Abstract
Summary: JCVI Metagenomics Reports (METAREP) is a Web 2.0 application designed to help scientists analyze and compare annotated metagenomics datasets. It utilizes Solr/Lucene, a high-performance scalable search engine, to quickly query large data collections. Furthermore, users can use its SQL-like query syntax to filter and refine datasets. METAREP provides graphical summaries for top taxonomic and functional classifications as well as a GO, NCBI Taxonomy and KEGG Pathway Browser. Users can compare absolute and relative counts of multiple datasets at various functional and taxonomic levels. Advanced comparative features comprise statistical tests as well as multidimensional scaling, heatmap and hierarchical clustering plots. Summaries can be exported as tab-delimited files, publication quality plots in PDF format. A data management layer allows collaborative data analysis and result sharing.
Availability: Web site http://www.jcvi.org/metarep; source code http://github.com/jcvi/METAREP
Contact: syooseph@jcvi.org
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Recent advances in sequencing technologies have boosted microbial ecology research by allowing cost-effective sequencing of microbial communities directly from their natural environment. As of today, 210 microbial metagenomes sampled from diverse environments such as the ocean, acid mine drainage, soil, bovine rumen and the human body have been reported (http://www.genomesonline.org/). Such studies promise to reveal new insights into how microbes adapt to their abiotic and biotic environment. To distill such information from metagenomic sequences, computational methods are being used to identify and analyze their functional and taxonomic signatures.
Metagenomic annotation pipelines have been developed that help researchers to identify such information (Markowitz et al., 2008; Meyer et al., 2008; Tanenbaum et al., 2010). To make higher level inferences from such annotated data, computational biologists have developed various analysis tools. MEGAN, for example, allows users to import BLAST outputs to generate taxonomic and functional summaries (Huson et al., 2007). Others allow statistical analysis, such as MetaStats and ShotgunFunctionalizeR (Kristiansson et al., 2009; White et al., 2009). While most of these tools provide generic interfaces that are independent of the annotation pipeline used, they do not allow to analyze function in the context of phylogeny and vice versa. For example, MG-RAST facilitates the analysis of either phylogenetic or metabolic content, but they cannot be related to one another. The same is true for MEGAN that provides individual taxonomic and functional summaries. Most importantly, with the ever increasing size of metagenomics datasets, performance and scalability of such tools becomes essential.
We describe JCVI Metagenomics Reports (METAREP), a new open source tool that addresses such shortcomings by providing a scalable yet flexible comparative metagenomics framework.
2 DATA IMPORT
Users can install METAREP and import annotation data obtained from reads or assemblies using METAREP's index generation scripts. The METAREP data format supports the most common metagenomics annotation data types including a free-text functional description, best BLAST hit information such as NCBI taxon, E-value, percent identity, percent sequence coverage, as well as GO ID, EC ID and protein domain ID (Supplementary Table S1). During the import process, auxiliary fields are populated using the NCBI Taxonomy and Gene Ontology (GO) to store all ancestors of a taxon or GO ID, respectively.
3 WEB ANALYSIS FEATURES
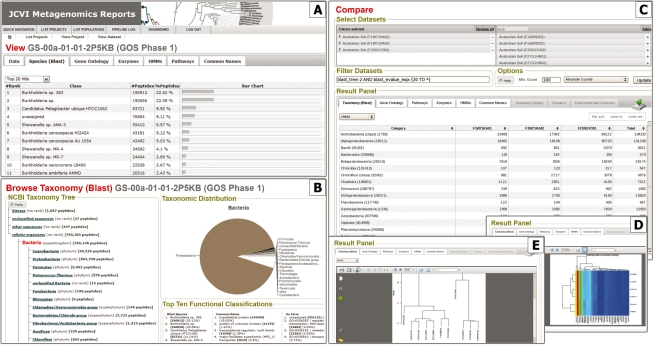
The METAREP View pages provide high-level summaries for a dataset (Fig. 1A). Each tab provides a ranked list and bar chart for the respective data type. The Species Tab, for example, summarizes top ranks for identified species. The Search pages let users specify fields, or logical combinations of fields (14 fields are supported) to filter datasets. The auxiliary fields blast_tree and go_tree may be used to select or exclude certain subsets of the NCBI Taxonomy or the GO tree. For example, one can search for all bacteria or exclude eukaryotes or search for a certain GO/taxonomic combination. The search returns lists and pie charts that summarize the top functional and taxonomic categories of the found subset. Similar summaries can be studied by using the Browse pages that are available for taxonomic, pathway, enzyme and GO classifications (Fig. 1B). The goal of the Compare pages is to help the user quickly find the interesting differences between datasets (Fig. 1C–E). Individual datasets can be grouped together to provide greater statistical resolving power. After selecting datasets using the searchable dataset-select box, users can specify a filter query that is applied to all datasets before the comparison. In addition, a minimum absolute count can be specified to filter out categories that are spurious. Various comparative options can be selected ranging from absolute and relative counts, to chi-square and non-parametric t-tests (MetaStats; White et al., 2009)) to multidimensional scaling, heatmap (Fig. 1D) and hierarchical cluster plots (Fig. 1E). Users can choose the annotation data type they wish to compare by choosing from several tabs (Taxonomy, GO, pathway, HMM, enzyme and common name are supported). The level of the comparison can be specified in each tab (for taxonomy the rank can be adjusted, for GO the distance from the root, etc.). Counts and statistics can be exported in tab-delimited, plots in PDF format.
Fig. 1.
The METAREP web interface is designed to be user-friendly and fast, allowing users to view, search, browse and compare metagenomics datasets.
4 IT INFRASTRUCTURE AND PERFORMANCE
METAREP uses the enterprise search platform Lucene/Solr served by a JETTY web server that runs on a Java HotSpot 64-Bit Server VM. Currently, we have indexed 68 million documents (46 GB) distributed over 330 index files. Much larger index volumes can be handled as shown by HAthi Trust, a digital library, which currently indexes 227 Tbytes of data (http://www.hathitrust.org/). User account information and dataset meta-information is stored in a MySQL database. The web logic is implemented in PHP using the CAKEPHP framework. Index files are served by two load balanced Dell Power Edge R710 servers each having eight cores (2.66 GHz), 72G RAM and 2x 600 GB HD. Query performance increases linearly with increasing workload until it peaks at 3100 search requests per second (Supplementary Fig. 1).
5 DISCUSSION
A variety of metagenomics analysis tools are provided either as standalone or via the web. The strength of our web-based approach is a generic data model indexed by a high-performance search engine in combination with a user-friendly Web 2.0 interface. JCVI's annotated metagenomes as well as other selected public datasets can be accessed at www.jcvi.org/metarep. Users that wish to analyze their own metagenomics data must install the software and import their own annotations. We invite interested programmers to contribute to the METAREP open-source project hosted at http://github.com/jcvi/METAREP. Finally, users are encouraged to suggest additional features of interest.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Cynthia Pfannkoch and Shannon Williamson for their critique and input.
Funding: US Department of Energy (#DE-FG02-02ER63453, #DE-FC02-02ER63446); National Institute of Allergy and Infectious Diseases (1U54AI084844); National Cancer Institute (UH2CA14023); Sloan Foundation (#2004-5-46EG); University of Illinois, Department of Primary Industries, Victoria, Australia.
Conflict of Interest: none declared.
REFERENCES
- Huson DH, et al. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansson E, et al. ShotgunFunctionalizeR: an R-package for functional comparison of metagenomes. Bioinformatics. 2009;25:2737–2738. doi: 10.1093/bioinformatics/btp508. [DOI] [PubMed] [Google Scholar]
- Markowitz VM, et al. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 2008;36:D534–D538. doi: 10.1093/nar/gkm869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum DM, et al. The JCVI standard operating procedure for annotating prokaryotic metagenomic shotgun sequencing data. Stand. Genomic Sci. 2010;2:2. doi: 10.4056/sigs.651139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JR, et al. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 2009;5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.