Abstract
Transforming growth factor-β1 (TGF-β1) expression in smooth muscle cells may play an important role in the pathogenesis of asthma. However, mechanisms that are involved in the regulation of TGF-β1 gene expression in human airway smooth muscle cells (HASMCs) remain elusive. Here, we show that mechanical stretch of HASMCs augmented TGF-β1 expression through a de novo RNA synthesis mechanism. Luciferase reporter assays revealed that stretch-induced TGF-β1 expression was mediated through the enhanced activation of TGF-β1 promoter. Interestingly, selective inhibitors of PTK, PI3K, or MEK1/2 attenuated TGF-β1 expression through blocking ERK1/2 phosphorylation and TGF-β1 promoter activity in response to stretch. In addition, stretch rapidly and transiently augmented GTP-bound RhoA and Rac1 but not Cdc42 GTPase. Either blockade of RhoA GTPase using C3 transferase, ROCK1/2 using Y27632, or knockdown of endogenous RhoA using RhoA siRNA attenuated stretch-induced TGF-β1 expression through the inhibition of ERK1/2 phosphorylation. Moreover, stretch augmented DNA binding activity of AP-1 in a time-dependent manner. Either treatment of HASMCs with the inhibitors of RhoA, ROCK1/2, PTK, PI3K, MEK1/2, or AP-1 or transfection of HASMCs with AP-1 decoy oligonucleotide attenuated stretch-induced TGF-β1 expression through repressing the DNA binding activity of AP-1. Site-directed mutagenesis demonstrated that two AP-1 binding sites in the TGF-β1 promoter region are responsible for stretch-induced TGF-β1 expression. Overall, in HASMCs, mechanical stretch plays an important role in TGF-β1 gene upregulation through a stretch-induced signaling pathway, which could be a potential therapeutic intervention for TGF-β1-induced pathogenesis in asthma.
Keywords: cyclic stretch, RhoA GTPase, ERK1/2, c-Jun
transforming growth factor-β1 (TGF-β1) is a multifunctional cytokine that is involved in many important biological functions (6). Many studies have shown overexpression of TGF-β1 in the airways of asthmatic individuals (19, 21, 22, 25, 30, 31, 32). In asthmatic airways, TGF-β1 is localized to airway smooth muscle cells, as well as mucous glands, endothelial cells, and other cells infiltrating the submucosa (32). TGF-β1 induces hypertrophy in human primary airway smooth muscle cells (11) and both hypertrophy and hyperplasia in bovine airway smooth muscle cells (5). These data suggest that airway smooth muscle cell may be a physiological target of TGF-β1 that might play a role in the pathogenesis of asthma.
During tidal breathing, the airway smooth muscle within airway walls is continuously exposed to mechanical strain. In particular, the airway walls in patients with asthma are exposed to excessive mechanical strain during bronchoconstriction (34). Thus, the cellular properties of airway smooth muscle such as contraction, stiffness, reorganization of the actin cytoskeleton, and alteration in gene expressions are influenced by mechanical strain in both normal and asthmatic conditions (27, 29). Therefore, it is important to characterize how mechanical strain triggers signal transduction in lung cells to fully understand the pathogenesis of asthma. Human airway smooth muscle cells (HASMCs) are particularly responsive to mechanical stretch as evident by observations from in vitro experiments documenting that airway smooth muscle cells respond to cyclic mechanical strain with increased proliferation, cell reorientation, protein production, reorientation of stress fibers, and additional recruitment of focal adhesions (27, 29). However, the precise cellular and molecular signaling mechanisms that underlie TGF-β1 expression in response to stretch in HASMCs remain unknown.
With regard to stretch-induced cellular signaling pathways, mitogen-activated protein (MAP) kinase pathways are well-established pathways that are activated in response to stretch (7, 13, 16). The extracellular signal-related kinase (ERK1/2), protein kinase 38 (p38), and c-Jun-NH2-terminal kinases (JNKs) have been widely studied in mammalian cells (20). In addition, small Rho family GTPase proteins, including RhoA, Rac1, and Cdc42 play important roles in the regulation of a variety of biological processes through their downstream protein kinases in cells (8, 24). The activation of Rho GTPases in response to stretch and their functions have been well established in cardiomyocytes (1, 14, 23). Moreover, many independent lines of evidence have shown that the induction of gene transcription in response to forces is mostly mediated through the activation of specific mechanosensitive transcription factors that bind on the specific sites in the target gene promoter (16, 26, 10). Characterization of TGF-β1 promoter region suggests that sequences located between nucleotides −453 to −323 are essential for its promoter activity, and deletion of this region completely abolished its transcriptional activity (15). Sequence analysis by TRANSFAC revealed that this 130-bp positive regulatory region contains binding sites for multiple transcription factors including two adjacent activator protein-1 (AP-1) binding sites and one NF-κB binding site. However, whether these transcription factors play a role in the stretch-induced regulation of TGF-β1 expression and its promoter activity in HASMCs remains largely unknown.
In this study, we identified the stretch-induced molecular signaling pathway that underlies TGF-β1 expression in HASMCs. Our data demonstrate that stretch-induced TGF-β1 expression and its promoter activity require protein tyrosine kinase (PTK) and phosphatidylinositol 3-kinase (PI3K) in a manner dependent on small RhoA GTPase, ROCK1/2, ERK1/2, and AP-1. Thus, stretch-augmented TGF-β1 expression in HASMCs may be distinct from other pathways mediating TGF-β1 expression. Furthermore, selective inhibitors of the above signaling proteins may have possible therapeutic benefits in TGF-β-induced airway remodeling in asthma (32).
METHODS
Materials.
HASMCs, basal medium, and supplemental mix were purchased from PromoCell (Heidelberg, Germany). Antibiotics, Easy-DNA kit, SuperScript III RTS first-strand cDNA synthesis kit, primers for RT-PCR, Lipofectamine 2000, Oligofectamine, genistein, and SP-600125 were purchased from Invitrogen (Carlsbad, CA). Actinomycin D and curcumin were purchased from Sigma (St. Louis, MO). C3 transferase was obtained from Cytoskeleton (Denver, CO). Rho-GDIα, NSC23766, LY-294002, and U0126 were obtained from Calbiochem (San Diego, CA). SB-203580, PCR master mix, pGL4.1 firefly luciferase, and pGL4.74 Renilla luciferase vectors, restriction enzymes, and Glo luciferase assay system were purchased from Promega (Madison, WI). RhoA siRNA, QIAprep miniprep, RNeasy mini kit, and QuantiTect SYBR green RT-PCR kit were purchased from Qiagen (Valencia, CA). QuikChange site-directed mutagenesis kit was obtained from Stratagene (La Jolla, CA). Quantikine human TGF-β1 kit was obtained from R&D Systems (Minneapolis, MN). Rho activation assay kit and RhoA, Rac1, and Cdc42 monoclonal antibodies were obtained from Cell Biolab (San Diego, CA). Phosphospecific anti-p44/42 (Thr202/Tyr204), anti-AP1 (c-jun), and goat anti-rabbit antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). NE-PER nuclear extraction reagent, biotin 3′-end DNA labeling kit, and LightShift chemiluminescent EMSA kit were purchased from Pierce (Rockford, IL). Phosphorothioate-modified sense and antisense single-stranded oligonucleotides were synthesized from Sigma Genosys (The Woodlands, TX).
Cell culture.
HASMCs were cultured in six-well cell culture plates at 37°C in a humidified atmosphere of 5% CO2 in smooth muscle cell basal medium supplemented with 5% fetal calf serum, 2 ng/ml basic fibroblast growth factor, 0.5 ng/ml epidermal growth factor, 5 μg/ml insulin, 100 U/ml penicillin, and 100 μg/ml streptomycin sulfate. After several subcultures, cells at passages between 5 and 7 were used in all experiments.
TGF-β1 promoter-reporter plasmid constructs and site-directed mutagenesis.
Human genomic DNA (gDNA) was isolated from 1 × 106 HASMCs using Easy-DNA kit following the manufacturer's directions. A 453-bp (−453 to +1) upstream promoter region from the first major transcription initiation site of human TGF-β1 promoter was amplified from 100 ng gDNA using 5′-GGTACCGTTGACAGACCCTCTTCTCC-3′ (forward) and 5′-GAGCTCTGGGAGGGAGATGGCCCAGG-3′ (reverse) primers and cloned into pGL4.1 luciferase reporter vector (Promega) and designated wild-type (WT) pGL4–453. KpnI and SacI restriction enzyme sites were engineered into forward and reverse primers, respectively. The site-specific mutations either in the proximal (P) (TGACTCT), distal (D) (TGTCTCA), or both (P and D) on the human TGF-β1 promoter were introduced using 5′-TCCCAGCCTactTCTCCTTCCG-3′ and 5′-CTCCCCTGTactTCATCCCCCG-3′ primers and Quick-Change site-directed mutagenesis kit according to the manufacturer's instructions. The pGL4–453 plasmid construct was used as a template to generate mutant (MT) P-MT-pGL4–453, D-MT pGL4–453, or PD-MT pGL4–453 constructs. DNA sequencing method confirmed the identity of the pGL4–453 constructs.
Transient transfection of plasmid DNA.
HASMCs were transfected with either WT pGL4–453, P-MT pGL4–453, D-MT pGL4–453, or PD-MT pGL4–453 promoter-reporter construct or pGL4 promoterless vector and cotransfected with pGL4.74 vector containing the Renilla luciferase gene to adjust the transfection efficiency using Lipofectamine 2000 in a serum-free medium according to the manufacturer's procedures. After 24 h of transfection, cells were switched to serum-containing medium and subjected to either stretch for the indicated time or maintained under static condition. Immediately after stretch, cell lysates were assayed for luciferase activity, and the firefly luciferase activity was normalized to that of Renilla luciferase using Dual-Glo Luciferase assay system according to the manufacturer's instructions.
RNA interference.
HASMCs were transfected with siRNA specific for RhoA according to the manufacturer's instructions. Briefly, HiPerFect transfection medium (3 μl) and a pool of 37.5 ng of RhoA or GAPDH siRNA (Qiagen, 5 nM) in 100 μl of serum-free medium were mixed and incubated at room temperature for 10 min. The cells in each well were then transfected with this mixture. Control cells were treated with the same amount of transfection medium without siRNA. After 12 h of transfection, fetal bovine serum was added to a final concentration of 5%. After 36 h, the cells were either subjected to stretch for the indicated time or maintained under the static condition.
Cyclic mechanical stretch.
In each experiment, HASMCs were seeded on type I collagen-coated Bioflex six-well plates at a density of 2 × 105 cells/ml medium per well and maintained as indicated above. Once 95% confluence was reached, cells were mechanically stretched to a maximum of 12% strain at a frequency of 0.3 Hz either for 1, 3, 6, 12, and 24 h or the indicated time periods, using a computer-controlled, vacuum-operated Flexercell Strain Unit (FlexCentral, FX-4000, FlexCell International). Control cells were maintained under the same conditions with no stretch unless otherwise indicated. In all experiments, cells cultured in every six-well plate were used for each time point period, and all experiments were repeated at least three times. After completion of the stretch regimen, cells were harvested and processed for either total RNA and protein extraction or other analyses.
RNA isolation and RT-PCR.
Total RNA was isolated from HASMCs using RNeasy mini kit and treated with DNase I according to the manufacturer's instructions. First-strand cDNA was synthesized from 1 μg of total RNA by reverse transcription in a 20-μl reaction using SuperScript III RTS first-strand cDNA synthesis kit following the manufacturer's directions. All PCR assays were performed in a 50-μl reaction mixture containing 2.5 μl cDNA and 200 nM of each primer using PCR master mix. The temperature cycle profile for the PCR reactions was 94°C for 2 min, 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min followed by 72°C for 5 min. GAPDH primers were used as internal control in all PCR samples. Agarose gel electrophoresis (2%) and direct DNA sequencing methods confirmed specificity of the PCR products. Real-time RT-PCR was performed in an Mx3005P QPCR system (Stratagene) using QuantiTect SYBR green RT-PCR kit according to the manufacturer's procedures. The temperature cycle profile for the RT-PCR reactions was 50°C for 30 min, 95°C for 15 min, and 40 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 30 s. Melting curve analysis was also included at one cycle of 95°C for 1 min, 55°C for 30 s, and 95°C for 30 s to verify the specificity of the amplified PCR products. The amount of TGF-β1 transcripts (2-ΔCT) was estimated by the comparative CT (ΔCT) method and normalized to an endogenous reference (GAPDH) relative to a calibrator. The sequence of primers used in the PCR strategy to amplify human TGF-β1 and GAPDH were 5′-ATTGAGGGCTTTCGCCTTAG-3′ (TGF-β1 forward primer), 5′-GAACCCGTTGATGTCCACTT-3′ (TGF-β1 reverse primer), 5′-GAAGGTGAAGGTCGGAGTCA-3′ (GAPDH forward primer), and 5′-TGGAAGATGGTGATGGGATT-3′ (GAPDH reverse primer).
ELISA.
ELISA was carried out using Quantikine human TGF-β1 kit according to the manufacturer's instructions. In brief, latent TGF-β1 from the cell culture supernatants was activated using 0.2 volume 1 N HCl at room temperature for 10 min and then neutralized to pH 7.2–7.6 by adding 0.2 volume 1.2 N NaOH/0.5 M HEPES solution. The activated TGF-β1 (50 μl) from each sample or standard was added into each well of a 96-well microplate coated with soluble TGF-β receptor type II and incubated at room temperature for 2 h. After three washings with wash buffer (0.05% Tween 20 in 1× PBS), 200 μl of horseradish peroxidase-conjugated polyclonal anti-TGF-β1 antibody (1:1,000 dilution) was added into each well and incubated at room temperature for 2 h, followed by three more washes in the wash buffer. Color development was performed by adding 200 μl of substrate solution (1:1 mixture of H2O2 and the chromogen tetramethylbenzidine solution) into each well and stopped by adding 50 μl 2 N sulfuric acid. The optical density of each well was determined within 30 min using a microplate reader (Bio-Rad, Hercules, CA) set to take dual endpoint readings at 450 nm and a reference wavelength of 540 nm. Activated TGF-β1 concentration in each sample was determined using a standard curve generated by four parameter logistic curve fit using SigmaStat 3.5 (Systat Software, San Jose, CA).
Rho-GTP pulldown assay.
Measurement of GTP-bound Rho proteins was performed using Rho activation assay kit following the manufacturer's directions. Briefly, cells were lysed in 1× ice-cold assay/lysis buffer (125 mM, pH 7.5, 750 mM NaCl, 5% Nonidet P-40, 50 mM MgCl2, 5 mM EDTA, 10% glycerol), incubated for 15 min on ice, and centrifuged at 14,000 g for 10 min. The supernatant from each sample (500 μl) was adjusted to 1 ml with 1× assay/lysis buffer and mixed with 40 μl of either rhotekin RBD or PAK PBD agarose bead slurry followed by incubation at 4°C for 1 h with gentle agitation. The agarose beads were pelleted by centrifugation at 14,000 g for 10 s, washed three times with 500 μl of 1× assay/lysis buffer, and resuspended in 40 μl of 2× reducing SDS-PAGE sample buffer. The precipitated GTP-bound Rho proteins were then detected by immunoblot analysis using a mouse monoclonal anti-RhoA, Rac1, or Cdc42 antibody. Total RhoA, Rac1, and Cdc42 in each sample lysate was determined by immunoblot analysis using respective antibody to normalize the activated GTP-bound Rho proteins.
Decoy oligonucleotide technique.
HASMCs were transfected with 20 μM ODN containing either consensus or scrambled sequences for AP-1 or NF-κB transcription factors for 6 h using Oligofectamine reagent in a serum-free medium according to the manufacturer's instructions. The sequences of single-stranded ODNs used in decoy oligonucleotide technique were WT AP-1: 5′-CGCTTGATGACTCAGCCGGAA-3′, scrambled AP-1: 5′-CGCTTGATTACTTAGCCGGAA-3′, WT NF-κB: 5′-CCTTGAAGGGATTTCCCTCC-3′, scrambled NF-κB: 5′-TTGCCGTACCTGACTTAGCC-3′.
EMSA.
Nuclear extract was isolated using NE-PER nuclear extraction reagent following the manufacturer's directions. The sequence of consensus complementary oligonucleotide for AP-1, 5′-CGCTTGATGACTCAGCCGGAA-3′ were 3′-biotinylated using the biotin 3′-end DNA labeling kit. The labeled both complementary oligonucleotides were mixed at an equimolar concentration in an annealing buffer (10 mM Tris, pH 7.5–8.0, 50 mM NaCl, 1 mM EDTA), heated at 95°C, and allowed to cool at room temperature for 2 h. Binding reactions were performed with 20 fmol of the biotin-3′end-labeled doubled stranded oligonucleotide and 4 μg of nuclear extract using LightShift chemiluminescent EMSA kit according to the manufacturer's procedures. After the binding reactions, samples were loaded onto native 6% polyacrylamide gels and electrophoresed at 100 V in 0.5× Tris borate/EDTA before being transferred onto a positively charged nylon membrane (BrightStar-Plus, Ambion, TX) using Trans-Blot SD semidry transfer cell (Bio-Rad) at 10 V for 30 min. Transferred DNAs were cross-linked to the membrane using a transilluminator for 10 min and detected using horseradish peroxidase-conjugated streptavidin (LightShift chemiluminescent EMSA kit) according to the manufacturer's instructions.
Chromatin immunoprecipitation assay.
Chromatin immunoprecipitation (ChIP) assays were performed to determine the binding activity of AP-1 on the human TGF-β1 promoter in HASMCs using EZ ChIP. The sequence of primers used in the PCR strategy to amplify human TGF-β1 promoter region bearing the AP-1 transcription factor binding sequence was 5′-GTTGACAGACCCTCTTCTCC-3′ and 5′-TGGGAGGGAGATGGCCCAGG-3′.
Statistical analyses.
Data were analyzed by one-way ANOVA. When the ANOVA identified differences among groups, a multiple comparisons test among means was performed with Tukey multiple range test. Analyses were performed using SigmaStat 3.0 (Systat Software).
RESULTS
Stretch increases TGF-β1 mRNA expression and protein release through de novo RNA synthesis mechanism in HASMCs.
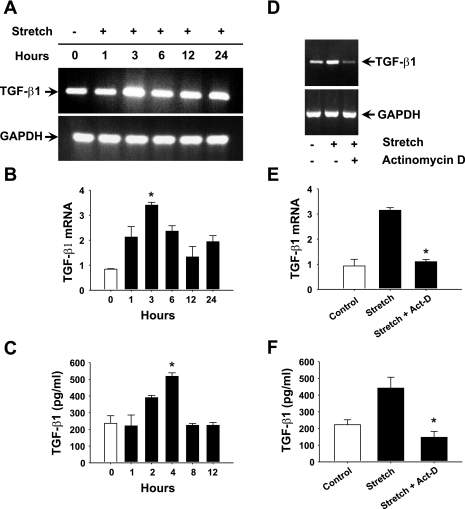
First, we explored whether stretch can modulate TGF-β1 expression in HASMCs. Our data showed that stretch increased TGF-β1 mRNA expression in a time-dependent manner (Fig. 1A). More precisely, real-time RT-PCR analyses demonstrated that stretch increased TGF-β1 mRNA levels 1 h after the application of stretch, significantly peaked at 3 h, and remained higher than control level at 24 h of stretch (Fig. 1B). The maximum induction ratio of TGF-β1 mRNA level was 3.4-fold at 3 h of stretch. Next, we sought to determine whether stretch-induced TGF-β1 mRNA expression modulates TGF-β1 protein release from HASMCs to the culture medium. The level of TGF-β1 protein increased at 2 h of stretch, significantly reached a 2.1-fold elevation at 4 h, and then dropped to control level at 12 h of stretch (Fig. 1C). These data indicate that 3-h stretch can significantly increase TGF-β1 mRNA level in HASMCs and protein release in the culture medium. To study whether stretch-induced TGF-β1 mRNA transcription was mediated through a de novo RNA synthesis, HASMCs were incubated in the presence or absence of 10 μg/ml actinomycin D, a transcriptional inhibitor, for 1 h followed by either stretch for 3 h or no stretch. Treatment of cells with actinomycin D significantly reduced the stretch-induced TGF-β1 mRNA expression compared with untreated cells (Fig. 1D). Real-time quantitative RT-PCR analysis showed that stretch-induced TGF-β1 mRNA level was significantly reduced when cells were treated with actinomycin D (Fig. 1E). The stretch-induced TGF-β1 protein level in the culture supernatant was also significantly reduced in the presence of actinomycin D (Fig. 1F). These results strongly demonstrate that a de novo mRNA transcription is required for the stretch-induced increase in TGF-β1 mRNA expression and for the maintenance of the protein level in HASMCs.
Fig. 1.
Stretch increases TGF-β1 expression through de novo RNA synthesis mechanism in human airway smooth muscle cells (HASMCs). Cultured HASMCs were subjected to cyclic mechanical stretch (12% strain, 0.3 Hz) for the indicated periods. A: stretch increases TGF-β1 mRNA expression in a time-dependent manner. The level of TGF-β1 mRNA expression is highest at 3 h of stretch. B: real-time RT-PCR confirms that stretching of HASMCs significantly increased TGF-β1 mRNA expression. C: culture supernatants collected for the indicated periods and processed for ELISA indicate that TGF-β1 protein release in the culture supernatant is significantly increased at 4 h of stretch. In another experiment, HASMCs were treated in the presence or absence of 10 μg/ml actinomycin D (Act-D) for 1 h followed by stretch for 3 h or maintained under static condition. D: actinomycin D attenuated stretch-induced TGF-β1 mRNA expression. E: real-time RT-PCR confirms that actinomycin D significantly attenuated stretch-induced TGF-β1 mRNA expression relative to untreated cells. F: ELISA results show that stretch-induced TGF-β1 protein release in the culture medium is significantly blocked by actinomycin D. Gel pictures are representative of 3 independent experiments. Reported values in the bar graphs are means ± SE of 3 independent experiments. *P < 0.05.
Stretch-induced TGF-β1 expression in HASMCs is regulated through PTK, PI3K, and MEK1/2.
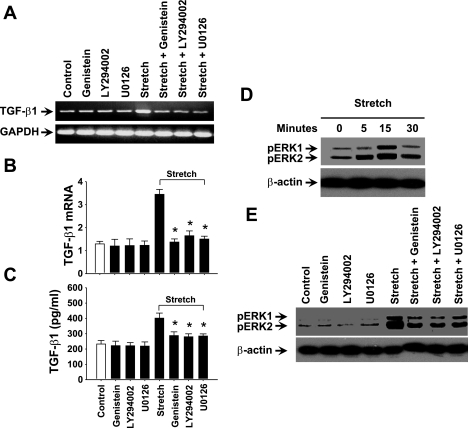
To investigate specific signaling molecules that could be involved in the regulation of TGF-β1 expression in response to stretch, we used selective pharmacological inhibitors to treat HASMCs. Interestingly, genistein (PTK inhibitor), LY-294002 (PI3K inhibitor), or U0126 (MEK1/2 inhibitor) inhibited the stretch-induced TGF-β1 mRNA expression (Fig. 2A). Real-time RT-PCR and ELISA studies showed a significant reduction in the stretch-induced TGF-β1 mRNA expression and protein release when the cells were treated with the above inhibitors (Fig. 2, B and C). However, when cells were treated with 20 μM JNK inhibitor (SP-600125) or 20 μM p38 inhibitor (SB-203580), the inhibitor had no effect to that of the control cells on TGF-β1 expression (data not shown) in response to stretch. These results suggest that stretch-induced TGF-β1 expression in HASMCs is regulated through PTK, PI3K, and MEK1/2 signaling pathway rather than JNK and/or p38 kinase.
Fig. 2.
Stretch-induced TGF-β1 expression in HASMCs is mediated through protein tyrosine kinase (PTK), phosphoinositide 3-kinase (PI3K), and MEK1/2. HASMCs were treated in the presence or absence of 50 μm of genistein (PTK inhibitor), LY-294002 (PI3K inhibitor), or U0126 (MEK1/2 inhibitor) for 1 h followed by 3-h stretch or no stretch. A: treatment of HASMCs with genistein, LY-294002, or U0126 attenuated stretch-induced TGF-β1 mRNA expression to that of untreated cells. B: real-time RT-PCR study shows that TGF-β1 mRNA expression is significantly reduced by treatment of cells with any of the above specific inhibitors in response to stretch. C: results of ELISA shows that inhibitors of PTK, PI3K, and MEK1/2 also significantly attenuated stretch-induced TGF-β1 protein release in the culture medium. D: HASMCs were stretched for the indicated times followed by isolation of cytoplasmic extract and processed for Western blot analysis with phospho-specific ERK1/2 antibody. Stretch induced phosphorylation of ERK1/2 in a time-dependent manner. Stretch-induced phosphorylation of ERK1/2 peaked at 15-min stretch. E: HASMCs were treated in the presence or absence of 50 μm genistein (PTK inhibitor), LY-294002 (PI3K inhibitor), or U0126 (MEK1/2 inhibitor) for 1 h followed by either 15-min stretch or no stretch. DMSO (1% final concentration) or water was used as vehicle for the appropriate inhibitor that dissolved in the same solvent. When cells were treated with the above specific inhibitors, we observed a significant block in the phosphorylation of ERK1/2 to that of untreated cells in response to stretch. Gel pictures are representative of 3 independent experiments. Reported values in the bar graphs are means ± SE of 3 independent experiments. *P < 0.05.
Next, we investigated the effect of stretch on the phosphorylation of ERK1/2 (a downstream target of MEK1/2). As we expected, stretch induced the phosphorylation of ERK1/2 in a time-dependent manner. The stretch-induced ERK1/2 phosphorylation was peaked at 15 min of stretch (Fig. 2D). Since PTK, PI3K, and MEK1/2 are involved in the regulation of TGF-β1 expression, we sought to determine whether these molecules are upstream of ERK1/2 in response to stretch. Western blot analyses showed that treatment of HASMCs with genistein, LY-294002, or U0126 completely blocked the stretch-induced phosphorylation of ERK1/2 (Fig. 2E). These data suggest that PTK, PI3K, and MEK1/2 are the upstream signaling molecules leading to stretch-induced phosphorylation of ERK1/2.
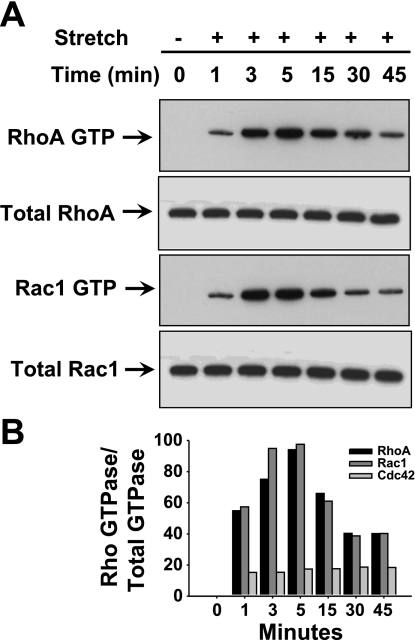
Stretch increases activation of Rho GTPases in HASMCs.
We found that stretch rapidly and transiently increased GTP-bound RhoA and Rac1 GTPases in a time-dependent manner. However, we could not detect a stretch-induced increase in Cdc42 GTPase in HASMCs (Fig. 3, A and B). More precisely, the activation of RhoA GTPase started to increase at 1 min after the onset of stretch, peaked at 5 min, and started to decline at 15 min of stretch. Like RhoA, Rac1 GTPase activity increased after 1 min of stretch, but remained high at 3 and 5 min, and declined at 15 min of stretch. These results demonstrate that stretch can induce the activation of RhoA and Rac1 GTPases in a time-dependent manner, whereas there was no effect of stretch on Cdc42 GTPase in HASMCs.
Fig. 3.
Stretching of HASMCs activates small Rho GTPase proteins in a time-dependent manner. HASMCs were stretched for the indicated time followed by isolation of cytoplasmic extract and used for pulldown assay. A: stretch rapidly and transiently induced the activation of RhoA GTPase and Rac1 in a time-dependent manner. The activation of RhoA and Rac1 is high as early as 5 min of stretch. However, stretch did not have any effect on the activation of Cdc42 GTPase. B: densitometry bar diagram indicates that the activated RhoA and Rac1 GTPases are higher at 5 min after onset of stretch. Western blot pictures are representative of 3 independent experiments. Reported values in the bar graphs are the values of a single experiment. P < 0.05.
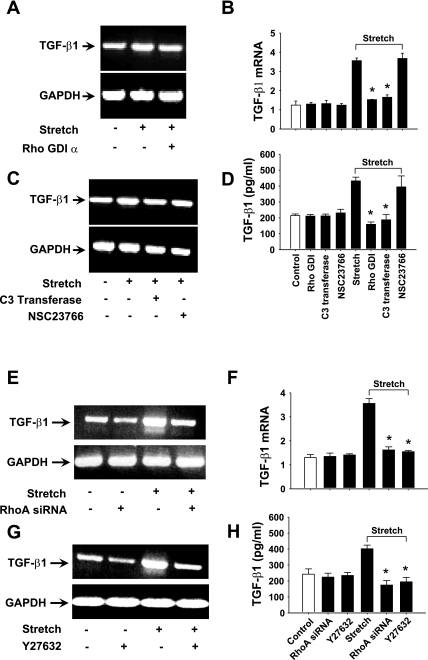
Stretch-activated RhoA GTPase and its downstream target Rho kinase ROCK1/2 are involved in the regulation of TGF-β1 expression in HASMCs.
To further investigate whether stretch-activated RhoA and Rac1 GTPases are involved in the regulation of TGF-β1 expression, we treated HASMCs with selective Rho GTPase inhibitors. Rho GDI-α (Rho GTPases inhibitor) significantly inhibited both stretch-induced TGF-β1 mRNA expression and protein release (Fig. 4, A, B, and D). Interestingly, C3 transferase (RhoA inhibitor) but not NSC23766 (Rac1 inhibitor) significantly blocked both stretch-induced TGF-β1 mRNA expression and protein release (Fig. 4, B–D). These results strongly suggest that the regulation of TGF-β1 expression in HASMCs is mediated through the activation of RhoA GTPase in response to stretch.
Fig. 4.
Rho GTPase RhoA regulates TGF-β1 expression in HASMCs in response to stretch. HASMCs were treated with either in the presence or absence of 5 μM Rho GTPases inhibitor Rho-GDIα for 1 h, 5 μg/ml RhoA inhibitor C3 transferase for 6 h, or 100 μM Rac1 inhibitor NSC23766 for 1 h followed by stretch for 3 h. A–D: Rho GDI-α and C3 transferase attenuated stretch-induced TGF-β1 expression, but the Rac1 inhibitor NSC23766 had no effect on TGF-β1 expression. B–D: real-time PCR and ELISA results reveal that Rho GDI-α and C3 transferase significantly blocked TGF-β1 mRNA expression and protein release to that of untreated cells in response to stretch. E–H: HASMCs were either transfected with RhoA siRNA for 36 h or treated with either in the presence or absence of 10 μM ROCK1/2 inhibitor Y-27632 for 1 h followed by stretch for 3 h. Transfection of cells with RhoA siRNA significantly blocked stretch-induced TGF-β1 mRNA expression and protein release. Y-27632 also significantly attenuated TGF-β1 mRNA expression and protein release in response to stretch. DMSO (1% final concentration) or water was used as vehicle for the appropriate inhibitor that dissolved in the same solvent. Gel pictures are representative of 3 independent experiments. Reported values in the bar graphs are means ± SE of 3 independent experiments. *P < 0.05.
To confirm the involvement of stretch-induced RhoA GTPase in TGF-β1 expression, we employed RNA interference technique using RhoA-specific siRNA. When cells were transfected with siRNA specific to RhoA, GTPase blocked stretch-induced TGF-β1 mRNA expression to that of control cells (Fig. 4E). Real-time RT-PCR and ELISA data showed that TGF-β1 mRNA expression was significantly blocked by RhoA siRNA followed by protein release in the culture medium (Fig. 4, F and H). These data confirmed that TGF-β1 expression was mediated through RhoA GTPase in response to stretch. SiRNA for GAPDH was used as positive control to examine transfection efficiency and toxicity (data not shown). Next, we studied the role of ROCK1/2 kinases, downstream target of Rho GTPase, in the regulation of TGF-β1 expression. The ROCK1/2 inhibitor Y-27632 inhibited TGF-β1 mRNA expression (Fig. 4G). Real-time RT-PCR and ELISA studies showed that TGF-β1 mRNA expression was significantly blocked by Y-27632 (Fig. 4F) followed by protein release (Fig. 4H) in the culture medium, suggesting that ROCK1/2 kinases are necessary for stretch-induced RhoA GTPase-mediated TGF-β1 expression in HASMCs.
Stretch-induced phosphorylation of ERK1/2 is mediated via RhoA and ROCK1/2 signaling in HASMCs.
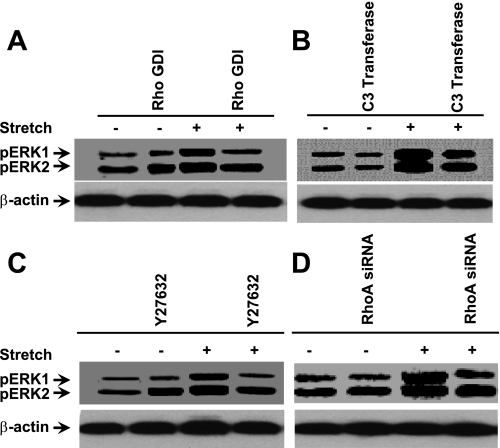
To study whether RhoA and ROCK1/2 signaling are important for the stretch-induced phosphorylation of ERK1/2, HASMCs were incubated either in the presence or absence of Rho GDI-α or C3 transferase (RhoA inhibitor) for 6 h followed by either stretch for 15 min or no stretch. As shown in Fig. 5, A and B, Rho GDI-α or C3 transferase attenuated the stretch-induced ERK1/2 phosphorylation. To confirm these results, we transfected HASMCs with RhoA siRNA. As expected, cells transfected with RhoA siRNA completely blocked stretch-induced phosphorylation of ERK1/2 (Fig. 5C). Moreover, to determine the role of ROCK1/2 in the stretch-induced phosphorylation of ERK1/2, HASMCs were treated with ROCK1/2 inhibitor. Stretching HASMCs in the presence of ROCK1/2 inhibitor Y-27632 blocked phosphorylation of ERK1/2 (Fig. 5D). These data strongly suggest that the stretch-induced phosphorylation of ERK1/2 is mediated through RhoA/ROCK1/2 signaling pathway.
Fig. 5.
Involvement of RhoA GTPase on the phosphorylation of ERK1/2 in HASMCs in response to stretch. HASMCs were treated with either in the presence or absence of 5 μM Rho GTPases inhibitor Rho-GDIα for 1 h, 5 μg/ml RhoA inhibitor C3 transferase for 6 h, 100 μM Rac1 inhibitor NSC23766 for 1 h, 10 μM ROCK1/2 inhibitor Y-27632 for 1 h, or transfected with RhoA siRNA for 36 h followed by stretch for 15 min. Total cytoplasmic proteins were isolated, and the phosphorylated ERK1/2 proteins were determined using Western blot analysis. A: HASMCs treated with Rho-GDIα attenuates stretch-induced phosphorylation of ERK1/2 to that of untreated cells. B: treatment of HASMCs with C3 transferase blocked phosphorylation of ERK1/2 to that of untreated cells in response to stretch. Cells treated with NSC23766 did not block stretch-induced phosphorylation of ERK1/2. C: the ROCK1/2 inhibitor Y-27632 inhibits phosphorylation of ERK1/2 in HASMCs in response to stretch. DMSO (1% final concentration) or water was used as vehicle for the appropriate inhibitor that dissolved in the same solvent. D: transfection of HASMCs with RhoA siRNA attenuates stretch-induced phosphorylation of ERK1/2 to that of nontransfected cells. Western blot pictures are representative of 3 independent experiments. Reported values in the bar graphs are means ± SE of 3 independent experiments. P < 0.05.
Transcription factor AP-1 regulates stretch-induced TGF-β1 expression in HASMCs.
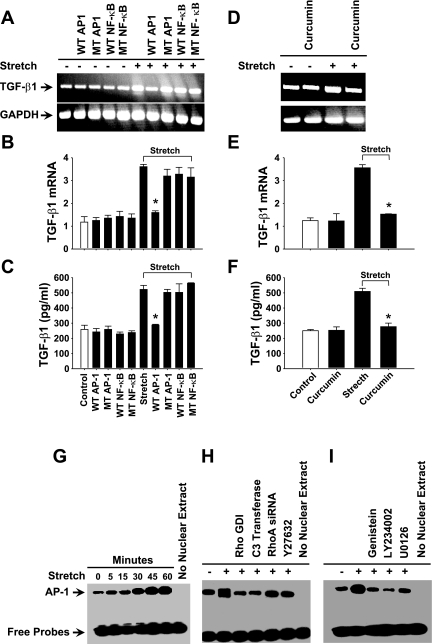
To investigate the specific transcription factors involved in the stretch-induced regulation of TGF-β1 expression in HASMC, we employed the ODN technique that is known to be effective in the inhibition of the activity of transcription factors (3). As shown in Fig. 6A, transfection of HASMCs with WT AP-1 but not WT NF-κB ODN noticeably blocked TGF-β1 mRNA expression. Real-time RT-PCR and ELISA studies confirmed the WT AP-1 mediated blockade of TGF-β1 mRNA expression and protein release (Fig. 6, B and C). On the other hand, transfection of HASMCs with the scrambled ODN for either AP-1 or NF-κB did not have any effect on the level of TGF-β1 mRNA or protein release. These data suggest that stretch-induced TGF-β1 expression in HASMCs is regulated by AP-1 transcription factor.
Fig. 6.
TGF-β1 gene expression in HASMCs is mediated by AP-1 transcription factor in response to stretch. HASMCs were transfected with 20 μM double-stranded ODN containing either in the presence or absence of consensus or scrambled sequences for AP-1 or NF-κB transcription factors for 6 h followed by stretch for 3 h. A: cells transfected with AP-1 ODN attenuates TGF-β1 expression in response to stretch to that of nontransfected cells. B: real-time RT-PCR study shows a significant reduction in stretch-induced TGF-β1 mRNA expression. C: ELISA result shows that release of TGF-β1 protein in the culture medium is significantly also blocked by AP-1 ODN rather than NF-κB ODN in response to stretch. HASMCs were incubated in the presence or absence of 75 μM curcumin, an AP-1 inhibitor, for 1 h followed by stretch for 3 h. D: HASMCs treated with curcumin attenuated stretch-induced TGF-β1 mRNA expression to that of untreated cells. E and F: real-time RT-PCR and ELISA studies show that stretch-induced TGF-β1 mRNA and protein are significantly attenuated by curcumin. HASMCs were stretched for the indicated periods. Nuclear proteins were isolated immediately after the stretch and processed to determine the DNA binding activity of AP-1 using EMSA. G: stretch activates DNA binding activity of AP-1 in a time-dependent manner. HASMCs were treated with either in the presence or absence of 5 μM Rho GTPases inhibitor Rho-GDIα for 1 h, 5 μg/ml RhoA inhibitor C3 transferase for 6 h, 100 μM Rac1 inhibitor NSC23766 for 1 h, 10 μM ROCK1/2 inhibitor Y-27632 for 1 h, or transfected with RhoA siRNA for 36 h followed by stretch for 30 min or no stretch. Total nuclear proteins were isolated, and the DNA binding activity of AP-1 was determined using EMSA. H: treatment of cells with Rho-GDIα, C3 transferase, Y-27632, or transfection with RhoA siRNA blocked DNA binding activity of AP-1 in response to stretch. HASMCs were treated with either in the presence or absence of 50 μm genistein (PTK inhibitor), LY-294002 (PI3K inhibitor), or U0126 (MEK1/2 inhibitor) for 1 h followed by 30-min stretch or no stretch. DMSO (1% final concentration) or water was used as vehicle for the appropriate inhibitor that dissolved in the same solvent. I: the representative EMSA image shows that the stretch-induced DNA binding activity of AP-1 transcription factor is attenuated by PTK, PI3K, and MERK 1/2 inhibitors to that of untreated cells. Gel and EMSA images are representative of 3 independent experiments. Reported values in the bar graphs are means ± SE of 3 independent experiments. *P < 0.05.
To further elucidate the role of AP-1 in the regulation of TGF-β1 expression, HASMCs were incubated in the presence or absence of 75 μM curcumin, an effective inhibitor of AP-1 transcription factor. Treatment of HASMCs with curcumin completely abolished TGF-β1 expression to that of control cells (Fig. 6D). Data from real-time RT-PCR and ELISA analysis showed that the TGF-β1 mRNA expression and protein release were significantly inhibited by curcumin treatment (Fig. 6, E and F). These results confirmed that the stretch-induced TGF-β1 expression in HASMCs is regulated by AP-1 transcription factor.
Next, we investigated the effect of stretch on the DNA binding activity of AP-1 transcription factor. HASMCs were subjected to stretch for the indicated time intervals, and nuclear extracts were isolated from stretched and control cells. EMSA was carried out using biotin-3′-end-labeled doubled-stranded oligonucleotide containing the putative binding sites for AP-1. As shown in Fig. 6G, stretch rapidly increased the DNA binding activity of AP-1 in a time-dependent manner. The activation of AP-1 increased at 5 min of stretch and peaked at 45 min of stretch. These results demonstrate that AP-1 is activated in a time-dependent manner in response to stretch of HASMCs.
Next, we determined the role of RhoA GTPase and its downstream kinases ROCK1/2 in the regulation of DNA binding activity of AP-1 in response to stretch. HASMCs were incubated in the presence or absence of Rho GDI-α for 1 h, C3 transferase for 6 h, or Y-27632 inhibitors for 1 h or transfected with RhoA siRNA for 36 h followed by either stretch for 45 min or no stretch. As shown in Fig. 6H, treatment of HASMCs with the above inhibitors or transfection with RhoA siRNA completely attenuated the stretch-induced DNA binding activity of AP-1. These data suggest that in HASMCs, stretch-induced DNA binding activity of AP-1 is mediated through RhoA GTPase and ROCK1/2 signaling pathway. Since the inhibition of PTK, PI3K, and MEK1/2 inhibited TGF-β1 expression, we next sought to determine whether these kinases are involved in the stretch-induced activation of AP-1 transcription factor. HASMCs were preincubated with either in the presence or absence of 50 μM genistein, LY-234002, or U0126 for 1 h followed by either stretch for 45 min or no stretch. Treatment of HASMCs with the above three inhibitors completely attenuated the stretch-induced DNA binding activity of AP-1 transcription factor (Fig. 6I). These data suggest that stretch stimulates the DNA binding activity of AP-1 through the activation of PTK, PI3K, and MEK1/2 kinases in HASMCs.
Stretch activates TGF-β1 promoter in HASMCs.
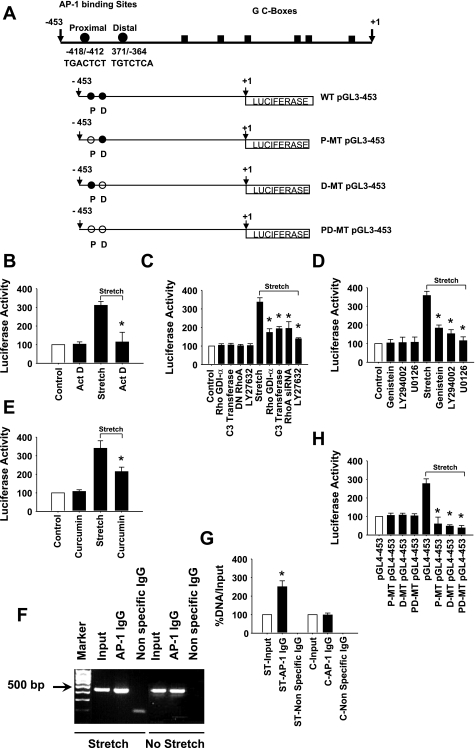
According to TRANSFAC analysis, the promoter of human TGF-β1 gene (GenBank acc. no. NM_000660) contains several response elements, including AP-1 transcription factor binding sites. To assess the molecular basis of TGF-β1 gene promoter in HASMCs, we used PCR strategy to clone a 453-bp 5′-flanking sequence upstream of the transcription start site of the TGF-β1 gene into pGL4-basic promoterless luciferase reporter vector and designated as WT pGL4–453. To evaluate whether stretch activates TGF-β1 promoter in HASMCs through a de novo RNA synthesis, we transfected cells with either WT pGL4–453 promoter-reporter construct (Fig. 7A) or pGL4 promoterless-reporter vector and cotransfected with pGL4.74 vector. After 24-h transfection, cells were treated with either in the presence or absence of actinomycin D for 1 h followed by either 1-h stretch or no stretch. Cells were lysed and assayed for luciferase activity. The transfection experiments revealed that stretch increased the activity of human TGF-β1 promoter fragment −453/+1 as assessed by luciferase activity. Actinomycin D blocked stretch-induced TGF-β1 promoter activity (Fig. 7B). The level of stretch-increased luciferase activity in cells transfected with TGF-β1 promoter-reporter construct without actinomycin D was nearly threefold higher compared with that of either untreated cells or those that were maintained under no stretch.
Fig. 7.
Stretch stimulates TGF-β1 promoter activity in HASMCs. A: schematic diagram of 453-bp human TGF-β1 promoter region corresponding to the transcription initiation site that cloned into promoterless pGL4basic vector that contains the firefly (Photinus pyralis) luciferase reporter gene and is designated as pGL4–453. Closed and open circles indicate wild-type (WT) and mutated (MT) AP-1 binding sites, respectively, and boxes show GC-rich regions. The WT pGL4–453 promoter-reporter plasmid construct was used as template for the site-directed mutagenesis as described in methods. Either mutated proximal (TGgtTCT), distal (gtTCTCA), or both (TGgtTCT/gtTCTCA) AP-1 binding sites of WT pGL4–453 construct is designated as P-MT pGL4–453, D-MT pGL4–453, and PD-MT pGL4–453, respectively. B: HASMCs were transiently transfected with WT pGL4–453 promoter-reporter plasmid construct and cotransfected with Renilla luciferase reporter plasmid (pGL4.74) for 24 h followed by treatment with actinomycin D for 1 h. Cells were either stretched or not stretched for 1 h. The firefly luciferase activity of the reporter gene was measured in the cell lysates and normalized to that of Renilla luciferase activity as described in methods. C: HASMCs were transiently transfected with WT pGL4–453 promoter-reporter plasmid construct and cotransfected with Renilla luciferase reporter plasmid (pGL4.74) for 24 h. In another experiment, RhoA siRNA was cotransfected also with the above plasmid constructs. After transfection, cells without RhoA siRNA were treated with either in the presence or absence of 5 μM Rho-GDIα (Rho GTPase inhibitor) for 1 h, 5 μg/ml C3 transferase (RhoA inhibitor) for 6 h, 100 μM NSC23766 (Rac1 inhibitor) or 10 μM Y-27632 (ROCK1/2 inhibitor) for 1 h followed by stretch for 1 h or no stretch. DMSO (1% final concentration) or water was used as vehicle for the appropriate inhibitor that dissolved in the same solvent. Luciferase activities of the reporter plasmids were estimated to determine the promoter activity of TGF-β1 gene. D: the plasmid transfected cells without RhoA siRNA were treated with either in the presence or absence of 50 μm genistein (PTK inhibitor), LY-294002 (PI3K inhibitor), or U0126 (MEK1/2 inhibitor) for 1 h followed by 1-h stretch. E: after 24 h of transfection, cells (no RhoA siRNA) were treated with or without 75 μM curcumin, an AP-1 inhibitor, for 1 h followed by 1-h stretch. F and G: HASMCs were either subjected to stretch for 1 h or kept under same conditions with no stretch. Chromatin of control and stretched HASMC was immunoprecipitated with either a nonspecific IgG or anti-AP-1 antibody. PCR was performed with the immunoprecipitated DNA using TGF-β1 primers encompassing the TGF-β1 promoter region bearing the AP-1 transcription factor binding. The binding activity of AP-1 transcription factor is higher in cells subjected to stretch than those not stretched. Real-time RT-PCR study shows that the stretch-induced DNA binding activity of AP-1 transcription factor is significantly higher in response to stretch than control cells. H: HASMCs were transiently transfected with either WT pGL4–453, P-MT pGL4–453, D-MT pGL4–453, or PD-MT pGL4–453 promoter-reporter plasmid construct and cotransfected with Renilla luciferase reporter plasmid (pGL4.74) for 24 h followed by 1-h stretch. The firefly luciferase activity of the reporter gene was measured to determine the stretch-induced TGF-β1 promoter activity. Cells transfected with either P-MT pGL4–453, D-MT pGL4–453, or PD-MT pGL4–453 promoter-reporter plasmid construct completely attenuates luciferase activity to that of cells transfected with WT pGL4–453 plasmid construct in response to stretch. Gel pictures are representative of 3 independent experiments. Reported values in the bar graphs are means ± SE of 3 independent experiments. *P < 0.05.
Role of RhoA GTPase, ROCK1/2, PTK, PI3K, MEK1/2, and AP-1 in stretch-induced TGF-β1 promoter activity in HASMCs.
Next, we explored whether the stretch-activated RhoA GTPase is involved in the TGF-β1 promoter activation. In the first experiment, HASMCs were transfected with either WT pGL4–453 promoter-reporter construct with or without RhoA siRNA or pGL4 promoterless vector and cotransfected with pGL4.74 vector into HASMCs. After 36-h transfection, cells were subjected to 1-h stretch or no stretch, lysed, and assayed for luciferase activity. In the second experiment, cells were transfected with either WT pGL4–453 promoter-reporter construct or with pGL4 promoterless vector and cotransfected with pGL4.74 vector into HASMCs. After 24-h transfection, cells were preincubated with either in the presence or absence of 5 μM Rho GDI-α or 10 μM Y-27632 for 1 h or 5 μg/ml C3 transferase for 6 h, followed by 1-h stretch or no stretch. The cells were lysed and assayed for luciferase activity. As we expected, cells either transfected with RhoA siRNA or treated with Rho GDI-α, C3 transferase, or Y-27632, significantly attenuated stretch-induced TGF-β1 promoter activity (Fig. 7C). These data strongly suggest that stretch-activated RhoA GTPase and its downstream target ROCK1/2 are upstream signaling molecules that are involved in the regulation of TGF-β1 promoter activity.
Since PTK, PI3K, and ERK1/2 kinases are involved in the regulation of TGF-β1 expression, we sought to determine whether blocking the stretch-induced activation of these kinases attenuate TGF-β1 promoter activity. HASMCs were transfected with either WT pGL4–453 promoter-reporter construct or with pGL4 promoterless vector and cotransfected with pGL4.74 vector. After 24-h transfection, cells were preincubated with either in the presence or absence of 50 μM genistein, LY-294002, or U0126 for 1 h followed by either 1-h stretch or no stretch. The cells were lysed and supernatants were assayed for luciferase activity. As shown in Fig. 7D, the stretch-induced TGF-β1 promoter activity was significantly attenuated by genistein, LY-294002, or U0126 inhibitors. These results demonstrate that the stretch-induced regulation of TGF-β1 promoter is mediated through PTK, PI3K, and ERK1/2 kinases.
To determine whether the stretch-activated AP-1 is involved in the activation of TGF-β1 promoter, HASMCs were incubated in the presence or absence of curcumin for 1 h followed by either stretch or no stretch for 1 h. The cells were lysed and assayed for luciferase activity. As shown in Fig. 7E, curcumin significantly attenuated the stretch-induced activation of TGF-β1 promoter, suggesting that the transcription factor AP-1 activates TGF-β1 promoter in HASMCs in response to stretch.
Stretch increases the DNA binding activity of c-Jun (AP-1) on TGF-β1 promoter in HASMCs.
To verify the ability of stretch-activated AP-1 to interact with the TGF-β1 promoter, we employed ChIP assay. After 1 h of stretch of HASMCs, the cross-linked chromatin was immunoprecipitated using a monoclonal antibody against AP-1. As negative controls, we included a nonspecific rabbit IgG that does not have a binding site on the TGF-β1 promoter. After immunoprecipitation and reversal of the cross-links, the enriched endogenous TGF-β1 promoter fragment in each sample was monitored using RT-PCR. Our results indicated that only the anti-AP-1 antibody anti-c-jun-immunoprecipitated chromatin contained the TGF-β1 promoter (Fig. 7F). Real-time RT-PCR showed that the level of anti-AP-1-immunoprecipitated DNA was 2.5-fold higher than the corresponding input level (Fig. 7G). There was no change in the control samples that were subjected to no stretch. These results confirm that the stretch-activated AP-1 directly binds on TGF-β1 promoter region and is involved in the regulation of TGF-β1 expression in HASMCs.
Localization of the stretch responsible cis-regulatory elements of on TGF-β1 promoter.
To study whether the two AP-1 binding sites −418/−412 and −371/−364 (Fig. 7A) in the TGF-β1 promoter sequence mediate the stretch-induced promoter activity in HASMCs, we utilized site-directed mutagenesis to mutate both proximal and distal AP-1 binding sites in the TGF-β1 promoter regions. HASMCs were transfected with either WT pGL4–453, P-MT pGL4–453, D-MT pGL4–453, or PD-MT pGL4–453 promoter-reporter construct or pGL4 promoterless vector and cotransfected with pGL4.74 vector followed by 1-h stretch or no stretch. The luciferase reporter assays showed that when cells were transfected with either P-MT pGL4–453, D-MT pGL4–453, or PD-MT pGL4–453, promoter-reporter construct showed a significant reduction in the luciferase activity to that of WT pGL4–453-transfected cells (Fig. 7H). These changes in the luciferase activity are due to mutation in either one or both AP-1 binding sites. These data suggest that both proximal and distal cis-regulatory elements for AP-1 binding sites on the TGF-β1 promoter region are necessary for stretch-induced TGF-β1 promoter activity in HASMCs.
DISCUSSION
In this study, we have investigated a signaling pathway by which cyclic mechanical stretch regulates TGF-β1 gene expression in smooth muscle cells of human airways. Our data demonstrated that cyclic stretching of HASMCs increased TGF-β1 mRNA expression and protein release in a time-dependent manner via a de novo RNA synthesis mechanism. Our data also showed that stretch increased the activity of luciferase gene through the induction of TGF-β1 promoter in HASMCs. The stretch-induced TGF-β1 expression and its promoter activity were mediated through PTK, PI3K, and MEK1/2 signaling pathway. Furthermore, stretch activated small RhoA GTPase and its downstream targets ROCK1/2 kinases and AP-1 transcription factor. Our data provide the first experimental evidence demonstrating that AP-1 played important roles in the mechanical regulation of TGF-β1 expression and its promoter activity. The presence of two adjacent AP-1 cis-acting elements on the TGF-β1 promoter region was essential for AP-1-mediated TGF-β1 promoter activity in response to stretch.
TGF-β1 is produced by various cell types in airways including airway smooth muscle cells (32). Other investigators have shown that TGF-β1 expression in airway structural cells is directly related to airway physiology and pathophysiology (2). For example, TGF-β1 induces airway smooth muscle cells to release collagen and glycosaminoglycans and is thus involved in the regulation of extracellular matrix. Airway remodeling thickens the airway wall, reduces baseline airway caliber, and exaggerates airway narrowing that causes airway obstruction, a characteristic feature of asthma. Excessive force generation by airway smooth muscle is the main culprit in airway narrowing during asthma attack. In the present study, we applied 12% of uniaxial stretch in terms of length to the silicone chamber on which HASMCs were cultured using a cell stretch apparatus. Under physiological conditions, airway smooth muscle is exposed to 4–5% of strain during tidal breathing (12, 17). When lung volume is doubled by a deep inspiration, airway smooth muscle is expected to be stretched by as much as 25–30% (29, 12). Thus, stretch amplitude employed in our study appears to represent the physiological levels in vivo.
In our previous work, we have demonstrated that stretching of smooth muscle cells leads to the activation of ERK1/2, JNK1, and p38 MAP kinase (17). However, the role of MAP kinases in the stretch-induced regulation of TGF-β1 expression remains largely unknown. In this study, we have uncovered a role of MEK1/2 in the stretch-induced expression of TGF-β1 in HASMCs.
Many studies have been conducted in an attempt to improve our understanding of the process of mechanical signal transduction. However, in the absence of specific mechanosensitive cell surface receptors, it is still not clear how mechanical forces are transmitted into intracellular signals that underlie gene expression. However, mechanical force could trigger the deformation of the cell membrane, which may directly or indirectly cause conformational changes in protein (and subsequent activation of them) that are anchored to the inner surface of cell membranes or in transmembrane proteins. Several effector proteins including small Rho family GTPase proteins are examples of membrane-bound proteins that might be affected by mechanical forces. Rho GTPases are members of the Ras super family of monomeric GTP-binding proteins. Our results suggested that the stretch-induced regulation of TGF-β1 gene in HASMCs is mediated through RhoA/ROCK1/2 signaling pathway. Our study also implicated that small Rho GTPase proteins such as RhoA and Rac1 are mechanosensitive GTPase proteins by which stretch may regulate the expression of mechanosensitive genes in HASMCs. It is important to point out that our data do not prove whether Rho kinase activates the ERK pathway directly. It is possible that Rho kinase acts on a different pathway upstream of ERK leading to the activation of ERK.
Many independent lines of evidence have shown that the induction of gene transcription in response to forces is mostly mediated through the activation of specific mechanosensitive transcription factor(s), which binds on the specific binding regions in the target gene promoter or enhancer region (10, 26). Characterization of TGF-β1 promoter region suggested that sequences located between nucleotides −453 and −323 are essential for its promoter activity, and deletion of this region completely abolished its transcriptional activity (15). Sequence analysis by TRANSFAC revealed that this 130-bp positive regulatory region contains binding sites for multiple transcription factors including two adjacent AP-1 binding sites and one NF-κB binding site. In the present study, our data showed that stretch rapidly activated both AP-1 (Fig. 6) and NF-κB (data not shown) in HASMCs. However, we found that the regulation of TGF-β1 expression and its promoter activity was mediated through AP-1 rather than NF-κB. The stretch-induced NF-κB activation appeared to be dispensable for the stretch-induced TGF-β1 expression. This is because the transfection of HASMCs with consensus AP-1 but not NF-κB ODN completely blocked stretch-induced TGF-β1 expression and its promoter activity. Moreover, treatment of HASMCs with the AP-1 inhibitor curcumin abolished stretch-induced TGF-β1 expression and its promoter activity. Our findings are consistent with previous studies in which the regulation of TGF-β1 expression and/or its promoter activity is mediated primarily through AP-1 rather than NF-κB in a variety of cells (4, 9, 33). However, a recent study in human lung carcinoma cells has shown that IL-1β-induced regulation of TGF-β1 gene is mediated through direct requirement of both NF-κB and AP-1 to the corresponding binding sites in the TGF-β1 promoter (18). In the present study, even though stretch induced the DNA binding activity of NF-κB, transfection of HASMCs with consensus NF-κB ODN did not alter stretch-induced TGF-β1 expression and stretch-induced activity of its promoter, suggesting that stretch-activated NF-κB may regulate genes other than TGF-β1 in HASMCs. Furthermore, the ChIP assay confirmed that the stretch-induced TGF-β1 gene regulation is mediated through at least one of the component of AP-1 complexes, c-jun. In addition, our data showed that transfection of HASMCs with either P-MT pGL4–453, D-MT pGL4–453, or PD-MT pGL4–453 promoter-reporter construct dramatically reduced stretch-induced luciferase activity. These results suggested that stretch-induced TGF-β1 expression and its promoter activity were mediated through the stretch-activated AP-1 transcription factor. In addition, the presence of proximal and distal AP-1 binding sites in the TGF-β1 promoter was necessary for the stretch-induced TGF-β1 gene regulation in HASMCs.
In summary, our study provided data on basic mechanisms and specific mechanosensitive signaling pathways that are involved in the regulation of stretch-induced TGF-β1 expression and its promoter activity in HASMCs.
GRANTS
This work was supported by National Heart, Lung, and Blood Institute Grants HL-63134 and HL-072839 and a grant from the National Science Foundation.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
REFERENCES
- 1.Aikawa R, Komuro I, Yamazaki T, Zou Y, Kudoh S, Zhu W, Kadowaki T, Yazaki Y. Rho family small G proteins play critical roles in mechanical stress-induced hypertrophic responses in cardiac myocytes. Circ Res 84: 458–466, 1999 [DOI] [PubMed] [Google Scholar]
- 2.Aubert JD, Dalal BI, Bai TR, Roberts CR, Hayashi S, Hogg JC. Transforming growth factor-beta 1 gene in human airways. Thorax 49: 225–232, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bielinska A, Shivdasani RA, Zhang LQ, Nabel GJ. Regulation of gene expression with double-stranded phosphorothioate oligonucleotides. Science 250: 997–1000, 1990 [DOI] [PubMed] [Google Scholar]
- 4.Birchenall-Roberts MC, Ruscetti FW, Kasper J, Lee HD, Friedman R, Geiser A, Sporn MB, Roberts AB, Kim SJ. Transcriptional regulation of the transforming growth factor beta 1 promoter by v-src gene products is mediated through the ap-1 complex. Mol Cell Biol 10: 4978–4983, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black PN, Young PG, Skinner SJ. Response of airway smooth muscle cells to TGF-β1: effects on growth and synthesis of glycosaminoglycans. Am J Physiol Lung Cell Mol Physiol 271: L910–L917, 1996 [DOI] [PubMed] [Google Scholar]
- 6.Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor-β1 in human disease. N Engl J Med 342: 1350–1358, 2000 [DOI] [PubMed] [Google Scholar]
- 7.Chang L, Karin M. Mammalian MAP kinase signaling cascades. Nature 410: 37–40, 2001 [DOI] [PubMed] [Google Scholar]
- 8.Cox D, Chang P, Zhang Q, Reddy PG, Bokoch GM, Greenberg S. Requirements for both Rac1 and Cdc 42 in membrane ruffling and phagocytosis in leukocytes. J Exp Med 186: 1487–1494, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhandapani KM, Khan MM, Wade FM, Wakade C, Mahesh VB, Brann DW. Induction of transforming growth factor-beta1 by basic fibroblast growth factor in glioma and astrocytes is medicated by MEK/ERK signaling and AP-1 activation. J Neurosci Res 85: 1033–1045, 2007 [DOI] [PubMed] [Google Scholar]
- 10.Gillespie PG, Walker RG. Molecular basis of mechanosensory transduction. Nature 413: 194–202, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Goldsmith AM, Bentley JK, Zhou L, Jia Y, Bitar KN, Fingar DC, Hershenson MB. Transforming growth factor-beta induces airway smooth muscle hypertrophy. Am J Respir Cell Mol Biol 34: 247–254, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes JMB, Hoppin FG, Jr, Mead J. Effects of lung inflation on bronchial length and diameter in excised lungs. J Appl Physiol 32: 25–35, 1972 [DOI] [PubMed] [Google Scholar]
- 13.Ingber DE. Cellular basis of mechanotransduction. Biol Bull 194: 323–327, 1998 [DOI] [PubMed] [Google Scholar]
- 14.Kawamura S, Miyamoto S, Brown JH. Initiation and transduction of stretch-induced RhoA and Rac1 activation through caveolae: cytoskeletal regulation of ERK translocation. J Biol Chem 278: 31111–31117, 2003 [DOI] [PubMed] [Google Scholar]
- 15.Kim SJ, Glick A, Sporn MB, Roberts AB. Characterization of the promoter region of the human transforming growth factor-beta 1 gene. J Biol Chem 264: 402–408, 1989 [PubMed] [Google Scholar]
- 16.Kumar A, Chaudhry I, Reid MB, Boriek AM. Distinct signaling pathways are activated in response to mechanical stress applied axially and transversely to skeletal muscle fibers. J Biol Chem 277: 46493–46503, 2002 [DOI] [PubMed] [Google Scholar]
- 17.Kumar A, Knox AJ, Boriek AM. CCAAT/enhancer-binding protein and activator protein-1 transcription factors regulate the expression of interleukin-8 through the mitogen-activated protein kinase pathways in response to mechanical stretch of human airway smooth muscle cells. J Biol Chem 278: 18868–18876, 2003 [DOI] [PubMed] [Google Scholar]
- 18.Lee KY, Ito K, Hayashi R, Jazrawi EPI, Barnes PJ, Adcock IM. NF-κB and activator protein 1 response elements and the role of histone modifications in IL-1β-induced TGF-β1 gene transcription. J Immunol 176: 603–615, 2006 [DOI] [PubMed] [Google Scholar]
- 19.Minshall EM, Leung DY, Martin RJ, Song YL, Cameron L, Ernst P, Hamid Q. Eosinophil-associated TGF-beta1 mRNA expression and airways fibrosis in bronchial asthma. Am J Respir Cell Mol Biol 17: 326–333, 1997 [DOI] [PubMed] [Google Scholar]
- 20.Nobes CD, Hall A. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J Cell Biol 144: 1235–1244, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nomura A, Uchida Y, Sakamoto T, Ishii Y, Masuyama K, Morishima Y, Hirano K, Sekizawa K. Increases in collagen type I synthesis in asthma: the role of eosinophils and transforming growth factor-β. Clin Exp Allergy 32: 860–865, 2002 [DOI] [PubMed] [Google Scholar]
- 22.Ohno I, Nitta Y, Yamauchi K, Hoshi H, Honma M, Woolley K, O'Byrne P, Tamura G, Jordana M, Shirato K. Transforming growth factor beta 1 (TGF beta 1) gene expression by eosinophils in asthmatic airway inflammation. Am J Respir Cell Mol Biol 15: 404–409, 1996 [DOI] [PubMed] [Google Scholar]
- 23.Pan J, Singh US, Takahashi T, Oka Y, Palm-Leis A, Herbelin BS, Baker KM. PKC mediates cyclic stretch-induced cardiac hypertrophy through Rho family GTPases and mitogen-activated protein kinases in cardiomyocytes. J Cell Physiol 202: 536–553, 2005 [DOI] [PubMed] [Google Scholar]
- 24.Qi M, Elion EA. MAP kinase pathways. J Cell Sci 118: 3569–3572, 2005 [DOI] [PubMed] [Google Scholar]
- 25.Redington AE, Madden J, Frew AJ, Djukanovic R, Roche WR, Holgate ST, Howarth PH. Transforming growth factor-beta 1 in asthma. Measurement in bronchoalveolar lavage fluid. Am J Respir Crit Care Med 156: 642–647, 1997 [DOI] [PubMed] [Google Scholar]
- 26.Sadoshima J, Takahashi T, Jahn L, Izumo S. Roles of mechano-sensitive ion channels, cytoskeleton, and contractile activity in stretch-induced immediate-early gene expression and hypertrophy of cardiac myocytes. Proc Natl Acad Sci USA 89: 9905–9909, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seow Chun Y, Schellenberg RR, Pare Peter D. Structural and functional changes in the airway smooth muscle of asthmatic subjects. Am J Respir Crit Care Med 158: 179S–186S, 1998 [DOI] [PubMed] [Google Scholar]
- 28.Smith PG, Garcia R, Kogerman L. Mechanical strain increases protein tyrosine phosphorylation in airway smooth muscle cells. Exp Cell Res 239: 353–360, 1998 [DOI] [PubMed] [Google Scholar]
- 29.Smith PG, Janiga KE, Bruce MC. Strain increases airway smooth muscle cell proliferation. Am J Respir Cell Mol Biol 10: 85–90, 1994 [DOI] [PubMed] [Google Scholar]
- 30.Takizawa H, Tanaka M, Takami K, Ohtoshi T, Ito K, Satoh M, Okada Y, Yamasawa F, Nakahara K, Umeda A. Increased expression of transforming growth factor-beta1 in small airway epithelium from tobacco smokers and patients with chronic obstructive pulmonary disease (COPD). Am J Respir Crit Care Med 163: 1476–1483, 2001 [DOI] [PubMed] [Google Scholar]
- 31.Tillie-Leblond I, Pugin J, Marquette CH, Lamblin C, Saulnier F, Brichet A, Wallaert B, Tonnel AB, Gosset P. Balance between proinflammatory cytokines and their inhibitors in bronchial lavage from patients with status asthmaticus. Am J Respir Crit Care Med 159: 487–494, 1999 [DOI] [PubMed] [Google Scholar]
- 32.Vignola AM, Chanez P, Chiappara G, Merendino A, Pace E, Rizzo A, la Rocca Antonio M, Bellia V, Bonsignore G, Bousquet J. Transforming growth factor-beta expression in mucosal biopsies in asthma and chronic bronchitis. Am J Respir Crit Care Med 156: 591–599, 1997 [DOI] [PubMed] [Google Scholar]
- 33.Weigert C, Brodbeck K, Klopfer K, Häring H, Schleicher E. Angiotensin II induces human TGF-β1 promoter activation: similarity to hyperglycaemia. Diabetologia 45: 890–898, 2002 [DOI] [PubMed] [Google Scholar]
- 34.Wiggs BR, Hrousis CA, Drazen JM, Kamm RD. On the mechanism of mucosal folding in normal and asthmatic airways. J Appl Physiol 83: 1814–1821, 1997 [DOI] [PubMed] [Google Scholar]