot. Gastrulae derived from one-cell stage MO 13-1 injected embryos had significantly reduced levels of endogenous ADAM13, as compared to uninjected embryos and those injected with control MO (Figure 1C). Similar KD of ADAM13 was observed with MO 13-3 (data not shown).
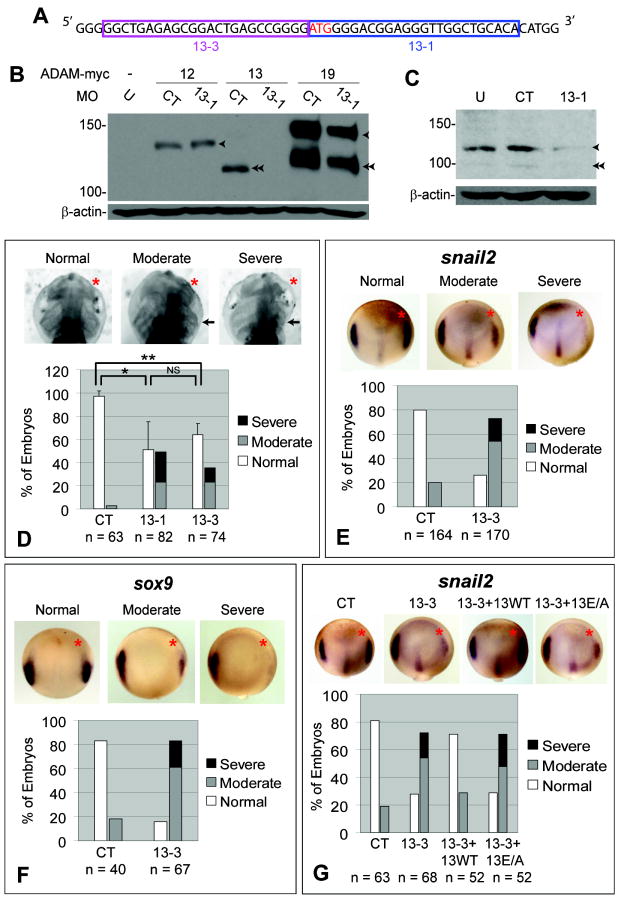
Figure 1. The metalloproteinase activity of ADAM13 is required for Xenopus CNC induction.
(A) cDNA sequence of adam13 showing the translation start site (red) and targets of MOs 13-1 and 13-3. (B) One blastomere of 2-cell stage embryos was injected with MO 13-1 (6 ng) and transcripts encoding myc-tagged ADAMs (ADAM12 or 13: 200 pg; ADAM19: 100 pg). Embryos were cultured for ~20 hrs and Western blot of whole-embryo lysates carried out using an anti-myc antibody. (C) Western blot of stage 11-12 embryo lysates showing KD of endogenous ADAM13 by injection of MO 13-1 at one-cell stage (12 ng MO/embryo; 20 embryos/lane). (D) Head cartilage phenotypes of ADAM13 KD. One anterior-dorsal blastomere of 8-cell stage embryos was injected with the indicated MO (1.5 ng). Embryos were allowed to develop to stage ~46 and scored for head cartilage defects. Images were grey-scale inverted for easier visualization of head cartilage. One example of each phenotype is shown in the upper panels, and results of 3 independent experiments are graphed in the lower panel (error bars represent standard deviations). *: P = 0.03; **: P = 0.005; NS, not significant (P = 0.43). See Figures S1D and Experimental Procedures for details on phenotype scoring and statistics. (E, F) Four-cell stage embryos were injected in one anterior-dorsal blastomere with 3 ng MO 13-3, allowed to develop to stage 12.5/13, and then processed by in situ hybridization for snail2 (E) or sox9 (F). (G) Embryos were injected with the indicated MO (3 ng) and “rescue” transcripts (100 pg each), and in situ hybridization performed as in (E). Red asterisks in (D-G) denote injected side. Single and double arrowheads in B and C point to the pro- and mature forms of ADAMs, respectively, and arrows point to impaired head cartilage structures as compared with the uninjected side. U, uninjected; CT, control MO; n, number of embryos scored (same below).
When ADAM13 was knocked down in the anterior-dorsal region, nearly half of the embryos developed with reduced (moderate) or missing (severe) head cartilage structures; this phenotype was typical of both 13-1 and 13-3 morphants (Figures 1D and S1D). Because head cartilage structures are derivatives of the CNC (Sadaghiani and Thiebaud, 1987), we further analyzed the effects of ADAM13 MOs on the expression patterns of neural crest markers. KD of ADAM13 caused a dramatic decrease in snail2 and sox9 expression at the anterolateral border of the forming neural plate at late gastrula/early neural plate stages (Figures 1E and 1F). ADAM13 MO-specific reduction in snail2 expression in pre-migratory CNC, and twist expression in migrating CNC cells were also observed at later stages (Figures S1E and S1F). Together these data suggest that KD of ADAM13 interferes with CNC induction. We also observed other morphological defects, including impaired eye structures, in both 13-1 and 13-3 morphants (Figure S1D and Wei et al., manuscript in preparation).
ADAMs are multidomain proteins that have other functions not dependent on their protease activities (Edwards et al., 2008; White et al., 2005). To examine whether the CNC phenotype observed in ADAM13 morphants was caused by loss of ADAM13 protease activity, we generated rescue constructs corresponding to either the wild-type or a protease-dead (E/A) form of ADAM13. Co-injection of a transcript made from the wild-type construct reversed the effect of ADAM13 MOs on snail2 expression (Figures 1G and S1G), again confirming that this effect is specific for KD of ADAM13. In contrast, the E/A rescue transcript (Figures 1G and S1G) and a transcript encoding an ADAM13 mutant with the pro-and metalloproteinase domains deleted (ADAM13ΔPM; data not shown) did not rescue. Consistent with these results, the morphological defect in head cartilage was also rescued by wild-type ADAM13 but not the E/A rescue transcript (Figure S1H). Although we cannot rule out a possible involvement of the non-protease domains, our results demonstrate a clear requirement for ADAM13 metalloproteinase activity in normal CNC induction.
ADAM13 Cleaves Efns B1 and B2 in Cultured Cells and Developing Embryos
A major current challenge is to identify the in vivo target substrates of protease activity during specific developmental events (Overall and Blobel, 2007). We considered a number of Xenopus membrane proteins that are known to be involved in neural crest development, and asked if they could be cleaved when co-expressed with ADAM13 in cultured HEK293T cells. We were particularly interested in class B Efns because of their reported functions in CNC development (Adams et al., 2001; Davy et al., 2004; Smith et al., 1997). Unlike the GPI-anchored class A Efns (Efns A1-A6), which bind preferentially to the EphA receptors, class B Efns (Efns B1-B3) are type I transmembrane proteins that interact preferentially with the EphB receptors (with few exceptions; Himanen et al., 2007; Poliakov et al., 2004). Interestingly, the three class B Efns have a strikingly similar cytoplasmic tail that can transduce a reverse signal to ligand expressing cells. When cells expressing class B Efns come into contact with cells expressing cognate receptors, both the ligand and receptor cytoplasmic tails are phosphorylated on tyrosine residues, suggesting a bi-directional (i.e., forward and reverse) signaling (Holland et al., 1996). Additionally, EfnB1 was shown to activate Wnt/PCP signals cell autonomously through a receptor-independent pathway (Lee et al., 2006; Lee et al., 2009).
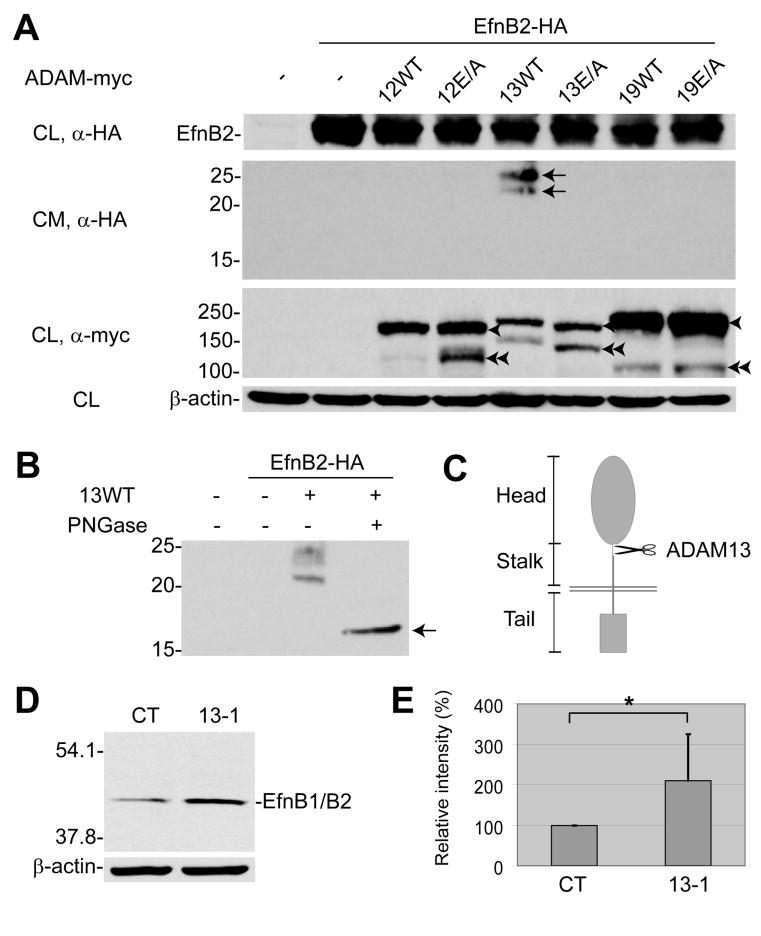
When expressed in 293T cells, Efns B1 and B2 appeared as multiple bands by SDS-PAGE due to post-translational modifications including phosphorylation (Xu et al., 2003). Co-transfection with wild-type ADAM13, but not the E/A mutant or the closely related ADAM12 or 19, resulted in cleavage of both Efns B1 and B2; the remaining membrane-bound “stubs” were further degraded (Figures S2A and S2B). We did not detect any cleavage of either EfnB1 or B2 by ADAM10 (data not shown), a distantly related ADAM that is known to cleave two class A Efns (Hattori et al., 2000; Janes et al., 2005). These data suggest that the cleavage activity for EfnB1/B2 reported here is specific for ADAM13. None of the ADAMs tested, including ADAM13, generated discrete cleavage products of EfnB3 in 293T cells (data not shown). To determine further the cleavage patterns for Efns B1 and B2, we generated additional constructs with an HA epitope tag attached to the distal part of the ectodomain. As seen in Figure 2A (second panel), several HA-tagged fragments were released into the media when 293T cells were co-transfected to express EfnB2 and wild-type ADAM13, but not ADAM12 or 19, or the E/A mutants. Upon treatment with PNGase, these fragments collapsed to a single band at ~16 kDa, suggesting that they represented different glycosylated forms of a single cleavage product (Figure 2B). Based on the size of the product and the sequence of EfnB2, we estimate that the ectodomain shed by ADAM13 encompasses the entire structural “head” (Figure 2C), which contains all known functional ectodomains (Himanen et al., 2001). Therefore the remaining membrane-associated stub is likely unable to interact with the cognate Eph receptors. Because Efn signaling usually requires membrane localization or artificial clustering of the ligands (Davis et al., 1994; Himanen et al., 2001), the shed EfnB2 ectodomain is also unlikely to be functional. Similar results were obtained for EfnB1 but not B3 (data not shown).
Figure 2. ADAM13 cleaves EfnB1/B2 in cultured cells and X. tropicalis embryos.
(A) Cultured HEK293T cells were transfected with DNA constructs (0.5 μg each) expressing EfnB2 (with an N-terminal HA tag) and wild-type or the E/A mutant of each ADAM (with C-terminal myc tags). Western blots of conditioned media (CM) and whole cell lysates (CL) were carried out using the indicated antibodies (α-HA or α-myc). Arrows indicate the ectodomain of EfnB2 shed by ADAM13; single and double arrowheads indicate the pro- and mature forms of ADAMs, respectively. (B) Conditioned media of HEK293T cells transfected to express EfnB2 and wild-type ADAM13, as in A, were treated with or without PNGase and processed for Western blot with an anti-HA antibody. The deglycosylated product is denoted with an arrow. The approximate location of the ADAM13 cleavage site in EfnB2 is indicated in (C). (D, E) Each of the two anterior-dorsal blastomeres of 8-cell stage X. tropicalis embryos was injected with control (CT) or 13-1 MO (1.5 ng). Embryos were allowed to develop to stage ~19, the heads collected by dissection, and Western blots of head lysates carried out using the C-18 antibody. Relative intensity of EfnB1/B2 was determined by normalizing the Western blot density of EfnB1/B2 band against that of β-actin, and by setting the amount calculated for control MO to 100%. Error bars represent standard deviations calculated for 7 independent experiments. *: P = 0.03.
We next explored whether ADAM13 can cleave EfnBs in developing X. tropicalis embryos. Co-injection of a transcript encoding EfnB1 with MO 13-3 resulted in an apparent accumulation of the expressed Efn at stage ~15 (Figure S2C). Similar effects were observed at stages 12/13 and ~22 and for EfnB2 (data not shown), suggesting a post-transcriptional “protective” effect for both Efns following ADAM13 KD. The cleavage products were not readily detected, probably due to rapid turnover of these products in developing embryos. To determine whether ADAM13 is required for cleavage of endogenous Efns B1/B2, MO 13-1 was injected into both sides of the anterior-dorsal region of 8-cell stage embryos, and Western blots of head lysates were carried out using an antibody that recognizes the cytoplasmic tails of Xenopus Efns B1 and B2, but not B3 (Figure S2D). As shown in Figures 2D and 2E, there was significantly more intact EfnB1/B2 in the heads of 13-1 morphants vs. embryos injected with control MO. Although we cannot rule out other possible mechanisms that may account for or contribute to the accumulation of EfnB1/B2, the most parsimonious explanation for these findings is that KD of ADAM13 protects EfnB1/B2 from cleavage in vivo.
ADAM13 Directs CNC Induction by Regulating EfnB and Wnt Signaling
A key question is whether cleavage of EfnB1/B2 is essential for ADAM13 function in CNC induction. Because KD of ADAM13 protects EfnB1/B2 in vivo, we hypothesized that excessive EfnB signaling could be causing the CNC defects in ADAM13 morphants. Consistent with this hypothesis, both efns b1 and b2 are expressed in the dorsal mesoderm in an overlapping pattern with adam13 during early gastrula stages (Figure S1A). Furthermore, ectopic expression of EfnB1 in the anterior-dorsal region of X. tropicalis embryos resulted in reduced snail2 expression at stage 12.5/13 (Figure 3A), a phenotype similar to ADAM13 KD (Figure 1E). Co-expression of wild-type ADAM13 but not the ΔPM mutant reversed this effect (Figure 3A), suggesting that ADAM13 can attenuate EfnB1 mediated inhibition of CNC induction through its protease activity.
Figure 3. ADAM13 controls CNC induction through regulation of EfnB and Wnt signaling.
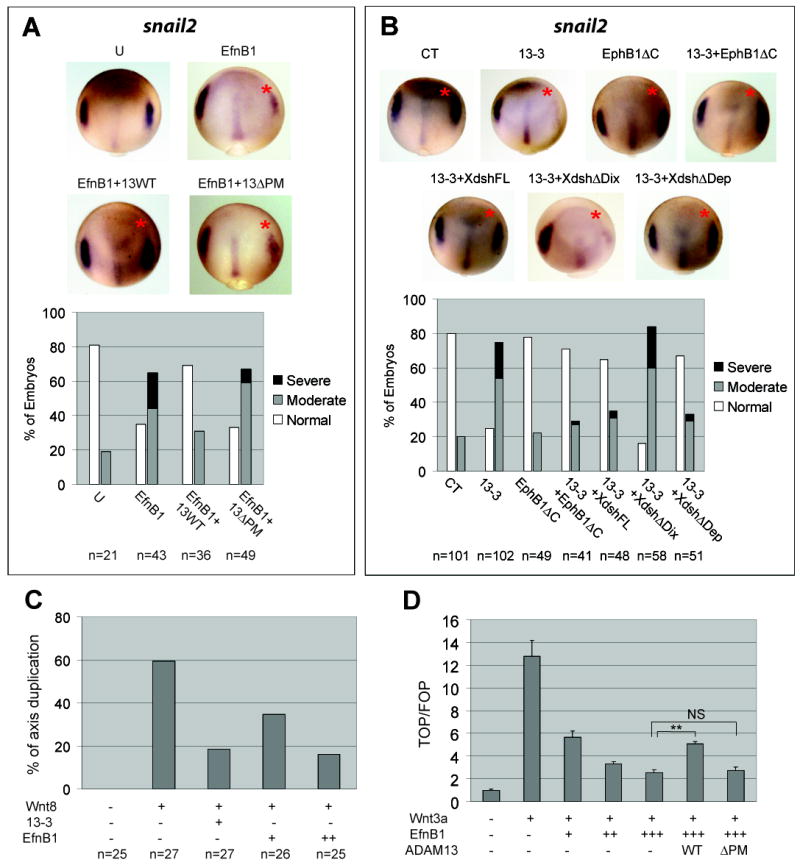
(A, B) Embryos were injected as in Figure 1E with transcripts as follows: 200 pg EfnB1 and 100 pg ADAM13 in (A), and 200 pg each in (B) along with 3 ng of control (CT) or 13-3 MO. In situ hybridization was processed for snail2 at stage 12.5/13; injected side is denoted with a red asterisk. (C) Embryos were injected in one ventral-vegetal blastomere at 16-cell stage with the indicated RNA (1.5 pg Wnt8; 100 or 200 pg EfnB1) and MO 13-3 (1.5 ng), and scored for secondary axis formation at tailbud stages. (D) HEK293T cells were transfected with pTopflash or pFopflash, pCMV-β-gal and the indicated constructs, and cultured for ~40 hrs. Cell lysates were processed for luciferase and β-galactosidase assays as controls for transfection efficiency. A representative experiment performed in triplicate is shown here. Results are presented as ratios of TOPFLASH vs. FOPFLASH luciferase activity (both were normalized for β-gal activity), and the value calculated for cells transfected with control plasmids only was set to 1. Error bars represent standard deviations. **: P < 0.001; NS: not signicicant (P = 0.71). U, uninjected.
To establish a functional link between class B Efns and ADAM13, we determined if the CNC phenotype caused by ADAM13 KD could be rescued by downregulating EfnB signaling. This was carried out using an EphB1 receptor construct with the kinase domain deleted (EphB1ΔC). In addition to known inhibitory activity for forward EfnB signaling (Durbin et al., 1998), this construct was recently shown to disrupt the receptor-independent EfnB signaling pathway (Lee et al., 2009). While a low dosage of the EphB1ΔC transcript alone did not cause any apparent alteration in snail2 expression, co-injection with ADAM13 MOs rescued the effect of ADAM13 KD on snail2 (Figures 3B and S3A). Similarly, the morphological defect in head cartilage was rescued (Figure S3B), indicating that ADAM13 functions in CNC induction by regulating EfnB signaling. Ectopic expression of a higher dosage of the EphB1ΔC transcript resulted in an anterior expansion of the snail2 expression domain (Figure S3C). This phenotype resembled that obtained by ectopic activation of canonical Wnt signaling (Hong et al., 2008; Li et al., 2009). Because Wnt is known to be essential for CNC induction (Wu et al., 2003), we went on to investigate whether Wnt signaling is a downstream target of EfnB signaling and ADAM13 protease activity.
It has been reported that the Xenopus dishevelled protein (Xdsh), a central component of both the canonical Wnt and PCP signaling pathways, can mediate forward and receptor-independent EfnB signaling (Lee et al., 2006; Lee et al., 2009; Tanaka et al., 2003). Moreover, disruption of Xdsh function interferes with CNC induction in X. laevis (De Calisto et al., 2005). Therefore we tested whether Xdsh function is required downstream of ADAM13. Ectopic expression of full-length Xdsh, as well as an Xdsh mutant with the Dep domain deleted (ΔDep), rescued the molecular and morphological defects caused by ADAM13 MOs. In contrast, an Xdsh mutant with the Dix domain deleted (ΔDix) was unable to effect a significant rescue at either early or later stages (Figures 3B, S3A and S3B). Because the Dix and the Dep domains of Xdsh mediate canonical Wnt and PCP signaling, respectively (Boutros and Mlodzik, 1999), these data suggest that the canonical Wnt pathway acts downstream of EfnBs to mediate ADAM13 function in CNC induction. To test the notion that ADAM13 and class B Efns are Wnt regulators, we assessed the effects of ADAM13 and EfnB signaling on the canonical Wnt pathway in both X. tropicalis embryos and cultured cells. When injected in a ventral-vegetal blastomere, mRNA encoding the soluble Wnt ligand Wnt8 induces a secondary dorsal axis through activation of the canonical Wnt pathway (Smith and Harland, 1991; Sokol et al., 1991). The axis duplication effect of Wnt8 was reduced by co-injection of ADAM13 MO or a transcript encoding EfnB1 (Figure 3C). To confirm that EfnB1 and ADAM13 modulate β-catenin dependent transcriptional activity, we took advantage of the TOP/FOPFLASH luciferase reporter constructs (Korinek et al., 1997). EfnB1 inhibited Wnt3a induced reporter expression in a dose-dependent manner in cultured HEK293T cells. Co-expression of wild-type ADAM13, but not the ΔPM mutant, partially reversed this inhibition (Figure 3D). Because HEK293T cells are known to express a high level of Eph receptors for EfnB1 (Xu et al., 2003), all three EfnB signaling pathways (i.e., forward, reverse and receptor-independent pathways) could be involved in the inhibition of canonical Wnt signaling by EfnB1. To differentiate between these possibilities, we used an EfnB1 mutant with the cytoplasmic tail deleted (EfnB1ΔC), which cannot transduce either reverse or receptor-independent EfnB signaling (Holland et al., 1996; Lee et al., 2006). As shown in Figure S3D, this mutant mimicked wild-type EfnB1 as a Wnt inhibitor, suggesting that forward EfnB signaling is at least partially responsible for the Wnt inhibitory activity displayed by the EfnB1 ligand. TOP/FOPFLASH assays using X. tropicalis embryos further demonstrated that Wnt/β-catenin dependent transcriptional activity can be downregulated by wild-type EfnB1 and upregulated by the dominant-negative EphB1ΔC receptor in vivo (Figure S3E).
Snail2 Is Essential for ADAM13 Function in Early CNC Induction
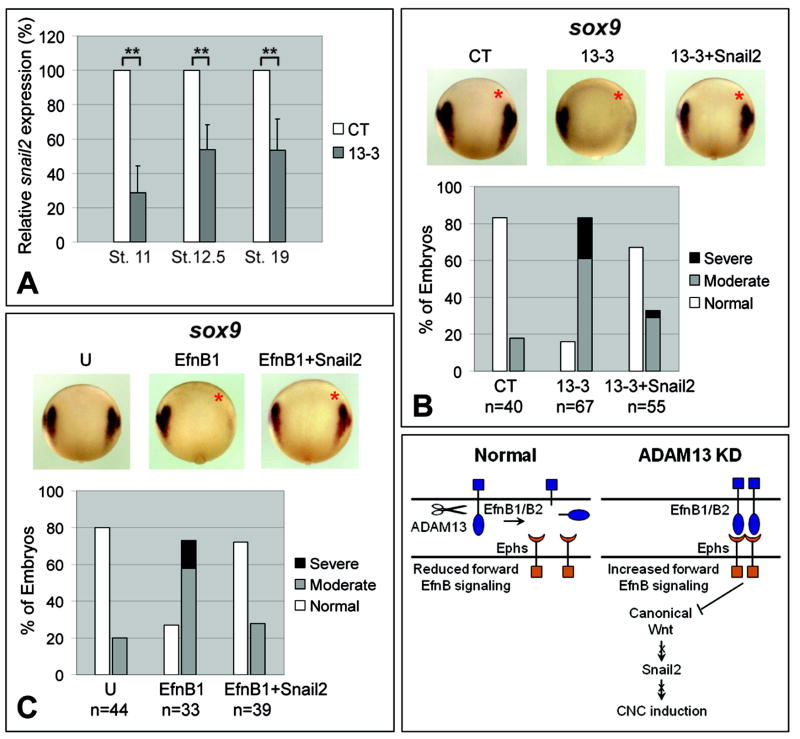
KD of ADAM13 suppresses CNC induction, as indicated by the downregulated expression of neural crest markers snail2, sox9 and twist (Figures 1E, 1F, S1E and S1F). However, Snail2 itself is required for CNC induction (LaBonne and Bronner-Fraser, 2000), and its expression is known to be directly controlled by Wnt/β-catenin signaling (Vallin et al., 2001). Therefore it is possible that the reduction in snail2 expression was a cause, instead of simply a consequence of the diminished CNC induction observed in ADAM13 morphants. To investigate this possibility, we carried out whole-embryo KD of ADAM13 and measured the expression levels of snail2 at different developmental stages by quantitative RT-PCR. As shown in Figure 4A, a reduction in snail2 expression was already apparent as early as stage ~11 (prior to CNC formation) in 13-3 morphants, and persisted through the onset of CNC migration (stage ~19). In normal late gastrula stage embryos, snail2 transcript is expressed bilaterally at the anterolateral border of the future neural plate, where the prospective CNC is induced (Steventon et al., 2009). There is also expression at this stage along the midline (Figure S4A, left panel). KD of ADAM13 caused a striking downregulation of snail2 transcript at the anterolateral neural plate boundary (note that adam13 is also expressed in this region; Figure S1B) but had little effect on snail2 expression along the midline, which is devoid of adam13 transcript by stage 13 (Figure S4A, right panel). These results indicate that ADAM13 MO inhibits early snail2 expression and the initial induction of prospective CNC during gastrulation. We further asked if the CNC phenotype caused by ADAM13 KD or overexpression of EfnB1 could be reversed by exogenous Snail2. As seen in Figure 4B (and Figures S4B and S4C), co-injection of snail2 transcript rescued sox9 and twist expression, as well as the head cartilage defects caused by ADAM13 MOs. Similar to ADAM13 KD, ectopic EfnB1 suppressed sox9 expression at early stages of CNC induction, a phenotype that was also effectively rescued by Snail2 (Figure 4C). Taken together, our results suggest that an EfnB-Wnt-Snail2 pathway mediates ADAM13 proteolytic function in early events that lead to CNC induction.
Figure 4. Snail2 is downstream of EfnB and canonical Wnt signaling to mediate ADAM13 function in CNC induction.
(A) Embryos injected at one-cell stage with the indicated MO (12 ng) were cultured to stages indicated, and quantitative RT-PCR carried out using embryo lysates. Relative levels of snail2 mRNA were determined by normalizing to amount of β-actin mRNA and by setting the levels calculated for control MO to 100%. Error bars represent standard deviations. **: P = 0.001 (N = 4) for stage 10.5, P = 0.005 (N = 3) for stage 12.5, and P= 0.008 (N = 3) for stage 19. (B, C) Embryos were injected with 3ng MO and/or 200 pg each transcript as indicated and in situ hybridization performed for sox9, as described in Figure 1F legend. The injected side is denoted with a red asterisk. (D) A model for ADAM13 function in CNC induction. See text for explanation.
DISCUSSION
Based on these results we propose a working model for ADAM13 function in early CNC induction. In normal embryos, ADAM13 cleaves EfnB1/B2 and attenuates EfnB signaling, thereby increasing the level of canonical Wnt signaling, which in turn upregulates snail2 and induces CNC cells at the neural plate boundary. When ADAM13 is knocked down, Efns B1 and B2 accumulate and inhibit canonical Wnt signaling. Downstream of this event, snail2 expression is suppressed, resulting in reduced CNC induction (Figure 4D).
In this study we present evidence demonstrating that Efns B1 and B2 are physiologically relevant substrates that mediate ADAM13 function in CNC induction. Previously we reported that ectopic expression of wild-type ADAM13 results in fused CNC streams and displaced trunk neural crest cells (Alfandari et al., 2001). These gain-of-function phenotypes could also be explained by ADAM13 cleavage of endogenous Efns B1 and B2. Complementary expression of EfnB2 and its cognate receptors is known to prevent intermingling of migrating CNC streams (Smith et al., 1997), whereas the positioning of EfnB1 provides a guidance cue for the migration of trunk neural crest cells, which express receptors for EfnBs (Krull et al., 1997). Ectopic overexpression of ADAM13 protease may eliminate functional Efns B1 and B2 in injected embryos and result in misoriented migration of both cranial and trunk neural crest cells, phenotypes reminiscent of disrupted EfnB1 and B2 signaling (Krull et al., 1997; Smith et al., 1997).
EfnB signaling has, thus far, only been linked to the Wnt/PCP pathway. Upon binding to EfnB ligands, EphB receptors are phosphorylated and can interact with Xdsh. In cultured cells such an interaction leads to activation of RhoA and Rho kinase, indicating that the Wnt/PCP pathway is activated (Tanaka et al., 2003). Alternatively, receptor-free EfnB1 can also activate Wnt/PCP signaling cell-autonomously by recruiting Xdsh to the plasma membrane (Lee et al., 2006; Lee et al., 2009). Because the canonical and non-canonical Wnt pathways are known to inhibit each other (Tahinci et al., 2007; Weidinger and Moon, 2003), it is possible that by cleaving EfnB1/B2, ADAM13 functions to shift the balance between Wnt/PCP and Wnt/β-catenin signaling to favor the latter. A similar role has been proposed for the Wnt co-receptor LRP6 and for MAK, a kinase that phosphorylates Dishevelled (Kibardin et al., 2006; Tahinci et al., 2007). Another question is which EfnB pathway(s) inhibit canonical Wnt signaling. In our cell-based assays, EfnB1ΔC effectively suppressed Wnt-induced TOPFLASH reporter expression (Figure S3D), pointing to forward EfnB signaling as the major pathway responsible for this inhibition. This is further supported by separate experiments that showed an increase in TOPLFLASH activity following expression of EphB1ΔC, which blocks signaling through the Eph receptor (Fig. S3E). However, a possible contribution from other Efn signaling pathways, such as the receptor-independent pathway (Lee et al., 2006; Lee et al., 2009), cannot be excluded based on these data. Future studies are needed to understand the mechanism(s) through which class B Efns inhibit canonical Wnt signaling.
The importance of Wnt/β-catenin signaling in neural crest induction has been well established (Wu et al., 2003). It was recently shown that the level of canonical Wnt signaling in prospective Xenopus CNC remains low during early gastrulation, but rises continuously starting from mid-gastrula stage (Steventon et al., 2009). We have observed a similar progressive increase in expression of the Wnt target gene snail2 in X. tropicalis embryos (data not shown). One possible explanation for this phenomenon is that CNC induction involves a transition from early suppression to later derepression/activation of canonical Wnt signaling. Our model (Figure 4D) provides a potential mechanism for this transition. The dorsolateral mesoderm is known to possess a neural crest inducing property (Hong et al., 2008; Steventon et al., 2009). Our in situ hybridization results show that both efns b1 and b2 are expressed in dorsal mesoderm of X. tropicalis gastrulae, with efnb2 predominantly expressed in the dorsal-most region and efnb1 also in the dorsolateral region (Figures S1A and S1A’). Therefore, it is likely that between the two class B Efns, EfnB1 plays a larger role in early suppression of canonical Wnt signaling in prospective CNC cells. In Xenopus embryos, adam13 is expressed in the mesoderm during gastrulation (Figure S1A), and may provide a driving force for the derepression/activation of canonical Wnt signaling in early CNC induction. Finally, several lines of evidence point to Wnt8 as the main (if not the only) Wnt ligand expressed in dorsolateral mesoderm and required for the initial induction of CNC (Hong et al., 2008; LaBonne and Bronner-Fraser, 1998; Steventon et al., 2009). We show here that the in vivo activity of Wnt8 can be modulated by both ADAM13 and EfnB1 (Figures 3C and S3E). This is in agreement with our proposed roles for ADAM13 and EfnBs in early CNC induction.
In summary, our data support a mechanism through which ADAM13 regulates early CNC induction. We propose that ADAM13 cleavage of EfnB1/B2 is required for derepression of canonical Wnt signaling, which leads to appropriate levels of snail2 expression and hence, normal CNC induction. Similar mechanisms may be used by ADAM13 or other ADAMs in other developmental contexts.
EXPERIMENTAL PROCEDURES
Embryos and Reagents
Wild-type X. tropicalis adults were purchased from NASCO. Embryos were prepared and injected as described in Extended Experimental Procedures. Embryo lysates were prepared as described below for cell lysates. RNA extraction, in vitro transcription and whole-mount in situ hybridization were carried out as described (Sive et al., 2000). Morpholino oligos were designed and synthesized by Gene Tools. Detailed information for plasmids and antibodies are described in Extended Experimental Procedures.
Cell Culture Based Cleavage Assays
HEK293T cells were cultured in 6-well plates, transfected at 50~70% confluency and allowed to grow for another 36 hr in serum-free medium before being harvested. Conditioned media were collected, centrifuged for 30 min at 12,000 g at 4°C to remove insoluble cell remnant, and concentrated using Amicon Microcon Ultracel YM-3 centrifugal filter devices. Deglycosylation was carried out using PNGase F (New England Biolabs). To prepare whole cell lysates, cells were lysed in ice-cold lysis buffer (20 mM Tris-HCl, pH 7.6, 100 mM NaCl, 1% Triton X-100, 1 mM PMSF, 5 mM EDTA) supplemented with 1x protease inhibitor cocktail (Sigma) and 10 mM o-phenanthroline, and centrifuged for 20 min at 12,000 g at 4°C. Supernatants were processed for SDS-PAGE and blotted with corresponding antibodies.
TOP/FOPFLASH Assays
HEK293T cells cultured in 24-well plates were transfected as described above (see Extended Experimental Procedures for amount of DNA transfected). About 40 hrs after transfection, cells were harvested, washed twice with PBS, resuspended in 1x lysis buffer (100 μl/well; provided with the assay kits), and frozen at -80°C. The cells were thawed later and centrifuged at 12,000 g at 4°C, and the supernatant was collected and used for luciferase and β-gal assays (both kits were from Promega). Luciferase activity was measured using a Femtomaster FB12 luminometer (Zylux). See Extended Experimental Procedures for details regarding embryo assays.
Phenotype Scoring and Statistics
Embryos allowed to develop to stage ~46 were scored for defects in head cartilage structures (see Figure S1D for examples of phenotypes). The % normal, moderate, and severe embryo phenotypes were calculated in every experiment, and averaged for multiple independent experiments. For phenotype comparisons, student’s t-tests were performed for the percentage of normal embryos.
Supplementary Material
Acknowledgments
We thank Dr. Ira Daar for kindly providing Efn and Eph receptor constructs and for useful suggestions and Dr. Xiaowei Lu for critical reading of the manuscript. We also thank the rest of the DeSimone lab for inspiring discussions and assistance with X. tropicalis husbandry. This work was supported by the NIH (DE14365 and HD26402 to D.W.D.) and March of Dimes Foundation (F405-140 to J.M.W. and 1-FY10-399 to D.W.D. and S.W.). S.W. was supported by an AHA postdoctoral fellowship and L.C.B. by an NIH postdoctoral fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams RH, Diella F, Hennig S, Helmbacher F, Deutsch U, Klein R. The cytoplasmic domain of the ligand ephrinB2 is required for vascular morphogenesis but not cranial neural crest migration. Cell. 2001;104:57–69. doi: 10.1016/s0092-8674(01)00191-x. [DOI] [PubMed] [Google Scholar]
- Alfandari D, Cousin H, Gaultier A, Smith K, White JM, Darribere T, DeSimone DW. Xenopus ADAM 13 is a metalloprotease required for cranial neural crest-cell migration. Curr Biol. 2001;11:918–930. doi: 10.1016/s0960-9822(01)00263-9. [DOI] [PubMed] [Google Scholar]
- Alfandari D, Wolfsberg TG, White JM, DeSimone DW. ADAM 13: a novel ADAM expressed in somitic mesoderm and neural crest cells during Xenopus laevis development. Dev Biol. 1997;182:314–330. doi: 10.1006/dbio.1996.8458. [DOI] [PubMed] [Google Scholar]
- Basch ML, Bronner-Fraser M. Neural crest inducing signals. Adv Exp Med Biol. 2006;589:24–31. doi: 10.1007/978-0-387-46954-6_2. [DOI] [PubMed] [Google Scholar]
- Boutros M, Mlodzik M. Dishevelled: at the crossroads of divergent intracellular signaling pathways. Mech Dev. 1999;83:27–37. doi: 10.1016/s0925-4773(99)00046-5. [DOI] [PubMed] [Google Scholar]
- Davis S, Gale NW, Aldrich TH, Maisonpierre PC, Lhotak V, Pawson T, Goldfarb M, Yancopoulos GD. Ligands for EPH-related receptor tyrosine kinases that require membrane attachment or clustering for activity. Science. 1994;266:816–819. doi: 10.1126/science.7973638. [DOI] [PubMed] [Google Scholar]
- Davy A, Aubin J, Soriano P. Ephrin-B1 forward and reverse signaling are required during mouse development. Genes Dev. 2004;18:572–583. doi: 10.1101/gad.1171704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Calisto J, Araya C, Marchant L, Riaz CF, Mayor R. Essential role of non-canonical Wnt signalling in neural crest migration. Development. 2005;132:2587–2597. doi: 10.1242/dev.01857. [DOI] [PubMed] [Google Scholar]
- Durbin L, Brennan C, Shiomi K, Cooke J, Barrios A, Shanmugalingam S, Guthrie B, Lindberg R, Holder N. Eph signaling is required for segmentation and differentiation of the somites. Genes Dev. 1998;12:3096–3109. doi: 10.1101/gad.12.19.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med. 2008;29:258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M, Osterfield M, Flanagan JG. Regulated cleavage of a contact-mediated axon repellent. Science. 2000;289:1360–1365. doi: 10.1126/science.289.5483.1360. [DOI] [PubMed] [Google Scholar]
- Himanen JP, Rajashankar KR, Lackmann M, Cowan CA, Henkemeyer M, Nikolov DB. Crystal structure of an Eph receptor-ephrin complex. Nature. 2001;414:933–938. doi: 10.1038/414933a. [DOI] [PubMed] [Google Scholar]
- Himanen JP, Saha N, Nikolov DB. Cell-cell signaling via Eph receptors and ephrins. Curr Opin Cell Biol. 2007;19:534–542. doi: 10.1016/j.ceb.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland SJ, Gale NW, Mbamalu G, Yancopoulos GD, Henkemeyer M, Pawson T. Bidirectional signalling through the EPH-family receptor Nuk and its transmembrane ligands. Nature. 1996;383:722–725. doi: 10.1038/383722a0. [DOI] [PubMed] [Google Scholar]
- Hong CS, Park BY, Saint-Jeannet JP. Fgf8a induces neural crest indirectly through the activation of Wnt8 in the paraxial mesoderm. Development. 2008;135:3903–3910. doi: 10.1242/dev.026229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes PW, Saha N, Barton WA, Kolev MV, Wimmer-Kleikamp SH, Nievergall E, Blobel CP, Himanen JP, Lackmann M, Nikolov DB. Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell. 2005;123:291–304. doi: 10.1016/j.cell.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Kibardin A, Ossipova O, Sokol SY. Metastasis-associated kinase modulates Wnt signaling to regulate brain patterning and morphogenesis. Development. 2006;133:2845–2854. doi: 10.1242/dev.02445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Krull CE, Lansford R, Gale NW, Collazo A, Marcelle C, Yancopoulos GD, Fraser SE, Bronner-Fraser M. Interactions of Eph-related receptors and ligands confer rostrocaudal pattern to trunk neural crest migration. Curr Biol. 1997;7:571–580. doi: 10.1016/s0960-9822(06)00256-9. [DOI] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Neural crest induction in Xenopus: evidence for a two-signal model. Development. 1998;125:2403–2414. doi: 10.1242/dev.125.13.2403. [DOI] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Snail-related transcriptional repressors are required in Xenopus for both the induction of the neural crest and its subsequent migration. Dev Biol. 2000;221:195–205. doi: 10.1006/dbio.2000.9609. [DOI] [PubMed] [Google Scholar]
- Lee HS, Bong YS, Moore KB, Soria K, Moody SA, Daar IO. Dishevelled mediates ephrinB1 signalling in the eye field through the planar cell polarity pathway. Nat Cell Biol. 2006;8:55–63. doi: 10.1038/ncb1344. [DOI] [PubMed] [Google Scholar]
- Lee HS, Mood K, Battu G, Ji YJ, Singh A, Daar IO. Fibroblast growth factor receptor-induced phosphorylation of ephrinB1 modulates its interaction with Dishevelled. Mol Biol Cell. 2009;20:124–133. doi: 10.1091/mbc.E08-06-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Kuriyama S, Moreno M, Mayor R. The posteriorizing gene Gbx2 is a direct target of Wnt signalling and the earliest factor in neural crest induction. Development. 2009;136:3267–3278. doi: 10.1242/dev.036954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overall CM, Blobel CP. In search of partners: linking extracellular proteases to substrates. Nat Rev Mol Cell Biol. 2007;8:245–257. doi: 10.1038/nrm2120. [DOI] [PubMed] [Google Scholar]
- Poliakov A, Cotrina M, Wilkinson DG. Diverse roles of eph receptors and ephrins in the regulation of cell migration and tissue assembly. Dev Cell. 2004;7:465–480. doi: 10.1016/j.devcel.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Sadaghiani B, Thiebaud CH. Neural crest development in the Xenopus laevis embryo, studied by interspecific transplantation and scanning electron microscopy. Dev Biol. 1987;124:91–110. doi: 10.1016/0012-1606(87)90463-5. [DOI] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. A Laboratory Manual. Cold Spring Harbor Laboratory Press; 2000. Early Development of Xenopus Laevis. [Google Scholar]
- Smith A, Robinson V, Patel K, Wilkinson DG. The EphA4 and EphB1 receptor tyrosine kinases and ephrin-B2 ligand regulate targeted migration of branchial neural crest cells. Curr Biol. 1997;7:561–570. doi: 10.1016/s0960-9822(06)00255-7. [DOI] [PubMed] [Google Scholar]
- Smith WC, Harland RM. Injected Xwnt-8 RNA acts early in Xenopus embryos to promote formation of a vegetal dorsalizing center. Cell. 1991;67:753–765. doi: 10.1016/0092-8674(91)90070-f. [DOI] [PubMed] [Google Scholar]
- Sokol S, Christian JL, Moon RT, Melton DA. Injected Wnt RNA induces a complete body axis in Xenopus embryos. Cell. 1991;67:741–752. doi: 10.1016/0092-8674(91)90069-b. [DOI] [PubMed] [Google Scholar]
- Steventon B, Araya C, Linker C, Kuriyama S, Mayor R. Differential requirements of BMP and Wnt signalling during gastrulation and neurulation define two steps in neural crest induction. Development. 2009;136:771–779. doi: 10.1242/dev.029017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahinci E, Thorne CA, Franklin JL, Salic A, Christian KM, Lee LA, Coffey RJ, Lee E. Lrp6 is required for convergent extension during Xenopus gastrulation. Development. 2007;134:4095–4106. doi: 10.1242/dev.010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Kamo T, Ota S, Sugimura H. Association of Dishevelled with Eph tyrosine kinase receptor and ephrin mediates cell repulsion. EMBO J. 2003;22:847–858. doi: 10.1093/emboj/cdg088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallin J, Thuret R, Giacomello E, Faraldo MM, Thiery JP, Broders F. Cloning and characterization of three Xenopus slug promoters reveal direct regulation by Lef/beta-catenin signaling. J Biol Chem. 2001;276:30350–30358. doi: 10.1074/jbc.M103167200. [DOI] [PubMed] [Google Scholar]
- Weidinger G, Moon RT. When Wnts antagonize Wnts. J Cell Biol. 2003;162:753–755. doi: 10.1083/jcb.200307181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JM, Bridges LC, DeSimone DW, Tomczuk M, Wolfsberg TG. Introduction to the ADAM family. In: Hooper NM, Lendeckel U, editors. The ADAM Family of Proteases. Springer; 2005. pp. 1–28. [Google Scholar]
- Wu J, Saint-Jeannet JP, Klein PS. Wnt-frizzled signaling in neural crest formation. Trends Neurosci. 2003;26:40–45. doi: 10.1016/s0166-2236(02)00011-5. [DOI] [PubMed] [Google Scholar]
- Xu Z, Lai KO, Zhou HM, Lin SC, Ip NY. Ephrin-B1 reverse signaling activates JNK through a novel mechanism that is independent of tyrosine phosphorylation. J Biol Chem. 2003;278:24767–24775. doi: 10.1074/jbc.M302454200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.