Abstract
Purpose
Notch, a type 1 transmembrane protein, plays a key role in the development of many tissues and organ types. Aberrant Notch signaling found in a wide variety of human cancers contributes to tumor development. Since Notch1 was found to be over-expressed in prostate cancer (PCa) cells, and human PCa tissue we, therefore tested our hypothesis that over-expression of Notch1 in PCa promotes tumor invasion.
Experimental design
Notch1 expression was evaluated in human PCa cells and in human PCa tissues. PCa cells were transiently transfected with Notch1 specific siRNAs in concentrations ranging from 30-120 nM and subsequently evaluated for effects on invasion and expression analysis for molecules involved in invasion.
Results
Small interfering RNA mediated knockdown of Notch1 in PCa PC3 and 22Rν1 cells dramatically decreased their invasion. Focused cDNA array revealed that Notch1 knockdown resulted in significant reduction in the expression of urokinase plasminogen activator (uPA) and matrix metalloproteinase (MMP)-9 gene transcripts. These data were further verified by RT-PCR, real- time RT-PCR and immunoblot analysis. Knockdown of Notch1 was also observed to significantly reduce the mRNA expression and protein levels of uPA and its receptor uPAR. A significant reduction in MMP9 expression in Notch1 knockdown cells suggested a role for Notch1 in augmenting MMP9 transcription.
Conclusions
Our data demonstrate involvement of Notch1 in human PCa invasion and that silencing of Notch1 inhibits invasion of human PCa cells by inhibiting the expression of MMP9 and uPA. Thus, targeting of Notch1 could be an effective therapeutic approach against PCa.
Introduction
Notch signaling has been known for decades to developmental biologists as a key player in cell fate determination (1, 2) and tissue homeostasis by maintaining the self-renewal potential of some tissues and inducing differentiation of others (3) including formation of the prostate gland (4). In humans, notch family of transmembrane proteins consists of four receptors Notch1 through Notch4 and five ligands, Jagged1 and 2 and Delta-like ligand (Dll) 1, 3 and 4 (5). Broadly, modular structure of notch consists of an extracellular ligand binding domain, a hydrophobic transmembrane domain and notch intracellular domain (NICD). Notch receptors undergo a series of programmed proteolytic events, first by α-secretase at the extracellular surface, which leads to liberation of the extracellular fragment, and then by intra-membraneous cleavage mediated by γ-secretase. NICD is then released from the inner surface of cell membrane and is translocated into nucleus where it activates transcription of the target genes (6). It has been shown that Notch1 receptor ligand Jagged1 is overexpressed in metastatic human prostatic tissue compared to localized PCa or benign prostatic tissue (7), implicating Notch1 in PCa progression. A recent study has shown that downregulation of Jagged1 inhibits growth of PCa cells (8). Elevated expression of Notch1 was observed in highly metastatic PCa cell lines as compared to normal prostate epithelial cells (9). Notch1 level was reported to be elevated in malignant prostatic epithelial cells of primary and metastatic tumors of transgenic mouse model of PCa (10, 11). Recent studies demonstrate involvement of notch signaling in cancer angiogenesis and metastasis (12-14) however, mechanisms for these effects remain unknown. In the present study, we provide evidence that Notch1 plays an important role in invasiveness of human PCa concomitant with decrease in the expression of matrix metalloproteinase 9 (MMP9) and urokinase plasminogen activator (uPA).
Materials and methods
Cell culture
Human PCa cell lines PC3, DU-145, LNCaP and 22Rν1 were obtained from American type cell culture (Manassas, VA) and cultured in RPMI-1640 medium supplemented with 10% FBS and 1% Penicillin-Streptomycin solution. Prostate epithelial cells (PrEC) and their growth media were procured from Cambrex BioScience (Walkersville, MD). Cells were grown in humidified incubator containing 5% CO2 at 37°C.
Immunohistochemistry
Human prostate tissues were obtained from the Department of Pathology, University of Wisconsin-Madison under an institutional review board approval. Immunohistochemical staining was done using an automated benchmark immunostainer (Ventana Medical Systems, Tucson, AZ). Tissue sections were subjected to antigen retrieval, incubation with specific primary antibody of full length Notch1 (Santa Cruz Biotechnology, Santa Cruz, CA), which is a rabbit polyclonal antibody raised against amino acids 20-150 mapping within an extracellular domain of Notch1 of human origin at a dilution of 1:50, followed by incubation with appropriate horseradish peroxidase conjugated secondary antibody. Immunoreactive complexes were detected using 3, 3′-diaminobenzidene and visualized in a Zeiss-Axiophot DM HT microscope and captured with an attached camera.
Histopathologic grading of PCa specimens
The Gleason system and the WHO grading system were used for evaluation of Notch1 expression in the prostate tissues. Prostate adenocarcinoma was first graded in Gleason patterns 2, 3, and 4. The intensity of immunoperoxidase staining for Notch1 was scored as 0 (negative), 1 (weak), 2 (moderate), and 3 (strong) based on the Gleason patterns. The primary Gleason pattern and the secondary Gleason pattern were added to arrive at a Gleason score, ranging from 6-8. A total of 318 core tissue samples were from 41 patients. Normal glandular tissues, HGPIN, and blood vessels were from the areas adjacent to the cancerous tissue.
Notch1 siRNA transfection
Validated Notch1 specific and scrambled siRNAs were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The Notch1 siRNA contains the pool of three independent sequences viz. 1. (sense 5′-CACCAGUUUGAAUGGUCAAtt-3′; antisence 5′-UUGACCAUUCAAACUGGUGtt-3′) 2. (sense 5′-CCCAUGGUACCAAUCAUGAtt-3′; anti-sence 5′-UCAUGAUUGGUACCAUGGGtt-3′) 3. (sense 5′-CCAUGGUACCAAUCAUGAAtt-3′; antisense 5′-UUCAUGAUUGGUACCAUGGtt-3′). Using electroporation (Amaxa Inc, Gaithersburg, MD) cells were transiently transfected with Notch1 siRNA in concentrations ranging from 30-120 nM and scrambled siRNA (120 nM). Cells were allowed to grow further in CO2 incubator for 24 h and later harvested for further analysis.
Western blot analysis
Forty micrograms of protein resolved over 4-20 % Tris-glycine gels (Invitrogen, Carlsbad, CA) was transferred onto nitrocellulose membranes and probed with appropriate primary antibody against Notch1 (Santa Cruz Biotech, Santa Cruz, CA) and Cleaved Notch1 (Val1744 Cell Signaling Technology, Bavaria, MA). The cleaved Notch1 antibody detects endogenous levels of the cytosolic domain of Notch1 only when cleaved between Gly1743 and Val1744. This antibody does not recognize full length Notch1 or cleaved Notch1 at other positions. Pro-MMP9 and uPA antibodies were purchased from Chemicon International Inc. (Temecula, CA), and uPAR antibody was obtained from R&D systems (Minneapolis, MN). Expression levels of proteins were analyzed as described (15). Densitometric measurements of the bands in Western blot analysis were done using digitalized Scientific Software program, UN-SCAN-IT, purchase from Silk Scientific Corporation (Orem, UT, USA).
Gene expression analysis
Gene expression analysis was performed using pathway focused Human Extracellular Matrix and Adhesion molecules oligo gene array (Superarray, Frederick, MD) containing 96 genes encoding proteins important for the attachment of cells to their surroundings. The array consisted of 96 cDNA in 8×14 grid of tetraspots printed on a nylon membrane. The array was hybridized with total cellular cDNA that was reverse transcribed from total cellular RNA obtained either from PC3 cells transfected with scrambled siRNA or from Notch1 siRNA. After hybridization, membrane was developed as per manufacturer's instructions. Data was analyzed using GEArray Expression Analysis Suite software (Superarray, Frederick, MD).
In vitro chemoinvasion assay
Notch1 siRNA or scrambled siRNA transfected cells were re-suspended in fresh culture medium and incubated in chemoinvasion chamber containing polycarbonate filter coated with Matrigel (Chemicon International, Temecula, CA) for 24 h. In the upper chamber 30,000 cells were seeded in FBS free culture media and the lower chamber contained culture media containing 10% FBS as a chemoattractant. The cells were allowed to migrate for 24 h following which the chamber was washed with PBS and cells visualized as per the manufacturer's instruction. To quantitate the migratory cells, the invasion chamber was dipped in 10% acetic acid, and the resultant solution was spectrophotometrically read at 540 nm according to Vendor's protocol (16).
Gelatin zymography
Equal number of PC3 cells was transiently transfected with Notch1-siRNA and control siRNA for 24 h. The conditioned media were collected, concentrated using amicon filter (Millipore, Billerica, MA) as per manufacturer's protocol and electrophoresed (40 μg protein) under non-reducing condition. The gelatinolytic activity of MMP9 was determined by employing zymography kit (Invitrogen, Carlsbad, CA) as per vendor's protocol.
Luciferase assay
2×106 cells were nucleofected with human MMP9 luciferase reporter plasmid (pGL3-MMP9, 1 μg), a gift from Dr. Dougles D. Boyden (M.D. Anderson Cancer Center, Houston, TX) alongwith 50 ng of renilla luciferase reporter plasmid pRL-TK (Promega, Madison, WI), which was used as an internal control to normalize transfection efficiency and varying concentrations of Notch1 siRNA (30, 60 and 120 nM). In parallel cells were also transfected with empty pGL3 reporter vector and a scrambled siRNA (120 nM) with no validated target sequence in the human genome. After nucleofection, 30,000 cells were distributed per well of a 24 well plate and allowed to grow for another 24 h. Dual luciferase assay reagent kit was procured from Promega, (Madison, WI) and luciferase activity was measured according to the manufacturer's protocol.
MMP9 ELISA assay
24 h post-transfection the cell culture media was collected and used for quantifying MMP9 levels by using MMP9 specific ELISA kit from Amersham Bioscience (Piscataway, NJ) and following the vendor's protocol.
Semi-quantitative polymerase chain reaction (PCR)
PCR reactions were carried out using forward and reverse primer combinations for uPA forward 5′-gtgaagaagggcgtccaaag-3′; reverse 5′-tcggcagtcaatgaggaaagt-3′), uPAR forward 5′tctatccggagcagctgaaaa-3′; reverse 5′-cgtgtagacgcctggcttgt-3′) and GAPDH forward 5′-aatcccatcaccatcttccaggag–3; reverse 5′-gcattgctgatgatcttgaggctg-3). PCR reaction standardization kits were obtained from Epicentre (Madison, WI). The cDNA was amplified with an initial denaturation at 94°C for 2 minutes followed by the sequential cycles of denaturation at 94°C for 45 seconds, annealing at 59°C for 45seconds, and extension at 72°C for 1 minute for 30 cycles, with final extension at 72°C for 7 minutes.
Quantitative RT-PCR
Real time amplification of MMP9, uPA and uPAR was performed from 2 μl cDNA prepared from 2 μg of total RNA. Folowing primers were used: MMP9 forward 5′-atttctgccaggaccgcttctact-3′; reverse 5′-cagtttgtatccggcaaactggct-3′), uPA forward 5′-tcacaccaaggaagagaatggcct-3′; reverse 5′-aatgacaaccagcaagaaagcggg-3′), uPAR forward 5′tgtggctcatcagacatgagctgt-3′; reverse 5′-ttgttgtggaaaccattggagccc-3′) and β-actin forward 5′-atctggcaccacaccttctacaatgagctgcg-3; reverse 5′-cgtcatactcctgcttgctgatccacatctgc-3). PCR reaction standardization kits were obtained from Epicentre (Madison, WI). The cDNA was amplified with an initial denaturation at 95°C for 10 seconds followed by the sequential cycles of denaturation at 94°C for 45 seconds, annealing at 55°C for 10 seconds, and extension at 72°C for 1 minute for 30 cycles, with final extension at 72°C for 7 minutes.
Statistical analysis
All measures were summarized as means±SE. Associations of categoric variables were evaluated using the Fisher's exact test. All tests were two-sided and conducted at the alpha = 0.05 significance level. All statistical analyses were performed with the S-plus, Professional Version 6.2 (Insightful Corp., Seattle, WA) software.
Results
Notch1 is overexpressed in human PCa cells
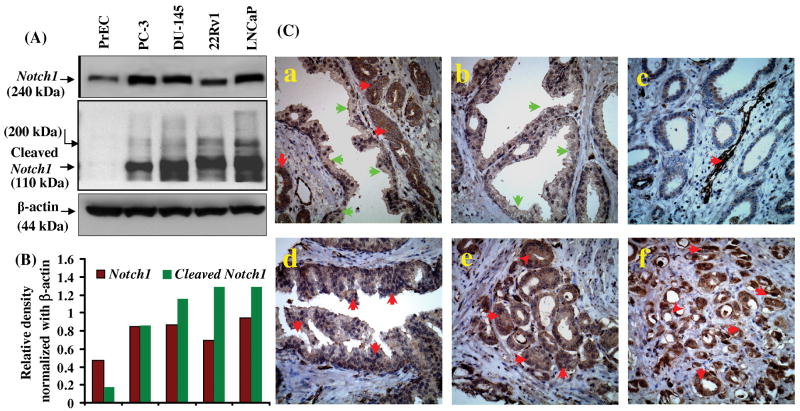
Notch1 has been implicated in many malignancies but its role in prostate carcinogenesis is not well defined. Therefore, we examined the expression of Notch1 in different PCa cell lines and also in normal PrEC. Western blot analysis demonstrated increased expression of Notch1 and cleaved Notch1 in human PCa cell lines. A significantly higher level of Notch1 expression was observed in androgen independent PC3, DU-145 and 22Rν1 cells and androgen-dependent LNCaP cells compared to PrEC (Fig. 1A). Notch1 protein in 22Rν1 cells migrated faster than in other PCa cells which could be a splice variant of the native Notch1 protein.
Figure 1. Expression of Notch1 in normal and prostate cancer cells and in prostate tumor tissue.
(A) Whole cell lysates were prepared and western blot analysis was performed to evaluate the protein levels of Notch1 and cleaved Notch1 in normal PrEC and PCa cell lines PC3, DU-145, LNCaP and 22Rν1. Blots were reprobed with β-actin antibody to analyze the equal loading of proteins. (B) Histogram represents density of bands in A normalized with β actin (C) Representative photomicrographs of prostate tumor biopsy specimen showing immunostaining for Notch1 in (a) cancerous as well as in normal adjacent tissue (b) normal prostate tissue (c) blood vessels in cancerous tissue (d) HGPIN (e) Gleason pattern 3 and (f) Gleason pattern 4. Green arrows indicate normal tissue with none to low staining and red arrows indicate cancerous tissue with moderate to high Notch1 expression.
Notch1 expression increases in human prostate tumor specimens with increasing tumor grade
To further establish the contextual role of Notch1 in PCa, immunohistochemistry was performed in human prostate tumor specimens of normal, high grade prostatic intraepithelial neoplasia (HGPIN) and PCa representing different tumor grades. A total of 318 specimens were examined. We observed enhanced immunoreactivity of Notch1 in tumor cells compared to normal epithelial cells in adjacent areas of the same tissue (Fig. 1C panel a). In addition, enhanced expression of Notch1 was also prominent in the cells surrounding blood capillaries in cancerous tissue (Fig. 1C panel c). Based on the scoring patterns, a significant difference was observed in Notch1 expression between cancer and normal tissues (Table 1A). The staining for Notch1 protein was moderate to strong in PCa specimens as compared to normal prostate specimens which exhibited either none or weak to moderate staining (Table 1A). In a total of 91 specimens of normal tissue obtained from adjacent regions of tumor tissues, the staining for Notch1 was weak in 33 (36%), moderate in 53 (58%) and negative in the remaining 1 (1%) specimens (Table 1A). HGPIN specimens (n=69) showed strong staining for Notch1 in 21 (30%), moderate staining in 41 (60%) and weak staining in the remaining 7 (10%) specimens (Table 1A). The percentage of HGPIN specimens exhibiting strong Notch1 staining was 7- fold higher than that in normal gland. Because HGPIN has been identified as the most significant risk stage for PCa development and the expression of Notch1 protein was found to be significantly increased both in HGPIN and PCa specimens, a strong link could be suggested between the expression of Notch1 protein and development of human PCa. Staining for Notch1 was observed in epithelial cells; however, in stromal cells the staining was either occasional or negative in normal as well as in cancer specimens. The immuno-reactivity of Notch1 was observed in a coarsely granular pattern in the cytoplasm of epithelial cells of normal, HGPIN, and of Gleason pattern 3 (Gleason Score 5-6) to Gleason pattern 4 (Gleason score 7-10) prostatic adenocarcinoma (Fig. 1B panel c-f). Accumulative analysis of all PCa specimens (n=158) suggested higher levels of Notch1, with moderate 67 (42%) to strong staining in 86 (54%) specimens, weak staining in 5 (3%) specimens and no staining in 0 (0%) specimens (Table 1A). These results indicate that Notch1 is overexpressed in human PCa.
Table 1.
| Table 1A. Expression of Notch1 in human normal prostate, HGPIN, and adenocarcinoma specimens. | ||||||
|---|---|---|---|---|---|---|
| Samples | Number | None | Weak | Moderate | Strong | P value |
| Normal glands | 91 | 1 (1%) | 33 (37%) | 53 (58%) | 4 (4%) | - |
| High grade PIN | 69 | 0 (0%) | 7 (10%) | 41 (60%) | 21 (30%) | 0.001* |
| Gleason pattern 3 | 130 | 0 (0%) | 5 (4%) | 57 (44%) | 68 (52%) | 0.001* |
| Gleason pattern 4 | 28 | 0 (0%) | 0 (0%) | 10 (36%) | 18 (64%) | 0.001* |
| Table 1B: List of genes modulated by knockdown of Notch1 in PC3 cells | ||||||
|---|---|---|---|---|---|---|
| List of genes downregulated by specific knockdown of Notch1 | ||||||
| UniGene | Ref Seq # | Symbol | Description | Fold change | ||
| Hs.77274 | NM_002658 | PLAU | Plaminogen activator, urokinase | 34.66 | ||
| Hs.514412 | NM_000442 | PECAM1 | Platelet/endothelial cell adhesion molecule (CD31 antigen) | 18.16 | ||
| Hs.643357 | NM_006988 | ADAMTS1 | ADAM metallopeptidase with thrombospondin type 1 motif, 1 | 11.00 | ||
| Hs.592171 | NM_002214 | ITGB8 | Integrin, beta 8 | 8.68 | ||
| Hs.632226 | NM_000213 | ITGB4 | Integrin, beta 4 | 8.12 | ||
| Hs.74034 | NM_001753 | CAV1 | Caveolin 1, caveolae protein, 22kDa | 6.81 | ||
| Hs.482077 | NM_002203 | ITGA2 | Integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) | 6.46 | ||
| Hs.55279 | NM_002639 | SERPINB5 | Serpin peptidase inhibitor, clade B (ovalbumin), member 5 | 5.02 | ||
| Hs.58488 | NM_003798 | CTNNAL1 | Catenin (cadherin-associated protein), alpha-like 1 | 4.58 | ||
| Hs.643447 | NM_000201 | ICAM1 | Intercellular adhesion molecule 1 (CD54), human rhinovirus receptor | 4.00 | ||
| Hs.133397 | NM_000210 | ITGA6 | Intregrin, alpha 6 | 3.82 | ||
| Hs.466871 | NM_002659 | PLAUR | Plasminogen activator, urokinase receptor | 3.40 | ||
| Hs.609663 | NM_002293 | LAMC1 | Laminin, gamma 1 (formerly LAMB2) | 2.93 | ||
| Hs.371147 | NM_003247 | THBS2 | Thrombospondin 2 | 2.43 | ||
| Hs.297413 | NM_004994 | MMP9 | Matrix metallopeptidase 9 gelatinase B, 92kDa gelatinase, 92kDA type IV collagenase | 4.20 | ||
| Hs.159581 | NM_016155 | MMP17 | Matrix metallopeptidase 17 (membrane-inserted) | 2.20 | ||
| Hs.411312 | NM_000419 | ITGA2B | Integrin alpha 2b (platelet glycoprotein llb of llb/llla complex, antigen CD41) | 2.08 | ||
| List of genes upregulated by specific knockdown of Notch1 | ||||||
| Hs.313 | NM_000582 | SPP1 | Secreted phosphoprotein 1 (osteopontin, bone sialoprotein I, early T-lymphocyte activator) | 9.07 | ||
| Hs.73800 | NM_003005 | SELP | Selectin P (granule membrane protein 140kDa, antigen CD62) | 6.66 | ||
| Hs.21422 | NM_005010 | NRCAM | Neuronal cell adhesion molecule | 5.07 | ||
| Hs.567417 | NM_006690 | MMP24 | Matrix metallopeptidase 24 (membrane-inserted) | 5.01 | ||
| Hs.300774 | NM_005141 | FGB | Fibrinogen beta chain | 4.38 | ||
| Hs.375129 | NM_002422 | MMP3 | Matrix metallopeptidase 3 (stromelysin 1, progelatinase) | 3.29 | ||
| Hs.652397 | NM_00362 | TIMP3 | TIMP metallopeptidase inhibitor 3 (Sorbsy fundus dystrophy, pseudoinflammatory) | 3.21 | ||
| Hs.2936 | NM_002427 | MMP13 | Matrix metallopeptidase 13 (collagenase 3) | 3.10 | ||
| Hs.161985 | NM_019894 | TMPRSS4 | Transmembrane protease, serine 4 | 2.67 | ||
| Hs.143434 | NM_001843 | CNTN1 | Contactin 1 | 2.64 | ||
| Hs.204732 | NM_021801 | MMP26 | Matrix metallopeptidase 26 | 2.60 | ||
| Hs.461086 | NM_004360 | CDH1 | Cadherin 1, type 1, E-cadherin (epithelial) | 2.50 | ||
| Hs.445981 | NM_001903 | CTNNA1 | Catenin (cadherin-associated protein), alpha 1, 102kDa | 2.16 | ||
Note. The expression of Notch-1 was evaluated as staining of the tissue as none (0), weak (1), moderate (2), and strong (3). Fisher exact test was used to examine the association between staining intensity and tissue type or staining intensity and tumor grade (for tumor specimen only).
P<0.001 was considered as significant.
Notch1 knockdown decreases cell invasion
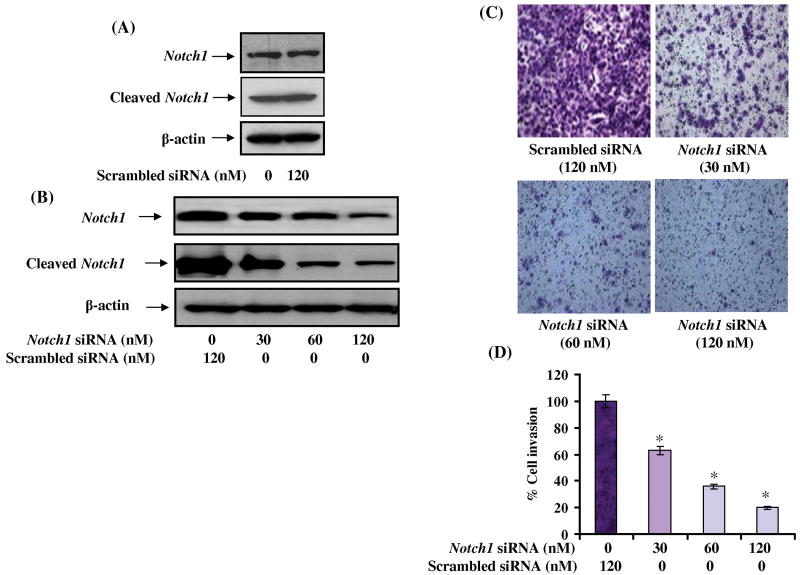
In order to address the role of Notch1 in PCa invasion, knockdown of Notch1 was achieved by transfecting PC3 cells with 3 independent pools of Notch1 specific siRNA compared to scrambled siRNA where no effect on Notch1 and cleaved Notch1 expression was observed (Fig. 2A). Since we had hypothesized that Notch1 is involved in prostate cancer invasion, and the fact that PC3 cells represent advanced metastatic cancer, prompted us to select this particular cell line for our studies. Notch1 siRNA dose-dependently decreased both Notch1 and cleaved Notch1 expression with maximum effect observed at a concentration of 120 nM 24 h post-transfection (Fig. 2B). To further demonstrate the effect of Notch1 knockdown, PC3 cells were subjected to invasion assay. Notch1 knockdown cells showed only a marginal invasion through the extracellular matrix compared to cells transfected with non-specific siRNA (Fig. 2C). Notch1 knockdown caused 40-80% decrease in cell invasion (Fig. 2D) suggesting an essential role of Notch1 in conferring invasive properties to PCa cells.
Figure 2. Effect of Notch1 knockdown on invasive behavior of PC3 cells.
(A) Notch1 expression in control and scrambled siRNA transfected cells. (B) Effect of Notch1 siRNA on the expression of Notch1 and cleaved Notch1. (C) Notch1 expression was knocked down in PC3 cells using Notch1 specific siRNA and subjected to invasion assay employing two-chambered invasion apparatus as described under materials and methods. The photomicrograph shows the number of migratory cells transfected with varying concentrations of Notch1 specific siRNA. (D) Histogram showing percent inhibition of PC3 cell invasion. The experiment was performed in triplicate and the value obtained from scrambled siRNA transfected cells was set as 100%.
cDNA array identifies decreased transcripts of genes implicated in tumor invasion
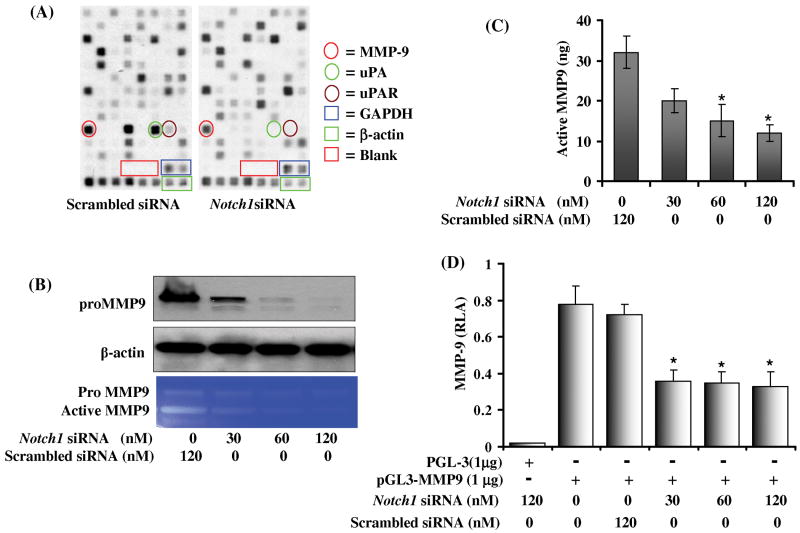
To define the role of Notch1 in PC3 cell invasion, a specifically designed cDNA gene array was employed. This array contained 96 cDNA fragments specifying genes with role in cell invasion and metastasis. A significant difference in gene expression of CD31, MMP9, uPA and uPAR (Fig. 3A) was observed in Notch1 knockdown cells. To ascertain fold changes between scrambled and Notch1 siRNA knockdown cells, we performed a detailed analysis of gene expression profile by clustergram and scatter plot indicated a 35-fold downregulation in the expression of uPA, 18-fold in CD31 and > 3-fold in the expression of MMP9 and uPAR (Table 1B). These results indicate that Notch1 regulates the expression of these downstream target genes which are involved in extracellular matrix degradation, suggesting that Notch1 might play a determining role in cell invasion.
Figure 3. Effect of Notch1 knockdown in PC3 cells on the expression of genes involved in extracellular matrix degradation and cell adhesion.
(A) Autoradiographic image of a cDNA array from scrambled siRNA transfected control (Left) and Notch1 knockdown cells (Right). Enriched tetraspots indicate the position of genes. Red encircle MMP9; green encircle uPA and brown encircle uPAR. (B) Western blot analysis to evaluate pro MMP9 protein expression in Notch1 knockdown cells. Blot was reprobed with β-actin antibody to analyze the equal loading of proteins. (C) Gelatin zymogram showing activity of pro MMP9 and active MMP9 in scrambled and Notch1 siRNA transfected cells. (D) Quantification of MMP9 secretion using MMP9 specific ELISA performed in the culture media of PC3 cells transfected either with scrambled or Notch1 specific siRNA. (E) Effect of Notch1 knockdown on the promoter activity of MMP9 gene expression in PC3 cells. 2×106 PC3 cells cotransfected with either scrambled or Notch1 specific siRNA along with 1 μg of pGL3 or 1μg MMP9 luciferase reporter plasmid and 50 ng of renilla luciferase reporter plasmid as an internal control as described in material and method section. The experiment was performed in quadruplet and the values are showing mean ± SD. Asterisk (*) represents p<0.01 as significant.
Notch1 knockdown decreases MMP9 expression
To further validate the results of microarray analysis, Notch1 knocked down cells were subjected to western blot analysis. We observed a decrease in pro MMP9 protein levels (Fig. 3B), suggesting that Notch1 may be involved in MMP9 activation either by enhancing its expression or by stabilizing the protein. Gelatin zymography was performed to assess MMP9 activity in cultured medium from Notch1 knockdown cells and we observed a significant decrease in MMP9 activity (Fig. 3B). To further strengthen our findings, MMP9 specific ELISA was performed to quantify secreted MMP9 protein levels. Results indicated a consistent decrease in MMP9 secretion compared with scrambled control (Fig. 3C) further emphasizing the role of Notch1 in regulation of MMP9. To further elucidate the involvement of Notch1 in MMP9 expression, luciferase assay was performed to evaluate the effect of Notch1 on MMP9 promoter activity. Notch1 knockdown cells showed a marked 2-fold decrease in reporter activity (P<0.01) indicating the involvement of Notch1 in MMP9 expression (Fig. 3D). Surprisingly, the decrease observed in MMP9 promoter activity was not dose-dependent. This suggests that although Notch1 is involved in MMP9 transcription, it may not be an exclusive mechanism which regulates MMP9 expression and indicates the existence of a post-translational stabilization mechanism. However, real time RT-PCR data showed a dose-dependent decrease in mRNA expression of MMP9 in Notch1 knockdown cells which further confirms our micro-array data. (supplementary figure 1).
Notch1 knockdown decreases the expression of uPA and its receptor uPAR
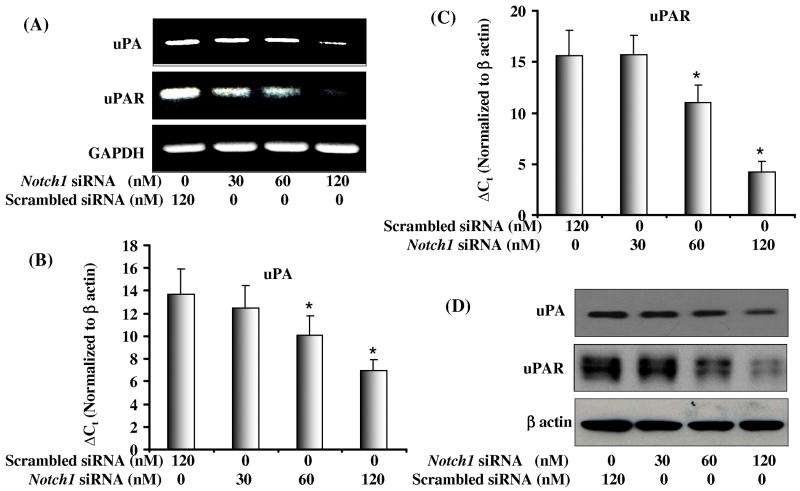
uPA is another marker for invasion and metastasis and has been shown to be upregulated in many malignancies including PCa. uPA regulates the conversion of pro-MMPs to their active forms, which are involved in extracellular matrix degradation. RT-PCR and real time RT-PCR were performed to confirm the micro-array data which showed downregulation of uPA and uPAR transcripts in Notch1 knockdown cells (Fig. 4 A-C). We also observed that knockdown of Notch1 in PC3 cells inhibited protein levels of the ligand uPA and its receptor uPAR (Fig. 4D). These data were in direct agreement with the micro-array data and suggest the involvement of Notch1 in regulation of uPA and uPAR.
Figure 4. Effect of Notch1 knockdown on mRNA and protein expressions of uPA and uPAR.
(A) Semiquantitative RT-PCR analysis shows mRNA expression of uPA and uPAR in cells transfected with increasing concentration of Notch1 siRNA. Scrambled siRNA was used as control in parallel. Lower panel showing GAPDH as an internal control. (B) Western imunoblot analysis of uPAR and uPA expression in PC3 cells transfected with increasing concentration of Notch1 specific siRNA. Blots were reprobed with β-actin antibody to analyze the equal loading of proteins. (C-D) Real time RT-PCR analysis showing mRNA expression of uPA and uPAR.
Notch1 knockdown decreases invasion and inhibits the expression of MMP9, uPA and uPAR in another human PCa 22Rν1 cell
To ascertain whether Notch1 knockdown produces similar effects in other PCa cells, we selected an androgen sensitive 22Rν1 cells and transiently transfected with scrambled siRNA and specific Notch1 siRNA. Knockdown of Notch1 in 22Rν1 cells significantly decreased protein expression of Notch1 and cleaved Notch1 (Fig. 5A), invasion of cells through the extracellular matrix (Fig. 5B), uPA expression (Fig. 5C), and MMP9 promoter activity (Fig. 5D) compared to cells transfected with non-specific siRNA. RT-PCR and real time RT-PCR were performed and showed downregulation of MMP9, uPA and uPAR transcripts in Notch1 knockdown cells (supplementary figure 5 A-C). We also observed that knockdown of Notch1 in 22Rν1 cells inhibited protein levels of the ligand uPA and its receptor uPAR (supplementary figure 5 D). These data suggest that effect of Notch1 knockdown in PCa cells invasion could be comprehensive as we observed similar mechanism in both the PCa androgen sensitive 22Rν1 as well as androgen insensitive PC3 cells.
Figure 5. Effect of Notch1 knockdown on invasion, mRNA expression of uPA, and MMP9 activity in 22Rν1 cells.
(A) Protein level of Notch1 and cleaved Notch1 in control, scrambled siRNA and Notch1 siRNA transfected 22Rν1 cells as determined by Western blot analysis. (B) The photomicrograph shows the number of migratory cells transfected with varying concentrations of Notch1 specific siRNA and scrambled siRNA, Lower panel showing quantification of migratory cells in scrambled and Notch1 specific siRNA transfected cells. The experiment was performed in triplicate and the value obtained from scrambled siRNA transfected cells was set as 100% migration. (C) Semiquantitative RT-PCR analysis shows mRNA expression of uPA in cells transfected with increasing concentration of Notch1 siRNA. Scrambled siRNA was used as control in parallel. Lower panel showing GAPDH as an internal control. (D) Effect of Notch1 knockdown on the promoter activity of MMP9 gene expression in PC3 cells. 2×106 22Rν1 cells cotransfected with either scrambled or Notch1 specific siRNA along with 1 μg of pGL3 or 1μg MMP9 luciferase reporter plasmid and 50 ng of renilla luciferase reporter plasmid as an internal control as described in material and method section. The experiment was performed in quadruplet and the values are showing mean ± SD. Asterisk (*) represents p<0.01 as significant.
Discussion
Aberrant expression of Notch1 has been detected in various types of human cancers including T-cell acute lymphoblast leukemia (17), breast carcinoma (18) and brain tumor (19). While some studies have shown the involvement of Notch1 in cancer progression, others have suggested anti-proliferative effect of Notch1 in some cancer types (3). Notch1 has also been reported to be overexpressed in malignant phenotype including moderately differentiated adenocarcinoma of TRAMP mice. (11). In spite of these studies, the role of Notch1 in prostate carcinogenesis remains poorly understood. In this study, we report overexpression of Notch1 in PCa cell lines consistent with previous findings of Zayzafoon et al (20). We also observed significantly elevated expression of Notch1 in human PCa tissues. Notch1 expression increased with increasing tumor grade with specimens of Gleason pattern 3 and 4 exhibiting significantly higher percentage of strong expression. An interesting observation was a significant induction of Notch1 in vascular endothelial cells of these tissues, consistent with earlier reports (21), suggesting that Notch1 may facilitate angiogenesis of PCa cells to neighboring and distant organs. We observed that targeted disruption of Notch1 in PC3 cells resulted in significant decrease in cell invasion across artificial matrix which mimics in vivo extracellular matrix. In cDNA array we observed significant modulation of genes which are involved in cell invasion and angiogenesis. We observed significant decrease in the expression of CD31, uPA, uPAR and MMP9. MMPs have been shown to be involved in extracellular matrix degradation and are overexpressed in advanced stage of PCa. We examined the functional activity of MMP9 in Notch1 knockdown cells and observed concentration dependent decrease in the MMP9 expression and activity indicating that Notch1 might directly regulate the expression of MMP9 via enhancing its promoter activity. Intracellular domain of Notch 1 vests a transactivation function and it is possible that it might help recruit transcriptional machinery to MMP9 regulatory element to enhance its expression. Although, a progressive decrease in Notch1 was observed with increasing concentration of Notch1 siRNA, the decrease in MMP9 promoter activity was not concentration-dependent in contrast to MMP9 protein expression, which rules in the possibility for the existence of a post-translational stabilization mechanism. We suggest that Notch1 controls MMP9 expression either directly by enhancing its transcriptional activity where it functions as transcription factor. Our data correspond to a previous report by Wang et al (22), which showed the regulation of MMP9 via Notch1 in pancreatic cancer cells. An additional mechanism that is commonly involved in promoting tumor cell invasion is uPA-uPAR system, which is one of the most frequent alterations observed in invasive type of PCa (23). We also observed decrease in uPA and uPAR expression in Notch1 knockdown cells suggesting that Notch1 can also regulate the expression of uPA and its receptor via a distinct gene expression mechanism. These findings suggest that Notch1 is involved in invasion and metastasis of PCa by regulating the expression of MMPs in addition to uPA-uPAR system, which might work in synergy to enhance tumor cell invasion and metastasis. In order to illustrate broader relevance of our hyopothesis we performed additional studies on androgen sensitive human PCa 22Rν1 cells, which also showed significantly higher expression of Notch1. Our results indicate that the disruption of Notch1 in these cells also leads to decrease in their invasiveness accompanied with the decrease in MMP9 and uPA expression and suggest the versatile role of Notch1 in PCa cell invasion. We suggest that Notch1 could be a target for intervention of human PCa.
Statement of Clinical Relevance.
In this study we have analyzed human prostate cancer cells and tissues for the expression of Notch1. Based on the experimental studies performed using specific depletion of Notch1 protein in PCa cells, it becomes evident that Notch1 is in fact overexpressed in PCa cells where it promotes cell invasion by a mechanism involving increased expression of MMP9 and uPA and its receptor uPAR. Extensive histochemical analyses involving both normal and prostate tumor tissue was undertaken to screen for Notch1 protein expression. In light of these findings our data strongly suggest that Notch1 protein is significantly overexpressed in a large cohort of PCa tissues as compared to normal human prostate tissue. Moreover, the expression was significantly concentrated in tumor areas situated near vasculature, which further indicates that Notch1 could augment prostate tumor cells invasion and metastasis. In summary, not only these findings suggest that Notch1 could potentially serve as a strong surrogate marker for PCa diagnoses but could also be developed to help screen for patients with high propensity for PCa metastasis. In addition, developing novel strategies to inhibit Notch1 signaling could pave way to effectively target PCa.
Supplementary Material
Real time RT-PCR showing the mRNA expression of MMP9 in PC3 cells transfected with specified concentration of scrambled and Notch1 specific siRNA. Data represents mean ± SD. * P<0.05.
(A-C) Real time RT-PCR showing the mRNA expression of MMP9, uPA and uPAR in PC3 cells transfected with specified concentration of scrambled and Notch1 specific siRNA. Data represents mean ± SD. * P<0.05. (D) Western imunoblot analysis of pro-MMP9, uPAR and uPA expression in 22Rn1 cells transfected with increasing concentration of Notch1 specific siRNA. Blots were reprobed with β-actin antibody to analyze the equal loading of proteins.
Acknowledgments
This work was supported by United States PHS grants RO1 CA 78809, RO1 CA 101039 and RO1CA120451. We thank Dr. Dougles D. Boyden (MD Anderson Cancer Center, Houston, TX) for providing pGL3-MMP9-luc reporter plasmid.
Abbreviations
- PCa
Prostate cancer
- siRNA
small interfering RNA
- MMP
Matrix metalloproteinase
- TIMP
Tissue inhibitor of matrix metalloproteinase
- ECM
Extra cellular matrix
- uPA
urokinase-type plasminogn actvator
References
- 1.Greenwald I. LIN-12/Notch signaling: lessons from worms and flies. Genes Dev. 1998;12:1751–62. doi: 10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–6. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 3.Radtke F, Raj K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nature Rev Cancer. 2003;3:756–67. doi: 10.1038/nrc1186. [DOI] [PubMed] [Google Scholar]
- 4.Wang XD, Shou J, Wong P, French DM, Gao WQ. The application of DNA sequencing in studying haplotypes of PAI-1 gene in patients with coronary artery disease. J Biol Chem. 2004;279:24733–44. [Google Scholar]
- 5.Leong KG, Karsan A. Recent insights into the role of Notch signaling in tumorigenesis. Blood. 2006;107:2223–33. doi: 10.1182/blood-2005-08-3329. [DOI] [PubMed] [Google Scholar]
- 6.Shih IeM, Wang TL. Notch signaling, gamma-secretase inhibitors, and cancer therapy. Cancer Res. 2007;5:1879–82. doi: 10.1158/0008-5472.CAN-06-3958. [DOI] [PubMed] [Google Scholar]
- 7.Santagata S, Demichelis F, Riva A, Varambally S, Hofer MD, Kutok JL, Kim R, Tang J, Montie JE, Chinnaiyan AM, Rubin MA, Aster JC. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res. 2004;64:6854–57. doi: 10.1158/0008-5472.CAN-04-2500. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Wang Z, Ahmed F, Banerjee S, Li Y, Sarkar FH. Downregulation of Jagged1 induces cell growth inhibition and S phase arrest in prostate cancer cells. Int J Cancer. 2006;119:2071–77. doi: 10.1002/ijc.22077. [DOI] [PubMed] [Google Scholar]
- 9.Scorey N, Fraser SP, Patel P, Pridgeon C, Dallman MJ, Djamgoz MB. Notch signalling and voltage-gated Na+ channel activity in human prostate cancer cells: independent modulation of in vitro motility. Prostate Cancer Prostatic Dis. 2006;9:399–406. doi: 10.1038/sj.pcan.4500894. [DOI] [PubMed] [Google Scholar]
- 10.Gipp J, Gu G, Crylen C, Kasper S, Bushman W. Hedgehog pathway activity in the LADY prostate tumor model. Mol Cancer. 2007;6:19. doi: 10.1186/1476-4598-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shou J, Ross S, Koeppen H, de Sauvage FJ, Gao WQ. Dynamics of notch expression during murine prostate development and tumorigenesis. Cancer Res. 2001;6:7291–97. [PubMed] [Google Scholar]
- 12.Zeng Q, Li S, Chepeha DB, Giordano TJ, Li J, Zhang H, Polverini PJ, Nor J, Kitajewski J, Wang CY. Crosstalk between tumor and endothelial cells promotes tumor angiogenesis by MAPK activation of Notch signaling. Cancer Cell. 2005;1:13–23. doi: 10.1016/j.ccr.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Hellström M, Phng LK, Hofmann JJ, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–80. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- 14.Noguera-Troise I, Daly C, Papadopoulos NJ, et al. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature. 2006 Dec 21;444(7122):1032–7. doi: 10.1038/nature05355. [DOI] [PubMed] [Google Scholar]
- 15.Saleem M, Adhami VM, Ahmad N, Gupta S, Mukhtar H. Prognostic significance of metastasis-associated protein S100A4 (Mts1) in prostate cancer progression and chemoprevention regimens in an autochthonous mouse model. Clin Cancer Res. 2005;11:147–53. [PubMed] [Google Scholar]
- 16.Pan Q, Bao LW, Merajver SD. Tetrathiomolybdate inhibits angiogenesis and metastasis through suppression of the NFkappaB signaling cascade. Mol Cancer Res. 2003;1:701–6. [PubMed] [Google Scholar]
- 17.Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–61. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 18.Stylianou S, Clarke RB, Brennan K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006;66:1517–25. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- 19.Dang L, Fan X, Chaudhy A, Wang M, Gaiano N, Eberhart CG. Notch3 signaling initiates choroid plexus tumor formation. Oncogene. 2006;25:487–91. doi: 10.1038/sj.onc.1209074. [DOI] [PubMed] [Google Scholar]
- 20.Zayzafoon M, Abdulkadir SA, McDonald MJ. Notch signaling and ERK activation are important for the osteomimetic properties of prostate cancer bone metastatic cell lines. J Biol Chem. 2004;279:3662–70. doi: 10.1074/jbc.M308158200. [DOI] [PubMed] [Google Scholar]
- 21.Taylor KL, Henderson AM, Hughes CC. Notch activation during endothelial cell network formation in vitro targets the basic HLH transcription factor HESR-1 and downregulates VEGFR-2/KDR expression. Microvasc Res. 2002;64:372–83. doi: 10.1006/mvre.2002.2443. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Banerjee S, Li Y, Rahman KM, Zhang Y, Sarkar FH. Downregulation of notch-1 inhibits invasion by inactivation of nuclear factor-kappaB, vascular endothelial growth factor, and matrix metalloproteinase-9 in pancreatic cancer cells. Cancer Res. 2006;66:2778–84. doi: 10.1158/0008-5472.CAN-05-4281. [DOI] [PubMed] [Google Scholar]
- 23.Pulukuri SM, Gondi CS, Lakka SS, Jutla A, Estes N, Gujrati M, Rao JS. RNA interference-directed knockdown of urokinase plasminogen activator and urokinase plasminogen activator receptor inhibits prostate cancer cell invasion, survival, and tumorigenicity in vivo. J Biol Chem. 2005;280:36529–40. doi: 10.1074/jbc.M503111200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Real time RT-PCR showing the mRNA expression of MMP9 in PC3 cells transfected with specified concentration of scrambled and Notch1 specific siRNA. Data represents mean ± SD. * P<0.05.
(A-C) Real time RT-PCR showing the mRNA expression of MMP9, uPA and uPAR in PC3 cells transfected with specified concentration of scrambled and Notch1 specific siRNA. Data represents mean ± SD. * P<0.05. (D) Western imunoblot analysis of pro-MMP9, uPAR and uPA expression in 22Rn1 cells transfected with increasing concentration of Notch1 specific siRNA. Blots were reprobed with β-actin antibody to analyze the equal loading of proteins.