Abstract
Transcriptional networks orchestrate complex developmental processes. Such networks are commonly instigated by master regulators of development. Considerable progress has been made in elucidating GATA factor-dependent genetic networks that control blood cell development. GATA-2 is required for the genesis and/or function of hematopoietic stem cells, whereas GATA-1 drives the differentiation of hematopoietic progenitors into a subset of the blood cell lineages. GATA-1 directly represses Gata2 transcription, and this involves GATA-1-mediated displacement of GATA-2 from chromatin, a process termed a GATA switch. GATA switches occur at numerous loci with critical functions, indicating that they are widely utilized developmental control tools.
Keywords: Chromatin Regulation, Erythrocyte, Hematopoiesis, Transcription Enhancers, Transcription Regulation, Erythropoiesis
Introduction
The actions of diverse signaling and regulatory factors must be intricately interwoven to ensure the flawless execution of complex developmental processes. Major efforts are ongoing to dissect how such networks are established and remodeled. This is particularly challenging given the large number of variables involved, as a complete inventory of relevant signals/factors is often unavailable. Furthermore, the necessity to consider the dynamics of the underlying mechanisms creates formidable obstacles. With the major progress in dissecting mechanisms underlying hematopoiesis, in which stem cells differentiate into multipotent progenitors, lineage-committed progenitors, and ultimately diverse blood cell types (Fig. 1), this is a particularly attractive system for addressing these issues.
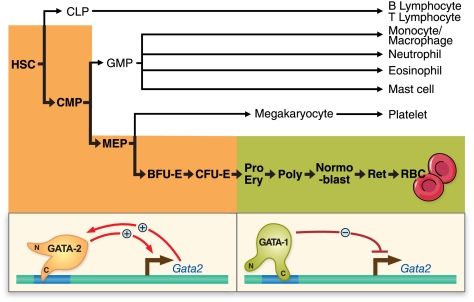
FIGURE 1.
GATA-1 directly represses the Gata2 locus during hematopoiesis. GATA-2 occupies multiple regulatory regions (GATA switch sites) at its own locus (71–73). Upon induction of GATA-1 expression, GATA-1 displaces GATA-2 (GATA switch), instigating transcriptional repression. Although the GATA switch is depicted to occur in the erythroid lineage, GATA-1 is also expressed in megakaryocytes, eosinophils, and mast cells, but mechanistic analyses of GATA switches are incomplete in these systems. CLP, common lymphoid progenitor; GMP, granulocyte-monocyte progenitor; BFU-E, burst-forming unit-erythroid; CFU-E, colony-forming unit-erythroid; CMP, common myeloid progenitor; MEP, megakaryocyte-erythroid progenitor; Pro Ery, proerythroblast; Poly, polychromatic normoblast; Ret, reticulocyte.
Hematopoiesis is controlled by numerous transcription and signaling factors with tightly integrated functions (1). Foundational studies defined the involvement of members of a family of developmental regulators, the GATA transcription factors (2–6). GATA-1, GATA-2, and GATA-3 are termed the hematopoietic GATA factors, based on their important activities to control distinct and overlapping aspects of hematopoiesis (7). GATA-1 is expressed in erythroid precursors, megakaryocytes, eosinophils, and mast cells, as well as testis (3, 4, 8–10). GATA-2 is expressed in hematopoietic stem cells (HSCs),3 multipotent hematopoietic progenitors, erythroid precursors, megakaryocytes, eosinophils, and mast cells (11–13). Beyond the hematopoietic system, GATA-2 is expressed in differentiated endothelial cells (5, 14) and in specific neurons (15–17). GATA-3 is expressed in HSCs, T-lymphocytes, neurons, kidney, and mammary gland (18–20). GATA-4, GATA-5, and GATA-6 are expressed in endoderm- and mesoderm-derived tissues and control such critical processes as heart and lung development (21, 22). Analogous to their shared and unique biological roles, GATA factors have both common and distinct biochemical attributes.
GATA Factor-dependent Transcriptional Control
Foundational Studies
GATA factors have a characteristic dual zinc finger module that is highly conserved among the six mammalian proteins (23). The C-terminal finger mediates sequence-specific DNA binding (24–26), whereas the N-terminal finger modulates DNA binding and contacts DNA with sequence specificity in certain contexts (Fig. 2) (24, 27, 28). The zinc fingers interact with multiple coregulators, which are discussed below. By contrast to the highly conserved fingers, GATA factor N and C termini are divergent. The N and C termini confer differential stabilities to GATA-1 and GATA-2, with GATA-1 being more stable than GATA-2 (29). GATA-1, but not GATA-2, contains sequences within the N terminus that bind the retinoblastoma protein (30). The GATA-1 N terminus is differentially required for the control of distinct target genes (31). Within the GATA-1 N terminus is a sumoylation site (Lys137) (32) that facilitates GATA-1-mediated activation and repression of select target genes (33). However, many questions remain unanswered regarding how the N and C termini of different GATA factors function.
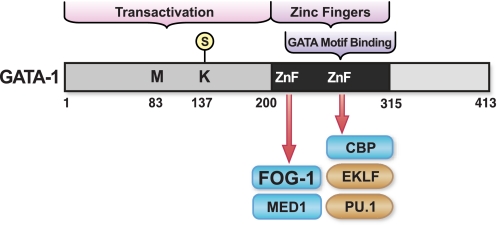
FIGURE 2.
Functional attributes of GATA-1. Murine GATA-1 sequences within its N terminus are implicated in conferring transactivation. These sequences include the sumoylation site Lys137 (32), which facilitates GATA-1 activity in a context-dependent manner (33). Met83 represents an alternative translation start site that yields a leukemogenic GATA-1 protein in acute megakaryoblastic leukemia (87). As described in the text, the C-terminal finger mediates binding to GATA motifs, whereas the N-terminal finger stabilizes DNA binding and mediates sequence-specific DNA binding in certain contexts. The zinc fingers (ZnF) mediate multiple protein/protein interactions, with the FOG-1 interaction with the N-terminal finger being particularly critical. EKLF, erythroid Krüppel-like factor.
The molecular principles underlying GATA factor function have emerged largely from studies of GATA-1 and GATA-2. Pioneering work by Evans and Felsenfeld identified a common DNA motif (WGATAR) present at cis-regulatory elements that activate transcription in an erythroid cell-specific manner (2). These studies laid the foundation for the cloning of GATA-1, the founding member of the GATA factor family (3, 4). Elegant functional analyses utilizing knock-out ES cells and mutant mouse strains unequivocally established a role for GATA-1 in controlling red blood cell, megakaryocyte, mast cell, and eosinophil development (34–41). The cloning of GATA-1 ushered in the discovery of additional GATA factor family members.
Coregulator Usage
Having established the GATA factor family and implicated GATA factors as critical regulators of specific developmental processes, a paramount question was whether they function through canonical transcriptional mechanisms, or if their mechanisms differ fundamentally from conventional transcription factors. Yeast two-hybrid screening revealed a cell type-restricted coregulator designated FOG-1 (friend of GATA-1), which, like GATA-1, is required for erythropoiesis and megakaryopoiesis (42, 43). Using a novel mammalian suppressor mutation approach, the functional importance of the GATA-1/FOG-1 interaction was established (44). Even mutation of a single residue (Val205) in the GATA-1 N-terminal zinc finger sufficed to reduce FOG-1 binding to GATA-1, and strikingly, Val205 mutations characterize a human hematopoietic disorder, familial dyserythropoietic anemia and thrombocytopenia (45). Although numerous GATA-1 target genes are activated or repressed via a FOG-1-dependent mechanism (44), several have been shown to be activated or repressed without FOG-1 (33, 46, 47).
Although FOG-1 has nine zinc fingers, they have not been shown to have intrinsic DNA binding activity. At least one of their functions is to mediate GATA-1 binding, as four fingers contact the GATA-1 N-terminal finger (48, 49). FOG-1 facilitates GATA-1 occupancy of a subset of its chromatin target sites (50, 51) and GATA-1-dependent chromatin looping (52). Because GATA-1-instigated looping requires GATA-1 chromatin occupancy, FOG-1-induced looping may be secondary to its facilitation of GATA-1 chromatin occupancy or may represent a novel pro-looping activity. In aggregate, it appears that GATA-1 utilizes a non-canonical transcriptional mechanism. However, FOG-1 contains an N-terminal sequence that binds the NuRD chromatin-remodeling complex (53), raising the possibility that FOG-1 also functions via a conventional mechanism to mediate recruitment of chromatin-remodeling and/or chromatin-modifying components. Analyses of knock-in mice lacking this NuRD-binding region indicated its importance for regulating erythropoiesis and megakaryopoiesis (54, 55). The mutant mice are viable, however, and defects are not as pronounced as those of GATA-1- and FOG-1-null mice. FOG-1 also contains a sequence that binds the corepressor CtBP (56), which associates with the histone demethylase LSD1 (lysine-specific demethylase 1) (57). A knock-in mouse bearing a deletion of the CtBP-binding sequence lacks a phenotype (58), although the CtBP interaction was not completely abolished by the mutation (59). Because FOG-1 translational isoforms differentially bind CtBP (59), a further assessment of the functional importance of this interaction is warranted.
Besides FOG-1 binding, the zinc finger module of GATA-1 binds the histone acetyltransferase CBP/p300 (60), which commonly mediates trans-acting factor function (61), and the Mediator complex subunit MED1 (62). GATA-1 recruits CBP/p300 (47, 63, 64), MED1 (65), and the chromatin remodeler BRG1 (66) to chromatin sites in a context-dependent manner. Unlike FOG-1 binding, the molecular underpinnings of these interactions are unresolved. Studies with BRG1 knock-out mice indicate that BRG1 mediates chromatin looping at the β- and α-globin loci and confers maximal transcriptional activity of the adult globin genes (47, 66–68). Knockdown studies suggest that MED1 modestly contributes to GATA-1 activity at endogenous loci (65), whereas the relative importance of CBP/p300 binding for GATA-1 function in vivo remains unclear.
Negotiating Chromatin
As literally millions of WGATAR motifs reside in complex genomes, this raises the question of whether GATA-1 occupies the majority or minority of these sites in vivo (69). With the development of ChIP assays to measure GATA-1 and GATA-2 chromatin occupancy, it became clear that only a small subset of the WGATAR motifs (even when evolutionarily conserved) are occupied in cells (70–73). Recent genome-wide analyses confirmed that GATA-1 (74–76) and GATA-2 (74) discriminate exquisitely among highly abundant WGATAR motifs in genomes. In erythroid precursor cells, GATA-1 and GATA-2 share many chromatin sites, although GATA-1- and GATA-2-selective sites also exist (74). GATA-1/2 occupancy sites commonly reside distal to promoters, indicative of a canonical long-range mechanism, although a minority (∼10%) of the sites are promoter-localized (74). De novo motif finding with GATA-1 genomic data sets led to the refinement of the GATA-1-binding sequence WGATAR to yield (CG)(AT)GATAA(GAC)(GAC) (74). Although GATA-1 occupies a significantly greater percentage of these extended versus WGATAR motifs, the percent of occupied motifs is still low, indicating the importance of parameters distinct from the motif itself.
What additional mechanisms facilitate and/or preclude GATA factor chromatin occupancy? FOG-1 facilitates GATA-1 chromatin occupancy (50, 51), but this mechanism is operational only at certain sites. Another parameter likely to be important is nearest-neighbor cis-elements. Genomic analyses revealed enrichments of several cis-elements at GATA-1 occupancy sites (74, 77), albeit the sequences lacked strong predictive power. Finally, the chromatin environment in which GATA motifs reside is almost certainly critical. The epigenetic landscape at GATA-2-occupied and GATA-2-unoccupied conserved GATA motifs differs greatly (77). Diacetylated H3 and H3-dimeK4 are enriched at occupied sites, whereas H3-trimeK9 and H3-trimeK27 are often under-represented. Unoccupied sites are characterized by high-level H3-trimeK9 or H3-trimeK27 (and occasionally both) and are often devoid of the active marks (77). Genome-wide comparisons of GATA-1 occupancy and histone marks also revealed distinct patterns at GATA-1-bound versus GATA-1-unoccupied sites. The distinctive histone marks may be primary determinants of GATA factor occupancy, perhaps through direct interactions with one or more GATA factor complex components. Alternatively, GATA factors may recruit the respective chromatin-modifying components that establish and/or maintain the specific patterns. Distinguishing between these possibilities will be essential to understand how GATA factors select chromatin sites.
Integrating GATA Factor Mechanisms via GATA Switches
GATA Switch Model
Studies of GATA factor mechanisms revealed compelling similarities between the actions of different GATA factors. GATA-1 and GATA-2 bind chromatin sites with high selectivity and share the majority of target sites in the cells studied (64, 71, 73, 74). Their mode of chromatin recognition appears to be nearly identical, at least based on the precise sequences of the GATA motif at the shared sites, consistent with their highly similar DNA-binding domains. As GATA-1 and GATA-2 function redundantly to support the genesis and/or survival of primitive erythroblasts (41), this is consistent with the finding that both factors share a large cohort of target genes. Furthermore, either the GATA-1 or GATA-2 interaction with FOG-1 is sufficient to support megakaryopoiesis (78).
In certain contexts, GATA-1 and GATA-2 exert distinct activities. GATA-2 uniquely regulates early hematopoiesis by controlling the genesis and/or survival of HSCs and/or multipotent progenitors (11, 12, 79). GATA-2, but not GATA-1, knock-out hematopoietic precursors are competent to undergo erythroid and myeloid terminal differentiation (12). In a GATA-1 knock-out context in which erythropoiesis is blocked, GATA-2 is up-regulated, and this is insufficient to support erythropoiesis (80, 81). GATA-2, but not GATA-1, is crucial for mast cell development (12). Based on these differences, it is attractive to propose that GATA-1 and GATA-2 function distinctly in different regulatory milieus. Given the common sharing of target sites, either GATA-1 and GATA-2 function distinctly through common sites, thereby yielding different biological activities, or GATA-1- and GATA-2-selective targets underlie the unique biological outputs.
The concept that GATA factors can function differently through shared chromatin sites emerged from studies on how GATA-1 represses transcription (23, 71). Although GATA-1 and GATA-2 were known to be expressed reciprocally during erythropoiesis (8, 80, 81), mechanisms underlying these patterns were unclear. We demonstrated that GATA-1 directly represses Gata2 transcription using a complementation assay in the GATA-1-null proerythroblast-like cell G1E (71). GATA-1-mediated Gata2 repression was associated with GATA-1 occupancy at five dispersed regions of the Gata2 locus (−77, −3.9, −2.8, −1.8, and +9.5 kb relative to the 1S promoter transcription start site), broad histone deacetylation, and RNA polymerase II expulsion (72, 73, 82). GATA-2 associates with these identical sites when Gata2 is transcriptionally active. Studies in zebrafish revealed that, although GATA-2 is expressed earlier than GATA-1 during embryogenesis, GATA-1 and GATA-2 are expressed in primitive blood precursors (83). Only loss of GATA-1 induces a fate switch involving conversion of primitive blood precursors into myeloid cells rather than predominant red blood cells (84). In addition, evidence was provided for GATA-1-specific and GATA-1/2-shared target genes in this system.
We proposed the GATA switch model, in which GATA-1 displaces GATA-2 from chromatin sites, often (but not always) instigating a distinct transcriptional output (Fig. 3) (23, 71, 85). Of note, we define “GATA switch” as the replacement of one GATA factor by another at a chromatin site and not the loss of expression of a GATA factor concomitant with expression of a distinct GATA factor. This is an important difference because the expression of specific GATA factors changes during development, and such changes are often measured in cell populations. Thus, a change in GATA factor expression in a cell population may not necessarily reflect a switch in the expression of specific GATA factors in the same cell, and therefore, in this scenario, a GATA factor would not have the opportunity to displace a distinct GATA factor from a common chromatin target site. Furthermore, although GATA factor concentrations are certainly important determinants of switches, as noted below, other factors can be critical regulators of the switch in chromatin site occupancy. A change in factor concentration therefore may be insufficient to instigate a bona fide switch at a target gene. GATA switches occur in primary cells and in vivo (55, 86). Three aspects of this proposed mechanism are of considerable interest. First, GATA factors have the capacity to function differently through identical chromatin sites. Second, because GATA switches occur at genes encoding critical regulators of hematopoiesis (e.g. Gata2), the switches may drive hematopoiesis. Finally, given mutations of GATA-1 and GATA-2 in human leukemias (87, 88), such alterations may impede GATA switches as a key step in hematologic malignancies.
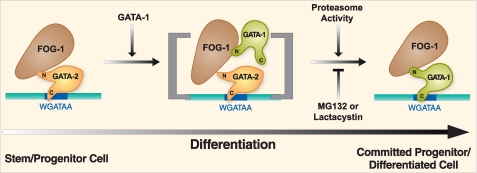
FIGURE 3.
GATA switch model. In early-stage erythroblasts, GATA-2 and FOG-1 colocalize at chromatin sites, and upon elevation of GATA-1 levels as erythropoiesis proceeds, GATA-1 displaces GATA-2. FOG-1 facilitates GATA-1 occupancy and GATA switches, but in certain contexts, GATA-1 chromatin occupancy appears to be FOG-1-independent. As the GATA-1 N-terminal zinc finger can contact multiple FOG-1 zinc fingers (48, 49), we predict that GATA-1 enters the GATA-2·FOG-1 complex, creating a transient biochemical intermediate in which both GATA factors engage distinct FOG-1 zinc fingers simultaneously. Proteasome inhibitors stabilize GATA-2 and impair GATA switches (29), and therefore, the low stability of GATA-2 appears to ensure efficient GATA switches. However, whether the proteasome acts on chromatin-bound GATA-2 in unknown. An additional key aspect of the mechanism that requires additional study is to establish the precise precursor during hematopoiesis in vivo in which GATA-1 levels rise sufficiently to gain a competitive advantage over GATA-2 bound to chromatin target sites. Given the role of FOG-1 in promoting switches, presumably mechanisms that increase or decrease FOG-1 activity and/or alter FOG-1 levels control GATA switches, but this possibility has not yet been experimentally tested.
Molecular Determinants of GATA Switches
In the context of naked DNA, the capacity of a protein to displace another from a common binding site is dictated by well defined biophysical parameters of the binding reactions. However, given the unpredictable influence of chromatin on DNA-binding site affinities, the residence of DNA-binding components in large multiprotein complexes, and poorly understood forces exerted by the subnuclear environment, defining the biophysical attributes of a displacement reaction, such as a GATA switch, in vivo is fraught with difficulties. Despite this formidable hurdle, progress has been made in dissecting GATA switch mechanisms.
The initial studies on GATA switches utilized a conditionally active GATA-1 protein containing the estrogen receptor ligand-binding domain expressed at a level similar to that of endogenous GATA-1 in erythroleukemia cells and erythroblasts (71, 73). Estrogen receptor/GATA-1 displacement of GATA-2 from chromatin increases in a graded fashion upon increasing the ligand concentration or time in the presence of a maximally effective ligand concentration. Thus, the ratio of the concentrations of active GATA-1 to GATA-2 is a critical determinant of GATA switches.
Although both GATA-1 and GATA-2 are degraded by the proteasome, GATA-2 is considerably less stable than GATA-1 (t½ = ∼1 h versus >4 h, respectively) (29). We reasoned that GATA-2 instability might be an important determinant of the GATA switch. Proteasome inhibitors increase GATA-2 stability, and under these conditions, GATA-1 has less efficacy to displace GATA-2 from chromatin (29). The proteasome inhibitor opposition to GATA switches may reflect a higher steady-state level of GATA-2, a requirement for rapid GATA-2 turnover to expel it from chromatin, or an indirect influence on another target that controls the GATA switch. To distinguish among these possibilities, it will be instructive to test whether stabilized GATA-2 mutants are refractory to GATA-1-mediated expulsion from chromatin.
Studies in FOG-1-null hematopoietic precursors expressing endogenous GATA-1 and GATA-2 revealed that expressing estrogen receptor/GATA-1 ∼30-fold higher than endogenous GATA-1 is insufficient to displace GATA-2 from chromatin (51). As GATA switches are rescued by expressing FOG-1, FOG-1 promotes GATA switches. This activity might reflect FOG-1 chromatin occupancy facilitator activity, which enhances GATA-1 chromatin occupancy at select sites (51), or its role in mediating GATA-1-dependent chromatin looping (52). Regardless, FOG-1 is a determinant of GATA switches.
Is the FOG-1 activity to promote GATA switches intrinsic to FOG-1, or does it require associated components, perhaps with specific enzymatic activities? Because FOG-1 co-purifies with the NuRD complex (53), the FOG-1 activity might require NuRD-dependent chromatin remodeling and/or other activities. Knock-in mice have been generated in which endogenous FOG-1 was replaced with N-terminal mutants defective in NuRD binding (e.g. FOG-1R3K5A) (54, 55). Although these mice do not precisely phenocopy GATA-1- or FOG-1-null mice, they exhibit defective erythropoiesis and megakaryopoiesis, consistent with impaired GATA-1 function. As GATA-2 expression is elevated in megakaryocytic and erythroid precursors from FOG-1R3K5A mice, the authors concluded that FOG-1 binding to NuRD is required for GATA switches that repress Gata2 transcription (55). The biochemical underpinnings of this requirement are unclear, but importantly, the mechanism by which a trans-acting factor utilizes coregulators to displace a related protein with distinct activities from a chromatin site represents a new paradigm.
In the context of molecular endocrinology, steroidal or non-steroidal small molecule ligands bind to nuclear receptors, inducing a switch in receptor binding to corepressors or coactivators (89). Although GATA switches are not instigated by ligands, whether GATA switches share common attributes with these ligand-dependent switches requires further investigation. As GATA-1 recruits additional coregulators, including MED1 (62), BRG1 (64, 66), and CBP/p300 (60), it will be important to determine whether the FOG-1/NuRD requirement for GATA switches is unique or if any coregulator that promotes GATA-1 activity also facilitates GATA switches.
GATA Switches beyond Red Blood Cell Development
Because multiple GATA factors can be expressed in the same cell type and/or distinct cells in a cell lineage and have a common naked DNA binding specificity, distinct GATA factors may commonly compete for identical chromatin sites. If different GATA factors exert qualitatively or quantitatively distinct activities, this would be analogous to the erythroid GATA switch. Although this is largely unexplored, evidence has emerged consistent with GATA switches in mast cells, megakaryocytes, and trophoblasts.
Analogous to the erythroid GATA-1/2 switch, a similar switch has been described in mast cells (90). FOG-1 expression in mast cell precursors blocks their maturation into mast cells and reprograms precursors into cells with erythroid, megakaryocytic, and granulocytic phenotypes (90). Similar to expressing FOG-1 in FOG-1-null erythroid precursors (51), FOG-1 expression in mast cell precursors induces a GATA switch at the Gata2 locus −2.8 kb site (90). This GATA switch-associated Gata2 repression was proposed to underlie the FOG-1-dependent reprogramming of mast cell precursors.
GATA switches can involve additional GATA factors besides GATA-1 and GATA-2. Evidence for a GATA-2/3 switch emerged from studies of placental development. GATA-2 and GATA-3 are expressed in trophoblast giant cells and regulate the expression of placental hormones (91). Although knock-outs of either gene individually do not block giant cell differentiation, both factors coordinately regulate genes involved in giant cell differentiation (92, 93). The GATA-2 knock-out exerted a greater inhibitory influence on placental angiogenesis versus the GATA-3 knock-out, highlighting quantitative differences in their activities to control angiogenesis. Studies in a cultured trophoblast cell line indicate that GATA-2 and GATA-3 can also exert qualitatively distinct functions (91). In trophoblast precursors, GATA-3 occupies two regulatory regions of the Gata2 locus, −3.9 and +9.5 kb relative to the promoter, which correlates with Gata2 repression (91). In a differentiation-dependent manner, GATA-2 displaces GATA-3, which is linked to Gata2 transcriptional activation. Thus, analogous to the erythroid GATA switch, GATA-2 occupancy appears to reflect positive autoregulation, whereas distinct GATA factors functioning through GATA switch sites confer repression (GATA-1 and GATA-3 in erythroid cells and trophoblast precursors, respectively). GATA-3 (followed by GATA-2) expression has also been observed during the ontogeny of the allantois (94), although the functional implications of this expression pattern are unclear.
Whereas there are compelling examples of cells expressing multiple GATA factors, additional work is required to assess whether GATA switches occur in these systems. GATA-2/3/4/6 are expressed in human hepatocytes (95). GATA-3 represses erythropoietin expression in murine hepatocytes, whereas GATA-4 induces erythropoietin expression in human hepatocytes (95). It was proposed that GATA-4 functions through the erythropoietin promoter in fetal hepatocytes to activate transcription, and the lack of GATA-4 in adult hepatocytes and/or the function of GATA-2/3 might confer repression in the adult. By contrast to these qualitatively distinct activities, GATA-4 and GATA-6 function cooperatively to induce expression of cardiac genes in cardiomyocytes (96).
Endothelial cells express multiple GATA factors, and GATA-3 is implicated in activating transcription of Tie2 (97), which encodes a tyrosine kinase receptor for the angiopoietin-1 ligand (98). The activation mechanism involves GATA-3 binding to sites within the 5′-untranslated region. GATA-2 has been implicated as a mediator of mechanosignaling-dependent angiogenesis, and this is associated with induction of VEGFR2 transcription (99). Elucidating the dynamics of GATA factor expression and chromatin occupancy in endothelial cells in vivo will provide important insights into the role of GATA switches in controlling angiogenesis and other critical endothelial cell-dependent processes.
Perspective
The further dissection of GATA switch mechanisms has potential to yield critical mechanistic, biological, and pathophysiological insights. Key unresolved issues include the following. 1) Because different GATA factors can exert qualitatively distinct functions through an identical chromatin site, what molecular attributes underlie the unique identities of individual GATA factor family members? 2) Although GATA factor concentrations, FOG-1, and proteasome-sensitive factor(s) control GATA switches, how important are these and other parameters in vivo? 3) What are the functional consequences of GATA switches at endogenous loci in vivo? Although cis-element functionality is commonly inferred based on transfection and transgenic studies, it is critical to assess function at their endogenous chromosomal sites. Using a cis-element knock-out approach, we demonstrated that the Gata2 −1.8 kb site is selectively required to maintain repression in late-stage erythroblasts in vivo (100). This conclusion would not have emerged from other modes of functional analysis. 4) Because GATA switches occur at a host of important regulatory genes, do GATA switches dominantly establish and/or orchestrate genetic networks that drive developmental processes? 5) Do GATA switches occur synchronously upon achieving the requisite concentration of a given GATA factor or asynchronously over a spectrum of GATA factor concentrations, dependent on the chromosomal environment and subnuclear neighborhood in vivo? 6) As noted above, GATA switches occur in distinct cellular contexts, raising the question as to what is the spectrum of GATA switch-dependent developmental processes, and does incorporating GATA switches into a developmental regulatory mechanism endow it with unique regulatory properties? 7) Although we have discussed binary GATA switches, because more than two GATA factors can be expressed in certain cell types, in principle, complex iterations of GATA switches may involve greater than two factors. Do ternary or even higher order GATA switches occur? 8) GATA-1 and GATA-2 function distinctly through certain common chromatin sites, raising the question as to what extent do GATA switches induce qualitative differences in the transcriptional and biological outputs? 9) Can the GATA switch concept be generalized to other transcription factor families, including the highly related Ets and/or Krüppel-like transcription factors? As exemplified by the shear number and broad scope of these unresolved issues, future efforts will almost certainly yield a rich set of discoveries that impact upon many fields.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants DK50107 and DK68034. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- HSC
- hematopoietic stem cell
- NuRD
- nucleosome remodeling and deacetylase
- CtBP
- C-terminal domain-binding protein
- CBP
- cAMP-responsive element-binding protein-binding protein.
REFERENCES
- 1.Orkin S. H., Zon L. I. (2008) Cell 132, 631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans T., Reitman M., Felsenfeld G. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 5976–5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans T., Felsenfeld G. (1989) Cell 58, 877–885 [DOI] [PubMed] [Google Scholar]
- 4.Tsai S. F., Martin D. I., Zon L. I., D'Andrea A. D., Wong G. G., Orkin S. H. (1989) Nature 339, 446–451 [DOI] [PubMed] [Google Scholar]
- 5.Wilson D. B., Dorfman D. M., Orkin S. H. (1990) Mol. Cell. Biol. 10, 4854–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamamoto M., Ko L. J., Leonard M. W., Beug H., Orkin S. H., Engel J. D. (1990) Genes Dev. 4, 1650–1662 [DOI] [PubMed] [Google Scholar]
- 7.Orkin S. H. (1992) Blood 80, 575–581 [PubMed] [Google Scholar]
- 8.Leonard M., Brice M., Engel J. D., Papayannopoulou T. (1993) Blood 82, 1071–1079 [PubMed] [Google Scholar]
- 9.Martin D. I., Zon L. I., Mutter G., Orkin S. H. (1990) Nature 344, 444–447 [DOI] [PubMed] [Google Scholar]
- 10.Ito E., Toki T., Ishihara H., Ohtani H., Gu L., Yokoyama M., Engel J. D., Yamamoto M. (1993) Nature 362, 466–468 [DOI] [PubMed] [Google Scholar]
- 11.Tsai F. Y., Keller G., Kuo F. C., Weiss M., Chen J., Rosenblatt M., Alt F. W., Orkin S. H. (1994) Nature 371, 221–226 [DOI] [PubMed] [Google Scholar]
- 12.Tsai F. Y., Orkin S. H. (1997) Blood 89, 3636–3643 [PubMed] [Google Scholar]
- 13.Minegishi N., Ohta J., Yamagiwa H., Suzuki N., Kawauchi S., Zhou Y., Takahashi S., Hayashi N., Engel J. D., Yamamoto M. (1999) Blood 93, 4196–4207 [PubMed] [Google Scholar]
- 14.Lee M. E., Temizer D. H., Clifford J. A., Quertermous T. (1991) J. Biol. Chem. 266, 16188–16192 [PubMed] [Google Scholar]
- 15.Nardelli J., Thiesson D., Fujiwara Y., Tsai F. Y., Orkin S. H. (1999) Dev. Biol. 210, 305–321 [DOI] [PubMed] [Google Scholar]
- 16.Zhou Y., Yamamoto M., Engel J. D. (2000) Development 127, 3829–3838 [DOI] [PubMed] [Google Scholar]
- 17.Craven S. E., Lim K. C., Ye W., Engel J. D., de Sauvage F., Rosenthal A. (2004) Development 131, 1165–1173 [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi-Osaki M., Ohneda O., Suzuki N., Minegishi N., Yokomizo T., Takahashi S., Lim K. C., Engel J. D., Yamamoto M. (2005) Mol. Cell. Biol. 25, 7005–7020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim K. C., Lakshmanan G., Crawford S. E., Gu Y., Grosveld F., Engel J. D. (2000) Nat. Genet. 25, 209–212 [DOI] [PubMed] [Google Scholar]
- 20.Kouros-Mehr H., Slorach E. M., Sternlicht M. D., Werb Z. (2006) Cell 127, 1041–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charron F., Nemer M. (1999) Semin. Cell Dev. Biol. 10, 85–91 [DOI] [PubMed] [Google Scholar]
- 22.Molkentin J. D. (2000) J. Biol. Chem. 275, 38949–38952 [DOI] [PubMed] [Google Scholar]
- 23.Bresnick E. H., Martowicz M. L., Pal S., Johnson K. D. (2005) J. Cell. Physiol. 205, 1–9 [DOI] [PubMed] [Google Scholar]
- 24.Martin D. I., Orkin S. H. (1990) Genes Dev. 4, 1886–1898 [DOI] [PubMed] [Google Scholar]
- 25.Ko L. J., Engel J. D. (1993) Mol. Cell. Biol. 13, 4011–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merika M., Orkin S. H. (1993) Mol. Cell. Biol. 13, 3999–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newton A., Mackay J., Crossley M. (2001) J. Biol. Chem. 276, 35794–35801 [DOI] [PubMed] [Google Scholar]
- 28.Pedone P. V., Omichinski J. G., Nony P., Trainor C., Gronenborn A. M., Clore G. M., Felsenfeld G. (1997) EMBO J. 16, 2874–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lurie L. J., Boyer M. E., Grass J. A., Bresnick E. H. (2008) Biochemistry 47, 859–869 [DOI] [PubMed] [Google Scholar]
- 30.Kadri Z., Shimizu R., Ohneda O., Maouche-Chretien L., Gisselbrecht S., Yamamoto M., Romeo P. H., Leboulch P., Chretien S. (2009) PLoS Biol. 7, e1000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson K. D., Kim S. I., Bresnick E. H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15939–15944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collavin L., Gostissa M., Avolio F., Secco P., Ronchi A., Santoro C., Del Sal G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8870–8875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee H. Y., Johnson K. D., Fujiwara T., Boyer M. E., Kim S. I., Bresnick E. H. (2009) Mol. Cell 36, 984–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pevny L., Simon M. C., Robertson E., Klein W. H., Tsai S. F., D'Agati V., Orkin S. H., Costantini F. (1991) Nature 349, 257–260 [DOI] [PubMed] [Google Scholar]
- 35.Simon M. C., Pevny L., Wiles M. V., Keller G., Costantini F., Orkin S. H. (1992) Nat. Genet. 1, 92–98 [DOI] [PubMed] [Google Scholar]
- 36.Pevny L., Lin C. S., D'Agati V., Simon M. C., Orkin S. H., Costantini F. (1995) Development 121, 163–172 [DOI] [PubMed] [Google Scholar]
- 37.Fujiwara Y., Browne C. P., Cunniff K., Goff S. C., Orkin S. H. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 12355–12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu C., Cantor A. B., Yang H., Browne C., Wells R. A., Fujiwara Y., Orkin S. H. (2002) J. Exp. Med. 195, 1387–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirasawa R., Shimizu R., Takahashi S., Osawa M., Takayanagi S., Kato Y., Onodera M., Minegishi N., Yamamoto M., Fukao K., Taniguchi H., Nakauchi H., Iwama A. (2002) J. Exp. Med. 195, 1379–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Migliaccio A. R., Rana R. A., Sanchez M., Lorenzini R., Centurione L., Bianchi L., Vannucchi A. M., Migliaccio G., Orkin S. H. (2003) J. Exp. Med. 197, 281–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fujiwara Y., Chang A. N., Williams A. M., Orkin S. H. (2004) Blood 103, 583–585 [DOI] [PubMed] [Google Scholar]
- 42.Tsang A. P., Visvader J. E., Turner C. A., Fujiwara Y., Yu C., Weiss M. J., Crossley M., Orkin S. H. (1997) Cell 90, 109–119 [DOI] [PubMed] [Google Scholar]
- 43.Tsang A. P., Fujiwara Y., Hom D. B., Orkin S. H. (1998) Genes Dev. 12, 1176–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crispino J. D., Lodish M. B., MacKay J. P., Orkin S. H. (1999) Mol. Cell 3, 219–228 [DOI] [PubMed] [Google Scholar]
- 45.Nichols K. E., Crispino J. D., Poncz M., White J. G., Orkin S. H., Maris J. M., Weiss M. J. (2000) Nat. Genet. 24, 266–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson K. D., Boyer M. E., Kang J. A., Wickrema A., Cantor A. B., Bresnick E. H. (2007) Blood 109, 5230–5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim S. I., Bultman S. J., Jing H., Blobel G. A., Bresnick E. H. (2007) Mol. Cell. Biol. 27, 4551–4565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fox A. H., Liew C., Holmes M., Kowalski K., Mackay J., Crossley M. (1999) EMBO J. 18, 2812–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cantor A. B., Katz S. G., Orkin S. H. (2002) Mol. Cell. Biol. 22, 4268–4279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Letting D. L., Chen Y. Y., Rakowski C., Reedy S., Blobel G. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pal S., Cantor A. B., Johnson K. D., Moran T. B., Boyer M. E., Orkin S. H., Bresnick E. H. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 980–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vakoc C. R., Letting D. L., Gheldof N., Sawado T., Bender M. A., Groudine M., Weiss M. J., Dekker J., Blobel G. A. (2005) Mol. Cell 17, 453–462 [DOI] [PubMed] [Google Scholar]
- 53.Hong W., Nakazawa M., Chen Y. Y., Kori R., Vakoc C. R., Rakowski C., Blobel G. A. (2005) EMBO J. 24, 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miccio A., Wang Y., Hong W., Gregory G. D., Wang H., Yu X., Choi J. K., Shelat S., Tong W., Poncz M., Blobel G. A. (2010) EMBO J. 29, 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao Z., Huang Z., Olivey H. E., Gurbuxani S., Crispino J. D., Svensson E. C. (2010) EMBO J. 29, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turner J., Crossley M. (1998) EMBO J. 17, 5129–5140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi Y. J., Matson C., Lan F., Iwase S., Baba T., Shi Y. (2005) Mol. Cell 19, 857–864 [DOI] [PubMed] [Google Scholar]
- 58.Katz S. G., Cantor A. B., Orkin S. H. (2002) Mol. Cell. Biol. 22, 3121–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Snow J. W., Orkin S. H. (2009) J. Biol. Chem. 284, 29310–29319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blobel G. A., Nakajima T., Eckner R., Montminy M., Orkin S. H. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2061–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goodman R. H., Smolik S. (2000) Genes Dev. 14, 1553–1577 [PubMed] [Google Scholar]
- 62.Stumpf M., Waskow C., Krötschel M., van Essen D., Rodriguez P., Zhang X., Guyot B., Roeder R. G., Borggrefe T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18504–18509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Letting D. L., Rakowski C., Weiss M. J., Blobel G. A. (2003) Mol. Cell. Biol. 23, 1334–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Im H., Grass J. A., Johnson K. D., Kim S. I., Boyer M. E., Imbalzano A. N., Bieker J. J., Bresnick E. H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17065–17070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pope N. J., Bresnick E. H. (2010) Nucleic Acids Res. 38, 2190–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim S. I., Bultman S. J., Kiefer C. M., Dean A., Bresnick E. H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2259–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bultman S. J., Gebuhr T. C., Magnuson T. (2005) Genes Dev. 19, 2849–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim S. I., Bresnick E. H., Bultman S. J. (2009) Nucleic Acids Res. 37, 6019–6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bresnick E. H., Johnson K. D., Kim S. I., Im H. (2006) Prog. Nucleic Acids Res. Mol. Biol. 81, 435–471 [DOI] [PubMed] [Google Scholar]
- 70.Johnson K. D., Grass J. A., Boyer M. E., Kiekhaefer C. M., Blobel G. A., Weiss M. J., Bresnick E. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11760–11765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grass J. A., Boyer M. E., Pal S., Wu J., Weiss M. J., Bresnick E. H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8811–8816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martowicz M. L., Grass J. A., Boyer M. E., Guend H., Bresnick E. H. (2005) J. Biol. Chem. 280, 1724–1732 [DOI] [PubMed] [Google Scholar]
- 73.Grass J. A., Jing H., Kim S. I., Martowicz M. L., Pal S., Blobel G. A., Bresnick E. H. (2006) Mol. Cell. Biol. 26, 7056–7067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fujiwara T., O'Geen H., Keles S., Blahnik K., Linnemann A. K., Kang Y. A., Choi K., Farnham P. J., Bresnick E. H. (2009) Mol. Cell 36, 667–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cheng Y., Wu W., Kumar S. A., Yu D., Deng W., Tripic T., King D. C., Chen K. B., Zhang Y., Drautz D., Giardine B., Schuster S. C., Miller W., Chiaromonte F., Zhang Y., Blobel G. A., Weiss M. J., Hardison R. C. (2009) Genome Res. 19, 2172–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu M., Riva L., Xie H., Schindler Y., Moran T. B., Cheng Y., Yu D., Hardison R., Weiss M. J., Orkin S. H., Bernstein B. E., Fraenkel E., Cantor A. B. (2009) Mol. Cell 36, 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wozniak R. J., Keles S., Lugus J. J., Young K. H., Boyer M. E., Tran T. M., Choi K., Bresnick E. H. (2008) Mol. Cell. Biol. 28, 6681–6694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chang A. N., Cantor A. B., Fujiwara Y., Lodish M. B., Droho S., Crispino J. D., Orkin S. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 9237–9242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ling K. W., Ottersbach K., van Hamburg J. P., Oziemlak A., Tsai F. Y., Orkin S. H., Ploemacher R., Hendriks R. W., Dzierzak E. (2004) J. Exp. Med. 200, 871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weiss M. J., Keller G., Orkin S. H. (1994) Genes Dev. 8, 1184–1197 [DOI] [PubMed] [Google Scholar]
- 81.Weiss M. J., Yu C., Orkin S. H. (1997) Mol. Cell. Biol. 17, 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wozniak R. J., Boyer M. E., Grass J. A., Lee Y., Bresnick E. H. (2007) J. Biol. Chem. 282, 14665–14674 [DOI] [PubMed] [Google Scholar]
- 83.Heicklen-Klein A., McReynolds L. J., Evans T. (2005) Semin. Cell Dev. Biol. 16, 95–106 [DOI] [PubMed] [Google Scholar]
- 84.Galloway J. L., Wingert R. A., Thisse C., Thisse B., Zon L. I. (2005) Dev. Cell 8, 109–116 [DOI] [PubMed] [Google Scholar]
- 85.Kim S. I., Bresnick E. H. (2007) Oncogene 26, 6777–6794 [DOI] [PubMed] [Google Scholar]
- 86.Lugus J. J., Chung Y. S., Mills J. C., Kim S. I., Grass J., Kyba M., Doherty J. M., Bresnick E. H., Choi K. (2007) Development 134, 393–405 [DOI] [PubMed] [Google Scholar]
- 87.Wechsler J., Greene M., McDevitt M. A., Anastasi J., Karp J. E., Le Beau M. M., Crispino J. D. (2002) Nat. Genet. 32, 148–152 [DOI] [PubMed] [Google Scholar]
- 88.Zhang S. J., Ma L. Y., Huang Q. H., Li G., Gu B. W., Gao X. D., Shi J. Y., Wang Y. Y., Gao L., Cai X., Ren R. B., Zhu J., Chen Z., Chen S. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2076–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Perissi V., Aggarwal A., Glass C. K., Rose D. W., Rosenfeld M. G. (2004) Cell 116, 511–526 [DOI] [PubMed] [Google Scholar]
- 90.Cantor A. B., Iwasaki H., Arinobu Y., Moran T. B., Shigematsu H., Sullivan M. R., Akashi K., Orkin S. H. (2008) J. Exp. Med. 205, 611–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ray S., Dutta D., Rumi M. A., Kent L. N., Soares M. J., Paul S. (2009) J. Biol. Chem. 284, 4978–4988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ng Y. K., George K. M., Engel J. D., Linzer D. I. (1994) Development 120, 3257–3266 [DOI] [PubMed] [Google Scholar]
- 93.Ma G. T., Roth M. E., Groskopf J. C., Tsai F. Y., Orkin S. H., Grosveld F., Engel J. D., Linzer D. I. (1997) Development 124, 907–914 [DOI] [PubMed] [Google Scholar]
- 94.Caprioli A., Minko K., Drevon C., Eichmann A., Dieterlen-Lièvre F., Jaffredo T. (2001) Dev. Biol. 238, 64–78 [DOI] [PubMed] [Google Scholar]
- 95.Dame C., Sola M. C., Lim K. C., Leach K. M., Fandrey J., Ma Y., Knöpfle G., Engel J. D., Bungert J. (2004) J. Biol. Chem. 279, 2955–2961 [DOI] [PubMed] [Google Scholar]
- 96.Charron F., Paradis P., Bronchain O., Nemer G., Nemer M. (1999) Mol. Cell. Biol. 19, 4355–4365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Song H., Suehiro J., Kanki Y., Kawai Y., Inoue K., Daida H., Yano K., Ohhashi T., Oettgen P., Aird W. C., Kodama T., Minami T. (2009) J. Biol. Chem. 284, 29109–29124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Davis S., Aldrich T. H., Jones P. F., Acheson A., Compton D. L., Jain V., Ryan T. E., Bruno J., Radziejewski C., Maisonpierre P. C., Yancopoulos G. D. (1996) Cell 87, 1161–1169 [DOI] [PubMed] [Google Scholar]
- 99.Mammoto A., Connor K. M., Mammoto T., Yung C. W., Huh D., Aderman C. M., Mostoslavsky G., Smith L. E., Ingber D. E. (2009) Nature 457, 1103–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Snow J. W., Trowbridge J. J., Fujiwara T., Emambokus N. E., Grass J. A., Orkin S. H., Bresnick E. H. (2010) PLOS Genet., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.