Abstract
A long-standing goal of computational protein design is to create proteins similar to those found in Nature. One motivation is to harness the exquisite functional capabilities of proteins for our own purposes. The extent of similarity between designed and natural proteins also reports on how faithfully our models represent the selective pressures that determine protein sequences. As the field of protein design shifts emphasis from reproducing native-like protein structure to function, it has become important that these models treat the notion of specificity in molecular interactions. Although specificity may, in some cases, be achieved by optimization of a desired protein in isolation, methods have been developed to address directly the desire for proteins that exhibit specific functions and interactions.
Keywords: Biophysics, Computer Modeling, Mathematical Modeling, Protein-Nucleic Acid Interaction, Protein-Protein Interactions
Introduction
The field of computational protein design pursues two goals. One is practical and regards the products of the design process: we desire the ability to engineer proteins for arbitrary functions. The second goal emphasizes scientific, rather than engineering, aims. This goal is concerned with the models that serve as input for the process: we desire a quantitative description of the principles that influence the selection of natural proteins. The extent to which designed proteins resemble natural proteins serves to evaluate how well our models simulate the selective pressures for natural proteins. This minireview is concerned with efforts to endow designed proteins with the specificity observed in naturally occurring proteins. Excellent reviews of other aspects of computational protein design have also appeared recently (1, 2).
Advances in molecular modeling have expanded the inventory of protein properties that can be reproduced by computational design. Early computational efforts sought only to replicate native-like packing arrangements for hydrophobic protein cores (3). It was found that this was achievable using a simple energy potential to enforce close packing without steric clashes. The structural representation used was a fixed backbone from an experimentally determined structure and a discrete set of commonly observed side chain conformations. More complete energy potentials enabled the complete redesign of a protein (4) and the construction of novel hydrogen-bonding networks (5). The incorporation of backbone relaxation techniques was important for the redesign of loop conformations (6). Finally, the design of the Top7 protein achieved the milestone goal of constructing a protein for which neither backbone topology nor amino acid sequence was derived from any naturally occurring protein (7). However, continuing differences in the properties of designed and natural proteins provide evidence (if any was needed) that the pressures applied by protein design potentials incompletely simulate those of natural selection (8).
The improvements in energy functions and conformational sampling described above enhance the precision and correctness with which single protein structures or complexes are evaluated. However, specificity requires consideration of multiple outcomes for a protein, both desired and undesired. The selection of a protein sequence that is optimal for a desired structure or interaction is termed “positive design,” whereas the selection of sequence elements to discourage undesired structures, or complexes, is termed “negative design” (9). Multistate design algorithms simultaneously consider both desired and undesired outcomes when selecting sequences. In the following, I describe the biological parallels for the functional specificity we seek to encode in the protocols of protein design, describe techniques for achieving specificity both with and without explicit multistate design, and finally discuss recent algorithmic advances likely to advance the field in the near future.
Natural Exemplars for Biomolecular Specificity
Specificity is crucial for the proper flow of information and energy through signal transduction, metabolic, and transcriptional pathways. At the same time, duplication and reuse of modular interaction domains are prevalent in the assembly of these systems. Consequently, a single cell may contain dozens of interacting partners drawn from a particular family of modular interactions. How is specificity maintained when interacting partners must coexist, but not interfere, with family members that possess significant structural and sequence similarity? Differing patterns of subcellular localization or developmental expression may ensure that undesired partners never meet, but often the specificity of the physical interaction alone is sufficient to restrict successful complex formation to cognate pairs.
In certain systems, specificity has been shown to be both independent of context and shaped by considerations of negative design. Zarrinpar et al. (10) studied the interactions between the 27 SH32 domains in the yeast proteome and the Pbs2 peptide (the ligand for the Sho1 SH3 domain). They found no binding between the Pbs2 peptide and the 26 non-cognate yeast SH3 domains. However, 6 of 12 non-yeast SH3 domains were able to bind the peptide. This suggests that the Pbs2 peptide has been optimized to maintain specificity only with respect to the relevant competing yeast SH3 domains. When confronted with “extra-proteomic” SH3 domains, the peptide was broadly cross-reactive. A large-scale study of PDZ domains in the mouse proteome suggests that negative selection has served to minimize cross-reactivity in this system as well (11).
There are two lessons here for protein design. The first is that specificity, for at least some families of interactions, is encoded solely in the sequences of the interacting partners themselves. For these interactions, specificity is not dependent on spatial or temporal co-localization. This indicates that the engineering of such a family falls within the scope of protein design. The second lesson is that it is crucial to understand and enumerate the relevant undesired partners in any interaction. This will be entirely dependent on the context in which a protein is expected to function. Interactions between a small number of purified components in a test tube will require fewer competing states than a protein that is expected to operate in a cell.
Specificity without Negative Design
As a rule of thumb, negative design considerations may be omitted when competing states are structurally distinct (Fig. 1A). There are many examples of computationally designed interactions that achieved specificity considering only the desired complex as a target. These include the stabilization of the open and closed forms of an integrin I domain (12), the construction of novel protein interfaces (13), the redesign of protein-DNA specificity (14), the design of small molecule-binding receptors (15), and the tightening of specificity for a protein with multiple partners (16).
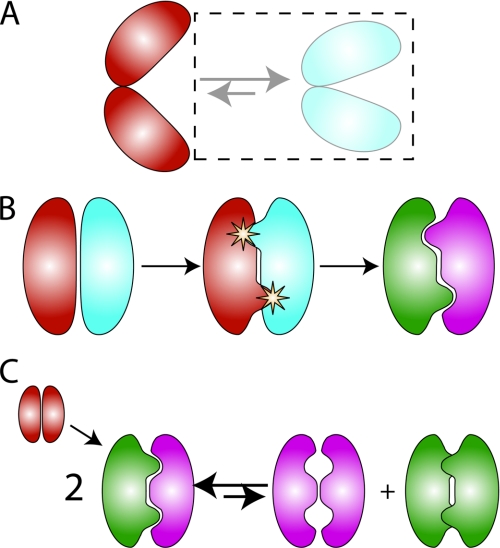
FIGURE 1.
Computational strategies for achieving specificity. A, positive design only. When the desired and undesired states are structurally distinct, as shown for hypothetical open and closed states for a protein, positive design may be sufficient, and negative design states may be ignored (dashed box on the right side of the equilibrium). B, second-site suppressor strategy for redesigning protein-protein interfaces. Negative and positive design elements may be added sequentially. Here, the design proceeds in two steps. First, point mutations are identified on both partners in the interaction that destabilize the native complex. Second, compensatory mutations are identified that restore affinity while accommodating the specificity-conferring amino acids. C, explicit multistate design. Here, a symmetric homodimer (shown in red) is converted into an obligate heterodimer (green and purple). The protein sequence is simultaneously selected both to stabilize the heterodimeric positive design state (left side of the equilibrium) and to destabilize the homodimeric negative design states.
In well characterized systems, a few key residues may suffice to enforce specificity. These may be selected by hand, with the remainder of the protein sequence determined in an automated fashion. Carefully placed charged residues have been used to destabilize undesired helical bundle arrangements (17), favor heterospecificity in meganuclease association (18), and convert an amyloid-like fibril into a monomeric protein (19). Ambroggio and Kuhlman (20) used a common zinc-binding motif to drive a conformational change between two alternate backbone folds for a single sequence.
However, some design tasks require explicit negative design states. For instance, Grigoryan et al. (21) sought to design coiled-coil inhibitors of basic leucine zipper (bZIP) proteins that were specific to individual bZIP proteins and bZIP families. Another common design goal requiring negative design is the conversion of a homodimeric protein to an obligate heterodimeric pair (22, 23). The related goal of converting one homodimer scaffold to two or more distinct, non-interacting homodimers cannot even be formulated in a single-state design framework. The neglected negative heterodimeric state is required to tie together what would be otherwise unconnected (and presumably identical) homodimeric designs.
Models for Specificity in Computational Protein Design
Specificity may be conferred upon a protein by a carefully constructed design process. For the limited goal of conformational, rather than functional, specificity, one approach is to iterate between sequence and structural optimization. This process converges upon mutually optimal sequence-structure pairs for which any local conformation change or mutation is predicted to decrease the value of the scoring function. This procedure has been quite successful, generating proteins with a novel backbone topology and novel loop conformations verified by structural characterization (6, 7).
Specific protein-protein interfaces can be designed using the “second-site suppressor” strategy (Fig. 1B) (24). In this two-step approach, both partners in a protein interface are mutated to generate a novel pair of proteins that do not interact with the wild-type proteins. First, destabilizing mutations across the interface are identified, ensuring that complexes between wild-type and designed partners are energetically unfavorable. Next, compensatory mutations are found in the interacting partner to construct a novel interface. By construction, the new complex is predicted to have both high affinity and specificity against the formation of wild-type/mutant hybrids. This approach has been successfully demonstrated in three experimental systems. The first application by Kortemme et al. (24) was the redesign and structural characterization of a novel colicin DNase-immunity protein pair with a specificity switch from the wild-type complex. Recently, Sammond et al. (25) applied the method to two systems: the G-protein component Gαi1-RGS14 GoLoco motif complex and the complex between UbcH7 and the ubiquitin ligase E6AP. A common finding for both groups is that the most successful designs utilize mutations to hydrophobic amino acids and that lowered affinities are observed in complexes containing engineered polar networks.
The prevalence of hydrophobic interactions over hydrogen-bonding interactions in these designed complexes could be due to at least three causes. First, the amino acids initially selected to destabilize the template interface are often large hydrophobic residues that create steric clashes. In the subsequent design step, these amino acids are more likely to nucleate the selection of hydrophobic clusters than polar networks, with which they can participate only weakly. Second, the introduction of polar networks may require a larger number of concerted mutations than are typically included in a design calculation. Finally, the conformational resolution provided by standard design protocols may be sufficient for modeling hydrophobic interactions but not for hydrogen-bonding interactions, resulting in a bias toward hydrophobic interfaces. Along these lines, it is noteworthy that the use of multiple-backbone models in the design of a colicin DNase-immunity protein interface (which results in an effectively higher resolution) yielded a novel hydrogen-bonding network, whereas single-backbone designs in the same system resulted primarily in mutations to hydrophobic amino acids (5).
Explicit Specificity via Multistate Design
Specificity may be explicitly demanded in a design calculation by including both positive and negative design states (Fig. 1C) (9). This was first demonstrated experimentally with coiled coils. Both the homodimeric and heterodimeric states for two distinct peptides were structurally modeled, and in separate designs, amino acid sequences were selected that shifted the equilibrium toward either state. Four pairs of obligate heterodimers and four pairs of non-interacting homodimers were experimentally validated (23). Barth et al. (26) combined this method with iterative structural relaxation to design a specific coiled-coil inhibitor targeting a metastable coiled-coil region in the yeast septin Cdc12p. Bolon et al. (22) studied the trade-offs between stability and specificity in the design and biophysical characterization of obligate heterodimeric mutants of the Haemophilus influenzae SspB protein. They concluded that negative design was necessary for association specificity in this system, where competing states possess significant structural similarity. Boas and Harbury (27) utilized a multistate approach to require both stability and affinity from a protein ligand-binding site. The most extensive use of negative design comes from a computational and experimental tour de force from Grigoryan et al. (21), who designed peptides to target each of 46 different human bZIP coiled-coil regions taken from 20 different bZIP families, using 20 negative design states (examples from each of 19 other families as well as the undesired homodimeric state). The experimental characterization was similarly comprehensive, as specificities were determined using a coiled-coil array consisting of all possible partners, demonstrating that most of the designs were successful.
Insights from Multistate Design
The scientific motivation for protein design is to codify and test our understanding of macromolecular function, in particular the pressures that determine the sequences of natural proteins. The limitations in current scoring functions and conformational sampling notwithstanding, the amino acid sequences selected by a design calculation report back upon the models that are used as input. By comparing designed proteins with natural proteins, we can assess the completeness (or otherwise) of our understanding of the requirements placed upon proteins. In what way does the inclusion of explicit models for specificity improve the match between designed and natural proteins?
One fruitful line of inquiry involves multispecific proteins, proteins that serve as hubs in regulatory networks and interact with multiple partners. In the parlance of computational design, these proteins must accommodate multiple positive design states. An interesting result is that when the interacting surfaces of these proteins are redesigned under the requirement that they bind well with multiple partners, the selected sequences are significantly more native-like than if they are required to bind only one of their partners (28). This indicates that the introduction of a model for (multi)specificity into protein design results in a more realistic description of how the sequences of proteins are selected by evolution and, by extension, that utilizing such a model for future de novo design efforts will generate proteins that behave in a more natural way. An experimental application of multispecificity is the design of the Sw2 protein, whose amino acid sequence was designed to be compatible with both the coiled-coil and zinc finger folds (with the protein switching between the folds in a zinc-dependent fashion) (20).
The insight into specificity gained from protein design can be contrasted with knowledge-based approaches. Data-derived recognition codes (29) and covariance data (30) can suggest ways to mix and match previously observed interactions to construct hybrid interaction specificities. However, structure-based protein design holds out the promise of generating genuinely novel proteins and interactions. For instance, the design of Top7, an engineered protein with a previously unobserved topology, demonstrated that Nature has not exploited the entirety of allowable fold space (7). Similarly, the redesign of specific interfaces has revealed motifs for specificity that have not yet been observed in naturally occurring proteins. This is even true for interaction families with many known examples, such as the coiled-coil regions of the bZIP transcription factor family (21, 23).
Future Directions
Computational protein design is a technology-driven field, and it is reasonable to survey current algorithmic development as a prelude to future experimental work. Recent advances in several areas will contribute to an improved ability to engineer specific proteins. Most directly, there has been an increased interest in new algorithms for explicit multistate design. Standard techniques for single-state design find optimal combinations of protein side chain conformations and are not transferrable to multistate design, where the desired protein sequences may adopt different conformations in positive and negative design states. Early work in multistate design used genetic algorithms and Monte Carlo optimization to search sequence space, with structural optimization performed for each state using standard repacking algorithms. An impressive large-scale demonstration of multistate design in the bZIP family of transcription factors has been reported using integer linear programming to select optimal protein sequences (31). In addition, novel multistate design methodologies based on dead-end elimination (32) and the FASTER algorithm for side chain optimization (33) have been described. The evaluation of these alternative approaches will require thorough experimental characterization of designed proteins but will be invaluable for identifying tractable and effective approaches for specific protein design.
A second area of recent activity with implications for protein design is the treatment of protein backbone flexibility. Both positive and negative designs benefit from conformational flexibility because the allowable sequence space is expanded as additional low-energy conformations become accessible. Algorithms that combine dead-end elimination with side chain “backrub” moves (34) and cluster expansions that incorporate template backbone diversity (35) have been reported to provide this benefit. An additional benefit of these methods is that the mismatch between the coarse sampling of protein structure and the steep spatial dependence of certain energy terms of molecular mechanics potentials is reduced. This is particularly important for negative design states, which are selectively destabilized during the design process. Often the interactions that are predicted to destabilize these states (such as steric clashes) are easily alleviated by modes of relaxation not permitted when the fixed backbone and side chain approximations are used.
Larger scale flexibility in loop regions is important primarily for positive design states. When the goal of a design is to repurpose a protein for a novel function, the conformation of the starting template protein is unlikely to be optimal. One remedy for this problem involves generating a set of native-like loops and selecting the best loop (or combination of loops) from this set. A robotics-inspired algorithm has been reported recently that can generate large numbers of native-like loops, frequently sampling loop reconstructions <1.0-Å Cα root mean square deviation from the native conformation (36). To take advantage of this development, the ability to identify optimal loops by energy and to efficiently incorporate loop sampling into design algorithms must be improved. Along these lines, Murphy et al. (37) were able to remodel a protein loop and switch the substrate specificity of an enzyme by requiring that the loop accommodate a specified functional amino acid in the active site. The use of functional constraints for individual amino acids, or sets of amino acids taken from a library of analogous interactions, is an attractive approach to guiding the selection of optimal loops from a large set of possibilities (38).
A third component of protein design that has received a second look is the scoring function. In the vast majority of design calculations, the scoring function for selecting an optimal protein sequence is based upon a molecular mechanics potential (39). Although additional knowledge-based terms are often included, the resolution of the energy potential in a design calculation is generally that of an all-atom model. The cluster expansion method for expressing macromolecular energies as a function of sequence alone breaks from this tradition. In this method, a sequence-based energy model is trained by evaluating a large set of random sequences using a structural model (31). A concern with sequence-only models is that they “average out” all structural information. In a design calculation, a combination of amino acids may be erroneously scored as favorable if it contains multiple sequence level interactions that cannot be realized by any single conformation. In the cluster expansion framework, higher order (e.g. three-body) interactions provide corrections for this type of error and have been shown to be necessary for describing proteins with compact folds (31). Nevertheless, the advantages of evaluating energies from sequence alone are striking: the decrease in computational burden enabled the inclusion of an unprecedented number of states in a multistate design calculation involving coiled coils (21). Whether the cluster expansion method proves as successful for other folds remains to be seen, but the intentional reassignment of computational resources from scoring functions to multiple negative design states underscores the growing importance of specificity in protein design.
Conclusions
A remarkable property of naturally occurring proteins is the specificity with which they process information, energy, and matter in living cells. In constructing models for protein design, we try both to mimic this property to engineer useful molecules and to understand how the underlying energetic and statistical principles combine to encode this property in protein sequences. A number of computational methods have been developed that have generated proteins that participate in some specific molecular interaction, and experimental results have been encouraging. The rising interest in this aspect of protein design and the current innovation in novel computational approaches will greatly increase our ability to design proteins as elegant as those in Nature.
This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- SH3
- Src homology 3.
REFERENCES
- 1.Karanicolas J., Kuhlman B. (2009) Curr. Opin. Struct. Biol. 19, 458–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mandell D. J., Kortemme T. (2009) Nat. Chem. Biol. 5, 797–807 [DOI] [PubMed] [Google Scholar]
- 3.Ponder J. W., Richards F. M. (1987) J. Mol. Biol. 193, 775–791 [DOI] [PubMed] [Google Scholar]
- 4.Dahiyat B. I., Mayo S. L. (1997) Science 278, 82–87 [DOI] [PubMed] [Google Scholar]
- 5.Joachimiak L. A., Kortemme T., Stoddard B. L., Baker D. (2006) J. Mol. Biol. 361, 195–208 [DOI] [PubMed] [Google Scholar]
- 6.Hu X., Wang H., Ke H., Kuhlman B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 17668–17673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhlman B., Dantas G., Ireton G. C., Varani G., Stoddard B. L., Baker D. (2003) Science 302, 1364–1368 [DOI] [PubMed] [Google Scholar]
- 8.Watters A. L., Deka P., Corrent C., Callender D., Varani G., Sosnick T., Baker D. (2007) Cell 128, 613–624 [DOI] [PubMed] [Google Scholar]
- 9.Hecht M. H., Richardson J. S., Richardson D. C., Ogden R. C. (1990) Science 249, 884–891 [DOI] [PubMed] [Google Scholar]
- 10.Zarrinpar A., Park S. H., Lim W. A. (2003) Nature 426, 676–680 [DOI] [PubMed] [Google Scholar]
- 11.Stiffler M. A., Chen J. R., Grantcharova V. P., Lei Y., Fuchs D., Allen J. E., Zaslavskaia L. A., MacBeath G. (2007) Science 317, 364–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimaoka M., Shifman J. M., Jing H., Takagi J., Mayo S. L., Springer T. A. (2000) Nat. Struct. Biol. 7, 674–678 [DOI] [PubMed] [Google Scholar]
- 13.Chevalier B. S., Kortemme T., Chadsey M. S., Baker D., Monnat R. J., Stoddard B. L. (2002) Mol. Cell 10, 895–905 [DOI] [PubMed] [Google Scholar]
- 14.Ashworth J., Havranek J. J., Duarte C. M., Sussman D. R. J., Monnat J., Stoddard B. L., Baker D. (2006) Nature 441, 656–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Looger L. L., Dwyer M. A., Smith J. J., Hellinga H. W. (2003) Nature 423, 185–190 [DOI] [PubMed] [Google Scholar]
- 16.Shifman J. M., Mayo S. L. (2002) J. Mol. Biol. 323, 417–423 [DOI] [PubMed] [Google Scholar]
- 17.Ghirlanda G., Lear J. D., Lombardi A., DeGrado W. F. (1998) J. Mol. Biol. 281, 379–391 [DOI] [PubMed] [Google Scholar]
- 18.Fajardo-Sanchez E., Stricher F., Pâques F., Isalan M., Serrano L. (2008) Nucleic Acids Res. 36, 2163–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W., Hecht M. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2760–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ambroggio X. I., Kuhlman B. (2006) J. Am. Chem. Soc. 128, 1154–1161 [DOI] [PubMed] [Google Scholar]
- 21.Grigoryan G., Reinke A. W., Keating A. E. (2009) Nature 458, 859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolon D. N., Grant R. A., Baker T. A., Sauer R. T. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12724–12729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Havranek J. J., Harbury P. B. (2003) Nat. Struct. Biol. 10, 45–52 [DOI] [PubMed] [Google Scholar]
- 24.Kortemme T., Joachimiak L. A., Bullock A. N., Schuler A. D., Stoddard B. L., Baker D. (2004) Nat. Struct. Mol. Biol. 11, 371–379 [DOI] [PubMed] [Google Scholar]
- 25.Sammond D. W., Eletr Z. M., Purbeck C., Kuhlman B. (2010) Proteins 78, 1055–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barth P., Schoeffler A., Alber T. (2008) J. Am. Chem. Soc. 130, 12038–12044 [DOI] [PubMed] [Google Scholar]
- 27.Boas F. E., Harbury P. B. (2008) J. Mol. Biol. 380, 415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Humphris E. L., Kortemme T. (2007) PLoS Comput. Biol. 3, e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benos P. V., Lapedes A. S., Stormo G. D. (2002) BioEssays 24, 466–475 [DOI] [PubMed] [Google Scholar]
- 30.Skerker J. M., Perchuk B. S., Siryaporn A., Lubin E. A., Ashenberg O., Goulian M., Laub M. T. (2008) Cell 133, 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grigoryan G., Zhou F., Lustig S. R., Ceder G., Morgan D., Keating A. E. (2006) PLoS Comput. Biol. 2, e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yanover C., Fromer M., Shifman J. M. (2007) J. Comput. Chem. 28, 2122–2129 [DOI] [PubMed] [Google Scholar]
- 33.Allen B. D., Mayo S. L. (2010) J. Comput. Chem. 31, 904–916 [DOI] [PubMed] [Google Scholar]
- 34.Georgiev I., Keedy D., Richardson J. S., Richardson D. C., Donald B. R. (2008) Bioinformatics 24, i196–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Apgar J. R., Hahn S., Grigoryan G., Keating A. E. (2009) J. Comput. Chem. 30, 2402–2413 [DOI] [PubMed] [Google Scholar]
- 36.Mandell D. J., Coutsias E. A., Kortemme T. (2009) Nat. Methods 6, 551–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy P. M., Bolduc J. M., Gallaher J. L., Stoddard B. L., Baker D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9215–9220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Havranek J. J., Baker D. (2009) Protein Sci. 18, 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boas F. E., Harbury P. B. (2007) Curr Opin. Struct. Biol. 17, 199–204 [DOI] [PubMed] [Google Scholar]