Abstract
Summary: Gene coexpression analysis was developed to explore gene interconnection at the expression level from a systems perspective, and differential coexpression analysis (DCEA), which examines the change in gene expression correlation between two conditions, was accordingly designed as a complementary technique to traditional differential expression analysis (DEA). Since there is a shortage of DCEA tools, we implemented in an R package ‘DCGL’ five DCEA methods for identification of differentially coexpressed genes and differentially coexpressed links, including three currently popular methods and two novel algorithms described in a companion paper. DCGL can serve as an easy-to-use tool to facilitate differential coexpression analyses.
Contact: yyli@scbit.org and yxli@scbit.org
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
From the perspective of systems biology, gene coexpression analysis is useful for investigating gene interconnection at the expression level. Differential coexpression analysis (DCEA), which examines the change in expression correlation of gene pairs between two conditions, helps to explore the global transcriptional mechanisms underlying phenotypic changes. Compared with traditional differential expression analysis (DEA), the development of DCEA tools is lagged. In this work, we developed an R package, DCGL, implementing three previously proposed DCEA methods and two new algorithms reported in a companion paper (Yu,H. et al., submitted for publication).
Log Ratio of Connections (LRC) calculates the logarithm of the ratio of the connectivities of a gene between two conditions (Reverter et al., 2006). Average Specific Connection (ASC) counts the ‘specific connections’ that exist in only one coexpression network (Choi et al., 2005). The weighted gene coexpression network analysis (WGCNA) weights links with correlation coefficients and compares the sums of the correlation coefficients of a gene (Mason et al., 2009; van Nas et al., 2009). In contrast, our two methods, differential coexpression profile (DCp) and differential coexpression enrichment (DCe), are designed based on the exact coexpression changes of gene pairs, and thus can differentiate significant coexpression changes from relatively trivial ones, and identify coexpression reversal between positive and negative (Yu,H. et al., submitted for publication). All the five methods are able to identify differentially coexpressed genes (DCGs) from microarray datasets, and DCe is also able to pick out differentially coexpressed gene pairs or links (DCLs).
2 DESIGN
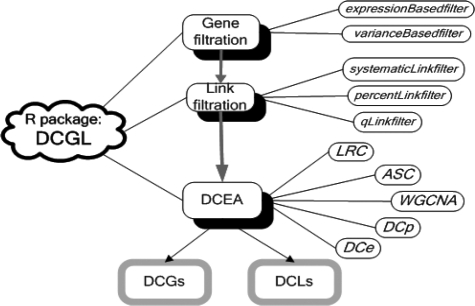
A typical DCEA workflow involves three successive procedures: gene filtration, link filtration, DCG and DCL identification. Correspondingly, DCGL consists of three parts of functions (Fig. 1). For gene filtration, one choice is based on the expression level (expressionBasedfilter) and the other based on its variability (varianceBasedfilter). For link filtration, we provide three functions for cutting off coexpression values (systematicLinkfilter, percentLinkfilter and qLinkfilter). A gene pair (link) is filtered out if both of its coexpression values for two conditions are lower than the cutoff.
Fig. 1.
DCGL design. Function names are shown in italic texts.
The third part, also the core of the package, includes five methods for identifying DCGs and DCLs, which mainly differ in the measure of differential coexpression (dC) of a gene. After the steps of gene filtration and link filtration, suppose gene i is associated with ni links whose coexpression values are projected to X = {xi1, xi2,…, xini}and Y = {yi1, yi2,…, yini} for two conditions. The dC measures of different methods are given in the following equations.
| (1) |
| (2) |
where N and K indicate the numbers of total links and total DCL links in the coexpression network, respectively, and ni and ki indicate the links and DCLs connected to gene i (see Yu,H. et al., submitted for publication).
 |
(3) |
with x′ and y′ are transformed from original x and y values with a ‘soft-thresholding’ strategy (Mason et al., 2009; van Nas et al., 2009).
| (4) |
Link sets C1(i) and C2(i) for two conditions are determined by screening the coexpression values according to a certain threshold.
 |
(5) |
3 IMPLEMENTATION
DCGL is released as an R package including two gene filtering functions, three link filtering functions and five DCEA functions (Fig. 1). These functions generally expect gene expression matrices (with genes in rows and samples in columns) as a major input, and the ultimate output are genes ranked by dC measure or P-value, from which one can obtain a DCG list. DCe has an additional output of classified DCLs. DCGL can be obtained from the supplementary data to this manuscript, or at http://cran.r-project.org/web/packages/DCGL/index.html.
We tested the five DCEA methods using dataset GSE3068 obtained from GEO (Table 1). Note that this test was carried out with the most time-efficient option of link filtration (setting thresholds on coexpression value directly). For the memory analysis, we tested DCp and DCe with the most memory-intensive filtration option ‘qLinkfilter’. We approached a memory limit of around 5.7 GB at a gene total of 7000. So it is anticipated that, if qLinkfilter is evoked, a gene expression matrix generally should undergo a gene filtration step beforehand so that the gene total is cut down to a few thousands or less.
Table 1.
Execution time (in seconds) of five DCEA methods in handling different subsets of GSE3068
| 1000 | 3000 | 5000 | 7000 | 8799 | |
|---|---|---|---|---|---|
| DCp | 1 | 10 | 6 | 50 | 82 |
| DCe | 6 | 38 | 88 | 161 | 257 |
| WGCNA | 1.2 | 9.6 | 26.4 | 51 | 82 |
| ASC | 1.2 | 9.6 | 26.4 | 53 | 86.2 |
| LRC | 1 | 8.4 | 24.6 | 48.8 | 78 |
Different subsets, with a reduced number of rows, were taken from GSE3068 by favoring genes with top-ranked expression variability. The computing platform is a Linux system with five nodes, each having a dual quad-core Intel Xeon 2.33 GHZ CPU and a RAM of 16 GB. Execution time was averaged over five repetitive runs.
4 EXAMPLE
Three simulated datasets are included in the package for exploring the functions. For example, ‘dataC’ gives expression values of 1000 genes in 20 samples divided equally into two groups corresponding to two conditions. Since this dataset contains a moderate number of genes, the gene filtration step can be skipped. The link filtration procedure is wrapped as a sub-function in the DCEA functions, so one can specify the link filtration choice in the arguments of DCEA functions.
If the DCEA function DCe is called, one can get a resulted variable with four components. The gene names ranked by the dC measure (P-value) make up the first ‘$DCGs’ component, while DCLs of different types are given in other three components.
Funding: Shanghai Institutes for Biological Sciences; Chinese Academy of Sciences (2008KIP207); the National ‘973’ Basic Research Program (2006CB0D1203, 2006CB0D1205); the National Natural Science Foundation of China (30770497, 31000380); National Key Technologies R&D Program (2007AA02Z331 and 2009ZX10603).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Choi JK, et al. Differential coexpression analysis using microarray data and its application to human cancer. Bioinformatics. 2005;21:4348–4355. doi: 10.1093/bioinformatics/bti722. [DOI] [PubMed] [Google Scholar]
- Mason MJ, et al. Signed weighted gene co-expression network analysis of transcriptional regulation in murine embryonic stem cells. BMC Genomics. 2009;10:327. doi: 10.1186/1471-2164-10-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverter A, et al. Simultaneous identification of differential gene expression and connectivity in inflammation, adipogenesis and cancer. Bioinformatics. 2006;22:2396–2404. doi: 10.1093/bioinformatics/btl392. [DOI] [PubMed] [Google Scholar]
- van Nas A, et al. Elucidating the role of gonadal hormones in sexually dimorphic gene coexpression networks. Endocrinology. 2009;150:1235–1249. doi: 10.1210/en.2008-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.