Abstract
Protein N-glycosylation and the Wnt/β-catenin signaling pathways play critical roles in development and cancer. Although N-glycosylation has been shown to influence Wnt signaling through its effects on Wnt ligands, it is unclear whether the Wnt/β-catenin pathway impacts protein N-glycosylation. In this study, we show that promoters of the first N-glycosylation gene, DPAGT1, from Chinese hamster ovary (CHO), Madin-Darby canine kidney (MDCK), and human epidermoid carcinoma (A253) cells contain the T-cell factor/lymphoid enhancer-binding factor (TCF/LEF) consensus sequence. Treatment of cells with a Wnt activator, lithium chloride, up-regulated DPAGT1 transcript levels that correlated with an increase in the β-catenin abundance. Furthermore, exposure of cells to a Wnt receptor ligand, Wnt3a, resulted in an increase in the DPAGT1 transcript levels that was abrogated by the Wnt inhibitor, Dickkopf-1. DNA mobility shift assays revealed specific protein complexes at the DPAGT1 TCF/LEF binding region that were competed off with antibodies to either Tcf3/4 or β-catenin. Chromatin immunoprecipitation analysis confirmed the presence of β-catenin at the DPAGT1 promoter in vivo. In addition, the DPAGT1 TCF/LEF sequence drove the expression of the luciferase reporter gene. Furthermore, up-regulation of DPAGT1 transcripts by Wnt3a led to altered N-glycosylation of E-cadherin. Interestingly, the DPAGT1 TCF/LEF sequence also interacted with γ-catenin, a close homologue of β-catenin, although not in a lithium chloride-dependent manner. Our results provide the first evidence that the Wnt/β-catenin signaling pathway regulates the metabolic pathway of protein N-glycosylation by targeting DPAGT1 expression. Moreover, they suggest the existence of another regulatory mechanism involving the interaction of Tcf with γ-catenin at the DPAGT1 promoter.
Keywords: Beta-Catenin, Glycosylation, Metabolism, T-cell factor (TCF), Transcription Target Genes, Wnt Pathway, DPAGT1, FOP Flash, TOP Flash, gamma-Catenin
Introduction
Protein N-glycosylation plays critical roles in development and homeostasis by affecting diverse cellular functions (1–4). In addition to the well acknowledged roles of N-glycans in protein folding, targeting, and secretion, N-glycosylation influences protein-protein interactions and cellular signaling events (1, 2, 5, 7–9). N-Glycosylation has been shown to be indispensable for development, and its dysregulation has been linked to various diseases, including cancer (3, 4, 10, 11). Expression of DPAGT1, the gene that initiates the synthesis of the lipid-linked oligosaccharide precursor for protein N-glycosylation in the endoplasmic reticulum and is a key determinant of the extent of protein N-glycosylation (12, 13), is down-regulated with cellular differentiation, suggesting that attenuation of N-glycosylation is required for the establishment of a differentiated phenotype (14). Indeed, recent studies have documented that DPAGT1 is an upstream regulator of N-glycosylation status of E-cadherin and that its diminished expression is required for the formation of mature intercellular adhesion complexes (15). Although the importance of N-glycosylation in cell and tissue function is well established, little is known about its upstream regulators.
The Wnt signaling pathway has long been known to be critical for a wide variety of developmental processes, including cell fate specification, proliferation, polarity, and migration (16). Although there are at least three Wnt signaling pathways involved in the signal transduction process, the canonical, or β-catenin-dependent, Wnt signaling pathway has been shown to be critical for cell fate decisions, proliferation, and differentiation of diverse cell types (17–19). In the canonical pathway, Wnt proteins bind to Frizzled receptors and trigger intracellular signaling cascades to activate Dishevelled family proteins (20, 21). Activation of Dishevelled inhibits the axin/GSK-3/adenomatous polyposis coli complex and prevents the phosphorylation of β-catenin at the N-terminal serine/threonine residues, leading to stabilization of its cytoplasmic pool. This enables β-catenin to enter the nucleus and interact with the Tcf2/Lef family of transcription factors to promote specific gene expression (22). Among the downstream targets of the canonical Wnt pathway are cyclin D1, c-Myc, c-Jun, and matrix metalloproteinases (16, 23, 24). Aberrant activation of the canonical Wnt pathway has been associated with a number of cancers, with inappropriate stabilization of β-catenin playing a key role in tumorigenesis (21). A close homologue of β-catenin, γ-catenin/plakoglobin, also functions as a transcriptional regulator in Wnt signaling and acts as an oncogene, although it appears to have a distinct role in Wnt signaling from that of β-catenin (25, 26).
In addition to its function in the canonical Wnt pathway, β-catenin is a key structural component of cadherin-mediated cell-cell adhesion complexes known as adherens junctions (AJs) (27, 28). Inappropriate phosphorylation of β-catenin at the tyrosine residues has been shown to lead to its dissociation from, and subsequent disassembly of, AJs (29, 30). Such diminished cadherin-mediated adhesion has been linked to cancer development and progression (10, 31, 32). Although γ-catenin has a well established role as a component of desmosomes (33), similar to β-catenin, it also plays a stabilizing role in cadherin junctions. However, little is known about the relationship between its role as an oncogene and a stabilizing component of AJs (34).
Wnts themselves are N-glycoproteins, and modification of some Wnts, like Wnt3a, with N-glycans has been shown to be required for their function and hence for the activation of the canonical Wnt pathway (35). We now report that the first N-glycosylation gene, DPAGT1, is a target of the canonical Wnt/β-catenin pathway, indicating that Wnt signaling affects protein N-glycosylation. We show that the DPAGT1 promoter from three different mammalian sources has the TCF/LEF-binding sequence that interacts with Tcf/β-catenin in vitro and in vivo. DPAGT1 transcription is activated by a stabilizer of β-catenin, lithium chloride (LiCl), and by the Frizzled ligand, Wnt3a, and is inhibited by the Wnt inhibitor, Dickkopf-1 (Dkk-1). We also show that the TCF/LEF binding region of the DPAGT1 promoter is functional, because it can drive the expression of the luciferase reporter gene. The DPAGT1 promoter also binds γ-catenin in vitro and in vivo, albeit in a manner distinct from that of β-catenin. Our studies provide the first evidence that protein N-glycosylation is regulated by the canonical Wnt signaling pathway through the DPAGT1 gene.
EXPERIMENTAL PROCEDURES
Reagents
Monoclonal antibodies to human E-cadherin, β-catenin, γ-catenin, and IgG isotype controls were obtained from BD Transduction Laboratories. Monoclonal antibody to Tcf3/4 was from Exalpha Biologicals. Polyclonal antibody to myosin II heavy chain isoform B was from Covance, and monoclonal antibody to actin (pan Ab-5, clone ACTN05) was purchased from NeoMarkers. Human, canine, and hamster biotin-derivatized DNA probes spanning the TCF binding region of the respective DPAGT1 promoters were prepared commercially (Integrated DNA Technologies). LiCl was purchased from Sigma. Rhodamine-phalloidin was obtained from Molecular Probes. Secondary antibodies included goat anti-mouse or anti-rabbit IgG derivatized with fluorescein isothiocyanate (FITC) (Molecular Probes).
Cell Culture and Preparation of Nuclear Extracts
MDCK, CHO, and A253 cells were from American Type Culture Collection and grown in McCoy's 5A, F-12K, and DMEM media, respectively, containing 10% FBS and 1% penicillin/streptomycin. In some cases, cells were treated with either 25 mm lithium chloride, Wnt3a (1 μg/ml), or Wnt antagonist Dkk-1 (1 μg/ml) for 24 h prior to isolation of RNA. To determine the effects of Wnt3a on the expression of DPAGT1, ALG1, and MGAT5 in MDCK cells, cells (passage 5) were grown to 80–90% confluence, serum-starved for 24 h, and then grown in the presence of 50% conditioned medium isolated from either L-mouse fibroblasts or L-mouse fibroblasts stably transfected with Wnt3a (ATCC) for 24 h. Total cellular RNA was extracted, reverse-transcribed, and quantitated using real time PCR. For studies of N-glycosylation of E-cadherin, MDCK cells were serum-deprived for 24 h and then grown in the presence of either 50% conditioned medium or 50% conditioned medium containing Wnt3a for 72 h. For the preparation of cell lysates, cells were extracted with 600 μl of ice-cold RIPA buffer (10 mm Tris, 150 mm NaCl, and 1% Triton X-100) containing 25 μg/ml soybean trypsin inhibitor, 100 μm benzamidine, 5 μg/ml leupeptin, and 0.5 μm PMSF. For analyses of E-cadherin N-glycosylation, whole cell lysates were prepared using Triton X-100/β-octylglucoside buffer, as described before (7). Protein concentrations were determined using the BCA protein assay (Pierce).
Plasmids
TOP Flash plasmid, which contains the wild type TCF-binding site, and FOP Flash vector, containing mutated TCF-binding sites, were purchased from Millipore. To determine functional significance of the DPAGT1 TCF sequence, three copies of the TCF-binding sequence from the human DPAGT1 promoter were cloned into the unique BamHI site upstream from the thymidine kinase promoter in the FOP Flash reporter plasmid using blunt end ligation.
Transient Transfection and Luciferase Assays
Plasmid DNA (2 μg), TOP Flash, FOP Flash, and FOP Flash containing 3× human DPAGT1 sequence (FOP DPAGT1) were transfected using Lipofectamine 2000 (Invitrogen) at 24 h after plating onto 35-mm plates. An empty pGL3-Basic vector was used as a control, and a reference plasmid, PSV-β-gal (0.1 μg, Promega), was used to normalize transfection efficiency. Luciferase assays were performed using a Luciferase kit according to the manufacturer's instructions (Promega). Briefly, cells were washed twice with PBS buffer and scraped with lysis reagent. The cells were centrifuged at 12,000 × g to pellet the debris. The cell extract was mixed with the luciferase assay reagent, and light emission was measured in a luminometer. The luciferase activity was assayed with duplicate samples within the linear range of the instrument. Values were normalized to β-galactosidase activity and to total protein as measured by the BCA assay using bovine serum albumin as a standard.
RNA Isolation and Real Time PCR
Total RNAs were extracted from CHO, A253, and MDCK cells using an RNeasy RNA isolation kit (Qiagen). Reverse transcriptase reactions for DPAGT1 were performed using a SuperScript first-strand synthesis system (Invitrogen) and SuperScript III reverse transcriptase. Reactions were carried out with ABI Prism 7300 sequence detection PCR machine (Applied Biosystems) using TaqMan gene expression system, as per the manufacturer's instructions. Statistical analysis was performed using real time PCR from three independent RNA preparations, with each experiment repeated twice (n = 6). The p values were calculated using an unpaired t test. The cDNAs from MDCK cells were also used for detecting ALG1 and MGAT5 steady state mRNA levels using gene-specific TaqMan probe and primers (Applied Biosystems). 18 S was used as an endogenous control.
Deglycosylation of E-cadherin
Total cell lysates were digested with 500 units of either PNGaseF or EndoH (New England Biolabs) for 1 h at 37 °C and analyzed by Western blot. For controls, samples were incubated without enzymes.
Western Blot
Total cell lysates (1–20 μg) were fractionated on either 10 or 7.5% SDS-PAGE and blotted onto PVDF membranes (Invitrogen). The membranes were blocked with 10% milk and incubated with primary antibodies in PBS/Tween (20 mm Tris, 137 mm NaCl, 0.1% Tween 20, pH 7.6) with 1% milk for 2 h at room temperature. Next, membranes were washed four times with PBS/Tween, followed by incubation with horseradish peroxidase-linked secondary antibody (1:3000). The results were visualized with ECL Plus detection reagents (Amersham Biosciences), and band analysis was performed using ImageJ (version 1.38) software.
Electrophoretic Mobility Shift Assay
Nuclear extracts were prepared according to a published procedure (36). Biotinylated oligonucleotides containing the human DPAGT1 promoter sequences (−336 to −319 bp) were synthesized (Integrated DNA Technologies) (Fig. 1). Complementary strands without biotin were annealed to generate a double-stranded oligonucleotide probe. For the DNA mobility shift assay, the binding reaction was performed for 30 min at room temperature in 20 μl of binding buffer containing 500 fmol of biotin-labeled probe, 1 μg of poly(dI·dC)·poly(dI·dC), and nuclear extract (3–5 μg). Double-stranded oligonucleotides (sense strand 5′-CTG GGT TGC CGG GCA ACT AAC-3′ and antisense strand 5′-GTT AGT TGC CCG GCA ACC CAG-3′) at 50-fold molar excess were used as nonspecific competitors. Separation of free radiolabeled DNA from DNA-protein complexes was carried out on a 6% nondenaturing polyacrylamide gel in a standard Tris borate electrophoresis buffer at 100 V at a 4 °C. The gels were transferred to a nitrocellulose membrane at 380 mA and cross-linked with UV light for 10 min. The membranes were blocked, incubated with stabilized streptavidin, and developed with the ECL detection reagent using a light shift assay (Pierce). In the experiments where antibodies were used to characterize the protein-DNA complex, nuclear extracts were preincubated with antibodies for 20 min at room temperature before the radiolabeled probe was added, followed by 20 min of incubation with the probe.
FIGURE 1.
Mammalian DPAGT1 promoter contains TCF/LEF-binding sequences. Promoter regions from three mammalian species, human, canine, and rodent, were scanned for TCF/LEF-binding sequences. Human and hamster TCF/LEF sequences are in the forward orientation, whereas in the canine promoter sequence it is in the reverse orientation. Numbers indicate distances from the transcription initiation sites.
Chromatin Immunoprecipitation (ChIP)
Chromatin in LiCl-treated A253 cells was cross-linked with 1% formaldehyde for 8 min at room temperature, sequentially washed with phosphate-buffered saline, solution I (10 mm HEPES, pH 7.5, 10 mm EDTA, 0.5 mm EGTA, 0.75% Triton X-100), and solution II (10 mm HEPES, pH 7.5, 200 mm NaCl, 1 mm EDTA, 0.5 mm EGTA). Cells were incubated in lysis buffer (150 mm NaCl, 25 mm Tris, pH 7.5, 1% Triton X-100, 0.1% SDS, 0.5% deoxycholate) supplemented with protease inhibitor tablet (Roche Applied Science) and PMSF. Genomic DNA was fragmented into <500-bp pieces using a Branson 250 sonicator. Aliquots of lysates containing 200 μg of protein were used for each immunoprecipitation reaction with anti-β-catenin, anti-γ-catenin (BD Biosciences), and mouse IgG antibodies followed by adsorption to protein A/G PLUS-agarose beads (Santa Cruz Biotechnology). Precipitated DNA-protein complexes were washed sequentially with RIPA buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 0.1% SDS, 0.5% deoxycholate, 1% Nonidet P-40, 1 mm EDTA), high salt buffer (50 mm Tris, pH 8.0, 500 mm NaCl, 0.1% SDS, 0.5% deoxycholate, 1% Nonidet P-40, 1 mm EDTA), LiCl buffer (50 mm Tris, pH 8.0, 250 mm LiCl, 0.1% SDS, 0.5% deoxycholate, 1% Nonidet P-40, 1 mm EDTA), and TE buffer (10 mm Tris, 1 mm EDTA pH 8.0), respectively. The DNA-protein complexes were eluted with 1% SDS and 0.1 m sodium bicarbonate solution, and DNA-protein cross-link was reversed by heating the samples to 65 °C overnight. Proteins were digested with proteinase K (Sigma), and DNA was phenol/chloroform-extracted and precipitated by 100% ethanol. DNA was dissolved in 50 μl of deionized distilled water, and 10 μl was used for each real time PCR. The primers and the probe, surrounding the Wnt-responsive element, were designed using the primer express program (Applied Biosystems). They were as follows: forward primer 5′-GCTTTAGAATTTACACCATCCTTCCA-3′, reverse primer 5′-AGCCAAGGTAAACACAACTCAGTTC-3′, and TaqMan probe 6FAMATC GTA GCT TTG TTT CCGMGBNFQ.
Microscopy, Immunofluorescence, and Imaging
For indirect immunofluorescence analyses, untreated and LiCl-treated cells were fixed in 3.7% paraformaldehyde, permeabilized with 0.1% Triton X-100, blocked with 10% goat serum, and incubated with primary antibodies to either β-catenin (0.25 μg/ml) or γ-catenin (0.25 μg/ml) at room temperature for 3 h. Cells were then incubated with FITC-tagged secondary antibodies (0.1 μg/ml) and counterstained with rhodamine-phalloidin for F-actin (1:100) and with TO-PRO-3 iodide for nuclei (1:1000), all obtained from Molecular Probes. The immunostained samples were analyzed with a Zeiss confocal laser scanning microscope LSM510 META (Fluar 5×/0.25, magnification Plan-Apochromat 40×/1.3 oil differential interference contrast). For visualization of individual optical sections and for generation of Z-stacks with optical slices of 1 μm, LSM510-expert mode acquisition software was used. To ensure valid comparison of fluorescence intensities between samples, settings were fixed to the most highly stained sample and all other images were acquired at those settings.
RESULTS
DPAGT1 Promoter Contains TCF/LEF-binding Sequences
Protein N-glycosylation has been shown to be regulated with cell proliferation and differentiation (8, 14, 37). Thus, we reasoned that as a key regulator of N-glycosylation, the DPAGT1 gene is likely a downstream target of signaling pathways with roles in development. Because Wnt/β-catenin signaling plays pivotal roles in many developmental processes, we analyzed the DPAGT1 promoter regions from three mammalian species, human, canine, and hamster, for the presence of TCF/LEF-binding sites. All three DPAGT1 promoters contained the TCF/LEF consensus sequences (Fig. 1). In human and hamster, the TCF/LEF-binding sites were in a forward orientation, although in canine, the consensus sequence was in a reverse orientation. Because TCF/LEF sequences function in both orientations, the presence of the TCF/LEF-binding sites in mammalian DPAGT1 promoters suggested that this gene was a target of the Wnt/β-catenin signaling pathway. In contrast, another gene in the N-glycosylation pathway, ALG1, that functions in the addition of the first mannose residue to the dolichol-pyrophosphate-GlcNAc2 intermediate in the endoplasmic reticulum did not contain TCF-binding sequences.
DPAGT1 Transcript Levels Are Regulated by Activators and Inhibitors of Wnt Signaling
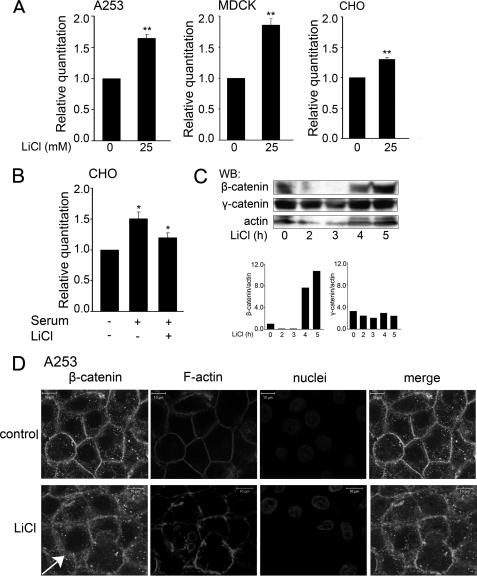
The Wnt/β-catenin signaling pathway is regulated by different input signals that serve as both its activators and inhibitors (16). Among the known activators of the canonical Wnt pathway is LiCl, which functions as an inhibitor of the GSK3β activity (38). When in complex with adenomatous polyposis coli and axin, GSK3β phosphorylates serine/threonine (Ser/Thr) residues in the N-terminal region of β-catenin, rendering it susceptible for degradation through the ubiquitin pathway. Upon inhibition of GSK3β, β-catenin accumulates in the cytoplasm and translocates to the nucleus where it interacts with TCF and activates target genes (22). To determine whether LiCl affected DPAGT1 transcript levels, we treated A253 cells with 5 and 25 mm LiCl and found a dose-dependent increase in DPAGT1 expression (data not shown). Because 25 mm LiCl had a more pronounced effect, we used this concentration in all subsequent studies. Treatment of A253, MDCK, and CHO cells with 25 mm LiCl resulted in the up-regulation of DPAGT1 transcript levels, although to different extents (Fig. 2A). The most pronounced induction was detected in MDCK cells (85%), followed by A253 cells (65%) and CHO cells (30%). These cell type variations in the LiCl induction of DPAGT1 transcription are likely to reflect cell type differences. Because LiCl has been shown to antagonize the effects of serum on the expression of early growth response genes, we examined DPAGT1 transcript levels in CHO cells treated with serum in the absence or presence of LiCl. Indeed, although serum alone up-regulated DPAGT1 transcripts by 50%, this effect was abrogated by LiCl (Fig. 2B). As expected, induction of DPAGT1 transcript levels in LiCl-treated A253 cells correlated with a 10-fold increase in β-catenin abundance within 5 h of LiCl addition (Fig. 2C). In contrast to β-catenin, 25 mm LiCl did not significantly affect the cellular levels of γ-catenin throughout the time course of the experiment (Fig. 2C). Immunofluorescence localization of β-catenin in A253 cells revealed that in addition to being present at cell-cell interfaces, it was also found in the cytoplasm. The cytosolic fraction of β-catenin was augmented in cells treated with LiCl, supporting its role as an activator of Wnt/β-catenin signaling (Fig. 2D, LiCl, arrow).
FIGURE 2.
Lithium chloride up-regulates DPAGT1 transcript levels. A, total RNAs were prepared from A253, MDCK, and CHO cells in the absence or presence of 25 mm LiCl and used for cDNA synthesis to assess DPAGT1 expression by real time RT-PCR. Gene expression profiles were generated by normalizing DPAGT1 expression with the 18 S ribosomal RNA gene and comparing it with the gene expression of untreated cells. Statistical analysis was performed using real time PCR from two independent RNA preparations, with each experiment being repeated twice (n = 4) (**, p < 0.01). B, serum counteracts the effects of LiCl on DPAGT1 expression. Total RNA lysates were prepared from CHO cells that were serum-starved for 24 h, followed by serum stimulation in the absence or presence of 25 mm LiCl. RNA lysates were used for cDNA synthesis to assess DPAGT1 expression by real time RT-PCR. Gene expression profiles were generated by normalizing DPAGT1 expression to housekeeping gene 18 S and comparing it with the gene expression of untreated cells. Statistical analysis was performed using RT-PCR from three independent RNA preparations, with each experiment being repeated twice (n = 6) (*, p < 0.05). C, Western blot (WB) analysis of β-catenin and γ-catenin expression following treatment of A253 cells with LiCl. Bar graph, fold changes in β-catenin and γ-catenin levels after addition of LiCl were determined by normalizing to actin in two different experiments. D, treatment of cells with LiCl results in an increased cytoplasmic localization of β-catenin. Untreated and LiCl-treated cells were grown in chamber slides to confluence and processed for indirect immunofluorescence staining using an antibody to β-catenin. Cells were counterstained for F-actin with rhodamine-phalloidin and for nuclei with TO-PRO-3 iodide. In control cells, β-catenin displayed more cell-cell border staining compared with LiCl-treated cells, where it was more cytoplasmic (arrow). Shown are 1-μm confocal x-y sections. Size bars, 10 μm. These data are representative of two different experiments.
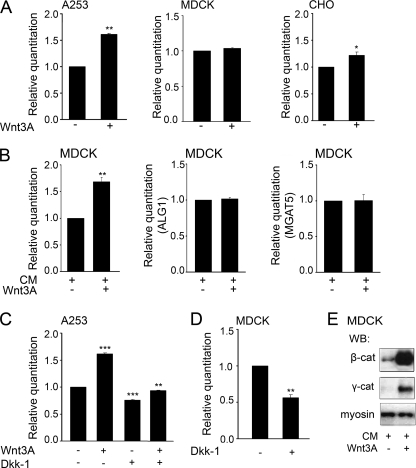
To confirm that the effects of LiCl on DPAGT1 expression were a consequence of the activation of the canonical Wnt pathway, we used one of the receptor ligands of Frizzled, Wnt3a, to activate Wnt signaling. Treatment of A253 cells with 3 μg of Wnt3a resulted in a 60% increase in the DPAGT1 transcript levels, suggesting that the Wnt pathway was an upstream activator of this gene (Fig. 3A). In CHO cells, Wnt3a caused a modest (22%) induction of DPAGT1 transcript abundance (Fig. 3A). In contrast, addition of Wnt3a had no discernable effect on the DPAGT1 transcript levels in MDCK cells. Because Wnt3a was added to cells at a single time point, the observed lack of response in MDCK cells was likely to reflect experimental conditions. MDCK cells have been shown to be sensitive to Wnt3a in co-cultures with NIH-3T3 cells expressing Wnts (39). Therefore, we tested whether growth of MDCK cells in the presence of 50% conditioned medium isolated from either L-mouse fibroblasts or L-mouse fibroblasts stably transfected with Wnt3a would lead to the induction of the DPAGT1 transcript. As shown in Fig. 3B, following growth for 24 h with 50% conditioned medium containing Wnt3a, DPAGT1 transcript levels increased by 70%. For a negative control, we used the N-glycosylation gene, ALG1, which functions downstream from DPAGT1 in the dolichol pathway in the endoplasmic reticulum and encodes the first mannosyltransferase and which lacks the TCF/LEF-binding sequence in its promoter. As expected, Wnt3a did not induce the ALG1 transcript (Fig. 3B). We also examined transcript levels of another N-glycosylation gene, MGAT5, which encodes N-acetylglucosaminyltransferase V (Mgat5 or GnT-V) that functions in the addition of branched N-glycans in the Golgi. The MGAT5 promoter has four sequences resembling TCF/LEF, although with different degrees of fidelity. They include a sequence beginning at −192 bp from the transcription start site with two mismatches, a site at −410 with one mismatch, a site at −1312 bp with one mismatch, and an exact TCF/LEF sequence at −1596 bp. Under the conditions of our study, however, Wnt3a did not induce MGAT5 transcript levels (Fig. 3B).
FIGURE 3.
Transcriptional expression of DPAGT1 is stimulated by Wnt3a and abrogated by Dkk-1. A, total RNAs isolated from A253, MDCK, and CHO cells treated with Wnt3a were used for cDNA synthesis to assess DPAGT1 expression by real time RT-PCR. Gene expression profiles were generated by normalizing DPAGT1 expression with housekeeping gene 18 S and comparing it with the gene expression of untreated cells. Statistical analysis was performed using real time PCR from three independent RNA preparations, with each experiment being repeated twice (n = 6) (*, p < 0.05; **, p < 0.01). Results represent one of three independent experiments. B, total RNAs were isolated from MDCK cells grown in CM in the presence or absence of Wnt3a. RNAs were used for cDNA synthesis to assess DPAGT1, ALG1, and MGAT5 expression by real time RT-PCR. Gene expression profiles were generated by normalizing DPAGT1, ALG1, and MGAT5 expression with housekeeping gene 18 S and comparing it with the gene expression of untreated cells. Statistical analysis was performed using real time PCR from three independent RNA preparations, with each experiment being repeated twice (n = 6) (**, p < 0.01). Results represent one of three independent experiments. C, total RNA lysates were prepared from A253 cells treated with either with Wnt3a, Dkk-1, or both and used for cDNA synthesis to assess DPAGT1 expression by RT-PCR. Gene expression profiles were generated by normalizing DPAGT1 expression with housekeeping gene 18 S and comparing it with the gene expression of untreated cells. Statistical analysis was performed using RT-PCR from three independent RNA preparations, with each experiment being repeated twice (n = 6) (**, p < 0.01; ***, p < 0.001). D, total RNAs were isolated from MDCK cells treated with Dkk-1 and used for cDNA synthesis to assess DPAGT1 expression by RT-PCR. Gene expression profiles were generated by normalizing DPAGT1 expression with housekeeping gene 18 S and comparing it with the gene expression of untreated cells. Statistical analysis was performed using real time PCR from three independent RNA preparations, with each experiment being repeated twice (n = 6) (**, p < 0.01). E, equal amounts of total cell lysate protein from MDCK cells, grown in CM in the presence or absence of Wnt3a, were analyzed for β-catenin and γ-catenin expression by Western blot (WB). Myosin served as a loading control.
In A253 cells, the Wnt3a-dependent increase in the DPAGT1 transcript levels was abrogated by the Wnt inhibitor, Dkk-1 (Fig. 3C). Treatment of A253 cells with Dkk-1 alone caused a 24% down-regulation of DPAGT1 transcripts, suggesting the existence of endogenous Wnt activity in these cells (Fig. 3C). Likewise, MDCK cells were sensitive to Dkk-1 treatment, which reduced DPAGT1 transcripts by 56% (Fig. 3D).
Similar to LiCl-treated A253 cells, incubation of MDCK cells with 50% conditioned medium containing Wnt3a was associated with an increased abundance of β-catenin (Fig. 3E, β-cat). The steady-state levels of γ-catenin were also augmented in Wnt3a-treated cells, although to a much lesser degree than β-catenin (Fig. 3E, γ-cat).
TCF/LEF-binding Sites in the DPAGT1 Promoter Interact with Tcf and β-Catenin in Vitro and in Vivo
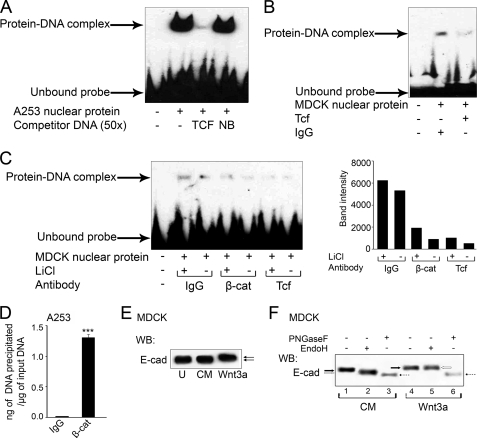
To validate the effect of Wnt activation on DPAGT1 expression, we examined the DPAGT1 TCF/LEF sequence for the ability to bind the Tcf-β-catenin protein complex. When a biotinylated human DPAGT1 promoter sequence spanning the TCF-binding sites was tested for interaction with nuclear extracts from A253 cells, a robust protein-DNA complex was detected (Fig. 4A). The complex was competed off by the TCF sequence but not by an unrelated sequence from the DPAGT1 promoter (Fig. 4A). Similar results were obtained with nuclear extracts from MDCK and CHO cells (data not shown). To determine whether Tcf and β-catenin proteins were present in complex with the TCF binding region of the DPAGT1 promoter, we carried out competition studies with antibodies to Tcf and β-catenin. Incubation of nuclear extracts from MDCK cells with the Tcf-specific antibody greatly diminished the abundance of the protein-DNA complex compared with the nuclear extracts incubated in the presence of IgG alone (Fig. 4B). Furthermore, the levels of protein-DNA complexes were greater when nuclear extracts were isolated from MDCK cells treated with LiCl compared with untreated cells (Fig. 4C, IgG). The abundance of protein-DNA complexes was diminished by competition with β-catenin and Tcf, either in the presence or absence of LiCl (Fig. 4C, IgG versus β-cat versus Tcf). As expected, LiCl did not affect the abundance of the protein-DNA complex in the competition study with the Tcf antibody (Fig. 4C, Tcf). Because both Tcf and β-catenin were able to compete off the nuclear complex at the TCF/LEF-binding site, this indicated that β-catenin interacted with Tcf at this site in the DPAGT1 promoter in vitro.
FIGURE 4.
TCF/LEF-binding sequence in the DPAGT1 promoter interacts with TCF and β-catenin in vitro and in vivo. A, DNA shift assay was performed using the nuclear extract (5 μg) from A253 cells that was incubated with a biotinylated (−336 to −319) sequence from the human DPAGT1 promoter. Where indicated, competitor TCF or unrelated probes (NB) were used (3rd and 4th lanes). Data shown represent one of four independent experiments. B, DNA mobility shift assay was carried out with a nuclear protein extract (5 μg) from MDCK cells and a canine-biotinylated (−244 to −221) DPAGT1 promoter sequence in the presence of either IgG (1 μg) or Tcf antibody (1 μg). Results are one of three independent experiments. C, gel shift assay was performed with a nuclear protein extract (5 μg) from MDCK cells, either untreated or treated with LiCl, and a canine biotinylated DPAGT1 promoter sequence in the presence of either IgG (1 μg), β-catenin (β-cat) antibody (1 μg), or TCF antibody (1 μg). These results are one of two independent experiments. Bar graph, band intensities were calculated in comparison to the IgG control in the presence of LiCl. The data are representative of three independent experiments. D, ChIP assay was performed on chromatin samples from A253 cells using either an antibody against β-catenin or IgG (negative control). Real time PCRs were carried out using primers and the TaqMan probe specific for the promoter region of DPAGT1. An unrelated sequence was used as a negative control. E, equal amounts of total cell lysate protein from untreated MDCK cells (U), and cells grown in CM in the presence or absence of Wnt3a, were analyzed for E-cadherin expression by WB. F, total cell lysates from cells, grown in CM in the presence or absence of Wnt3a, were treated with EndoH and PNGaseF and analyzed by Western blot (WB). The results depict one of three independent experiments. E-cad, E-cadherin.
To determine whether the Tcf-β-catenin complex was also present at the DPAGT1 promoter in vivo, we carried out ChIP assays with chromatin isolated from A253 cells treated with LiCl and an antibody to β-catenin. Relative to the IgG control, there was a 60-fold increase in the amount of β-catenin bound to the DPAGT1 TCF/LEF sequence (Fig. 4D). We conclude that the TCF/LEF-binding site in the DPAGT1 promoter interacts with Tcf and β-catenin in vitro and in vivo.
Up-regulation of DPAGT1 by Wnt3a Leads to Altered N-Glycosylation of E-cadherin
Our previous studies have shown that DPAGT1 impacts E-cadherin N-glycosylation, both quantitatively and qualitatively (10, 15). High levels of DPAGT1 expression correlate with extensive N-glycosylation of E-cadherin with complex N-glycans, whereas down-regulation of DPAGT1 leads to reduced N-glycosylation status of E-cadherin concomitant with the modification with high mannose/hybrid N-glycans. To align the Wnt-dependent transcriptional activation of DPAGT1 with N-glycosylation of E-cadherin, we examined sensitivity of E-cadherin to PNGaseF and EndoH following the induction of DPAGT1 with Wnt3a. PNGaseF is an amidase that removes most N-glycans from N-glycoproteins, although EndoH is an endoglycosidase that removes high mannose/hybrid N-glycans. Western blot analyses showed that E-cadherin from MDCK cells grown in the presence of 50% conditioned medium (CM) containing Wnt3a for 72 h migrated with a higher molecular size compared with E-cadherin from control cells, grown in the presence of 50% conditioned medium without Wnt3a or from untreated cells (Fig. 4E). PNGaseF treatment of E-cadherin from Wnt3a cells resulted in mobility shift that reduced its molecular size to that of PNGaseF-treated E-cadherin from CM cells (Fig. 4F, lanes 3 and 6, dashed arrows). Moreover, E-cadherin from control cells was sensitive to EndoH, indicating that, in addition to complex N-glycans, it had high mannose/hybrid N-glycans (Fig. 4F, lanes 1 and 2, arrows). In contrast, E-cadherin from Wnt3a-treated cells exhibited little sensitivity to EndoH (Fig. 4F, lanes 4 and 5, arrows), resembling the scenario of E-cadherin from proliferating MDCK cells and A253 cancer cells, where it is modified primarily with PNGaseF-sensitive, complex N-glycans (7, 10). We conclude that up-regulation of DPAGT1 in response to Wnt3a treatment results in increased N-glycosylation of E-cadherin with complex N-glycans.
TCF Sequence from the DPAGT1 Promoter Drives Expression of the Luciferase Reporter Gene
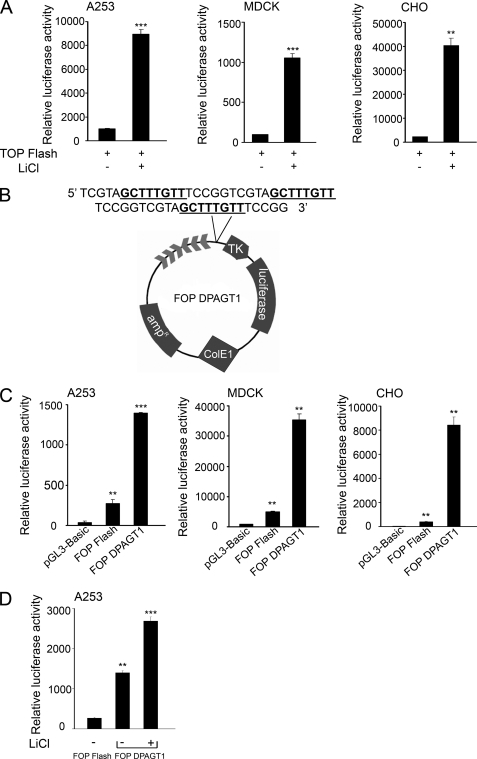
The results thus far strongly suggested that DPAGT1 expression was regulated in response to Wnt/β-catenin signaling through the binding of Tcf/β-catenin to the TCF/LEF sequence in its promoter. To validate that the DPAGT1 TCF/LEF promoter sequence was biologically functional, we examined whether it could influence expression of a luciferase reporter. As expected, transfection of CHO, MDCK, and A253 cells with a luciferase reporter vector containing three wild type TCF/LEF-binding sites (TOP Flash) caused a robust increase (10–20-fold) in the luciferase activity upon Wnt stimulation with LiCl (Fig. 5A). We next transfected these cells with either a vector that had three mutant TCF/LEF-binding sites (FOP Flash) or with a FOP Flash vector bearing three TCF/LEF-binding sites from the DPAGT1 promoter (FOP DPAGT1) upstream of the thymidine kinase promoter sequence (Fig. 5B). All three cell lines transfected with the FOP DPAGT1 vector showed significantly augmented luciferase activity compared with cells transfected with either FOP Flash alone or a negative control plasmid, pGL3-Basic, lacking TCF-binding sites (Fig. 5C). The variations in the luciferase activities among the cell lines most likely reflected the differences in cell context. Moreover, A253 cells transfected with FOP DPAGT1 displayed a further increase in the luciferase activity following stimulation with LiCl (Fig. 5D). These results confirmed that the Wnt/β-catenin pathway was a direct transcriptional activator of the DPAGT1 gene.
FIGURE 5.
TCF/LEF-binding sequence from the DPAGT1 promoter drives luciferase reporter gene expression. A, luciferase reporter gene under the regulation of three tandem copies of TCF/LEF-responsive elements in the TOP Flash vector was transfected into either A253, MDCK, or CHO cells, and cells were grown for 24 h in the absence or presence of LiCl, and extracts were analyzed for luciferase activity after 48 h. Error bars indicate S.E. (**, p < 0.01; ***, p < 0.001). B, schematic of FOP DPAGT1, the FOP Flash vector with three copies of the TCF/LEF-responsive element from the DPAGT1 promoter upstream of the luciferase reporter gene. C, either FOP DPAGT1, FOP Flash, or pGL3-Basic were transfected into A253, MDCK, or CHO cells, and cells were grown for 48 h. Extracts were analyzed for luciferase activity. Error bars indicate S.E. (**, p < 0.01; ***, p < 0.001). D, FOP DPAGT1 was transfected into A253 cells, and cells were grown in the presence of LiCl for 24 h. FOP Flash was used as a negative control. Transfections were normalized using the Psv-β-galactosidase control vector. Extracts were analyzed for luciferase activity after 48 h. The values depicted are S.E. of three independent experiments performed in duplicate (n = 6) (**, p < 0.01; ***, p < 0.001).
TCF/LEF-binding Site in the DPAGT1 Promoter Interacts with γ-Catenin
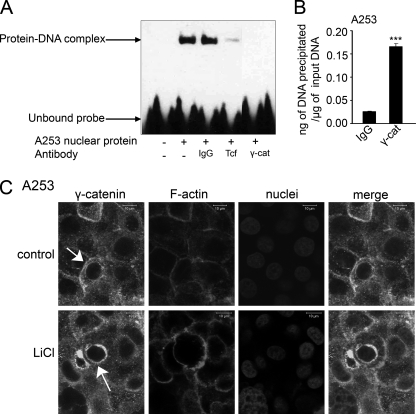
Another protein with the Tcf/Lef binding domain highly homologous to β-catenin is γ-catenin/plakoglobin. Although γ-catenin has been shown to interact with Tcf/Lef, its binding pattern differs from that of β-catenin (40). Thus, we sought to determine whether γ-catenin could also interact with the DPAGT1 TCF/LEF binding region. Antibodies to γ-catenin were effective in competing off the protein-DNA complex at the DPAGT1 TCF/LEF-binding site compared with IgG (Fig. 6A), indicating that γ-catenin was able to interact with this site in vitro. To assess if binding of γ-catenin to the DPAGT1 promoter occurred in vivo, we performed a ChIP assay with chromatin isolated from A253 cells treated with LiCl and an antibody to γ-catenin. As shown in Fig. 6B, there was a 7-fold increase in the amount of γ-catenin bound to the DPAGT1 TCF/LEF sequence compared with the IgG control. However, in contrast to β-catenin, this binding of γ-catenin to the DPAGT1 promoter was not inducible by LiCl (Fig. 2C). On the other hand, there was a detectable increase in the abundance of γ-catenin after stimulation with Wnt3a (Fig. 3E), confirming its role in Wnt signaling (41). Immunofluorescence staining of γ-catenin in A253 cells revealed that its cytoplasmic localization was not impacted by treatment with LiCl (Fig. 6C). Nonetheless, in A253 cells, γ-catenin displayed more pronounced cytoplasmic localization than β-catenin (Fig. 2). Interestingly, a significant fraction of γ-catenin was detected in a perinuclear region (Fig. 6C, arrow). These results suggest that γ-catenin interacts with the DPAGT1 promoter through a Wnt-dependent mechanism distinct from that of β-catenin.
FIGURE 6.
DPAGT1 promoter binds γ-catenin in vitro and in vivo. A, DNA mobility shift assay was carried out with a nuclear protein extract (5 μg) from A253 cells and a human DPAGT1 promoter sequence (−336 to −319) in the presence of either IgG (1 μg), Tcf antibody (1 μg), or γ-catenin (γ-cat) antibody (1 μg). Results are one of three independent experiments. B, ChIP analysis using an antibody against γ-catenin was performed on chromatin samples from A253 cells. Real time PCR was carried out using primers and the TaqMan probe specific for the promoter region of DPAGT1. Genomic DNA precipitated by IgG was used as a negative control. Statistical analysis was performed using real time PCR from two independent RNA preparations, with each experiment being repeated twice (n = 4) (**, p < 0.01; ***, p < 0.001). C, untreated and LiCl-treated cells were grown in chamber slides to confluence and processed for indirect immunofluorescence staining using an antibody to γ-catenin. Cells were counterstained for F-actin with rhodamine-phalloidin and for nuclei with TO-PRO-3 iodide. Shown are 1 μm confocal x-y sections. Size bars, 10 μm. These data are one of two independent experiments.
DISCUSSION
Our studies reveal that the first N-glycosylation gene, DPAGT1, is a target of the Wnt/β-catenin signaling pathway. Under conditions of Wnt activation, DPAGT1 transcription is up-regulated though the binding of β-catenin to Tcf at the TCF/LEF-binding site in the DPAGT1 promoter. Because Wnt signaling plays pivotal roles in cell proliferation and differentiation, and because DPAGT1 and the N-glycosylation status of proteins are regulated with growth and differentiation, our findings provide, at least in part, a molecular explanation for these observations.
Treatment of nonmalignant CHO and MDCK cells, as well as A253 cancer cells, with a β-catenin stabilizer, LiCl, resulted in an up-regulation of DPAGT1 transcript levels concomitant with an increase in the cytoplasmic pool of β-catenin (Fig. 2). Similar up-regulation of DPAGT1 transcription was obtained upon the addition of a Frizzled ligand, Wnt3a, and this effect was abrogated by the Wnt inhibitor, Dkk-1 (Fig. 3).
DNA binding assays using the TCF/LEF binding region from the DPAGT1 promoter validated this finding by showing the presence of both Tcf and β-catenin in vitro (Fig. 4). Likewise, the ChIP assay confirmed the presence of β-catenin at the DPAGT1 promoter in vivo (Fig. 4D). Finally, the TCF/LEF region from the DPAGT1 promoter was shown to drive the expression of the luciferase reporter gene in all three cell lines (Fig. 5). Collectively, our studies provide the first evidence that DPAGT1 is regulated by Wnt/β-catenin signaling.
We note that another key N-glycosylation gene, MGAT5, which functions in the Golgi in the addition of branched N-glycans (1) and which has been recognized to promote cancer metastasis (8, 9, 43, 44), is not a target of the Wnt/β-catenin pathway (Fig. 3B).
Interestingly, LiCl abrogated the up-regulation of DPAGT1 expression by serum (Fig. 2B). To date, the antagonistic effect of Wnt activation on serum response has been reported for early growth response genes (45). Although DPAGT1 has been shown to behave like an early growth response gene in yeast, in mammalian systems it does not respond to mitogenic stimulation in a manner characteristic of this class of genes.3 Thus, our findings extend the antagonistic effects of Wnt activation on mitogenic stimulation to a broader category of genes.
The Wnt/β-catenin pathway has been increasingly recognized to play critical roles in cellular metabolism (46). Specifically, Wnt signaling has been implicated in a number of distinct metabolic processes, including amino acid, lipid, and glucose sensing (47). Through its effects on expression of key metabolic proteins, genes, and transcription factors, Wnt signaling is thought to coordinate the metabolic flux under normal physiological conditions. In some cases, Wnt signaling regulates metabolism via its effects on a post-translational level. For instance, Wnt regulates a cluster of glycolytic enzymes, comprised of aldolase, phosphofructokinase, and hexokinase via the GSK-Axin destruction complex. In macrophages, activation of the Wnt/β-catenin pathway has been shown to depend on glucose availability and the hexose biosynthetic pathway. Moreover, N-glycosylation itself plays important roles in glucose sensing, as the initial substrate for the DPAGT1 gene product, N-acetylglucosamine-1-phosphate transferase, is UDP-GlcNAc, a product to the hexosamine pathway (47).
Although our studies show that Wnt/β-catenin signaling impacts N-glycosylation via DPAGT1, N-glycosylation itself has been shown to affect Wnt signaling via modification of Wnt ligands (35). Because DPAGT1 determines the N-glycosylation status of proteins (12, 13), and because Wnts require N-glycosylation for activity (35), the Wnt/β-catenin pathway is likely to be DPAGT1-dependent.
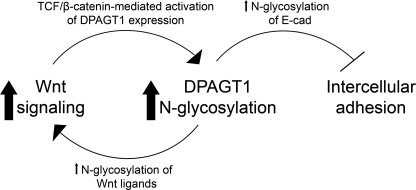
An additional interaction between Wnt/β-catenin signaling and N-glycosylation occurs at the level of E-cadherin-mediated cell-cell adhesion (48). DPAGT1 expression controls the N-glycosylation status of E-cadherin, which, in turn, impacts the molecular organization of AJs, tight junctions, and their cytoskeletal interactions (7, 15, 49). This study shows that the Wnt/β-catenin-dependent up-regulation of DPAGT1 transcription leads to altered N-glycosylation of E-cadherin, producing a shift from a mixture of high mannose/hybrid and complex N-glycans to almost exclusively complex structures (Fig. 4, E and F). This suggests that in addition to controlling the extent of protein N-glycosylation, DPAGT1 affects the types of N-glycans. We and others have shown that extensive N-glycosylation of E-cadherin with complex N-glycans inhibits cell-cell adhesion (7, 50). Thus, our present findings suggest that DPAGT1 coordinates pro-proliferative effects of Wnt/β-catenin signaling with weak cell-cell adhesion. The schematic depicting how DPAGT1 integrates Wnt signaling with E-cadherin adhesion is shown in Fig. 7. Furthermore, E-cadherin itself is a downstream target of Wnt/β-catenin signaling, which represses its expression on a transcriptional level via Twist, Snail, and Slug (6, 51, 52). Thus, activation of Wnt/β-catenin signaling suppresses intercellular adhesion on two levels as follows: through up-regulation of DPAGT1, leading to extensive N-glycosylation of E-cadherin, and through transcriptional inhibition of E-cadherin. In addition, functional AJs require interaction between E-cadherin and β-catenin, and once dissociated from AJs, β-catenin can feed into the Wnt pathway. Such a scenario may be the underlying cause of many epithelial cancers (27, 42).
FIGURE 7.
Schematic illustration of how DPAGT1 coordinates Wnt signaling with E-cadherin adhesion. In response to Wnt activation, β-catenin is stabilized and translocates to the nucleus where it binds to Tcf at the DPAGT1 promoter and activates DPAGT1 expression. Increased expression of DPAGT1 promotes N-glycosylation of Wnt ligands and Wnt activity, which further up-regulates DPAGT1 expression. A Wnt-dependent increase in DPAGT1 expression leads to an increased N-glycosylation of E-cadherin (E-cad) with complex N-glycans, which interferes with intercellular adhesion.
Our present studies also suggest that γ-catenin is a potential transcriptional activator of DPAGT1 through a mechanism that is different from that of β-catenin (Fig. 6). Although both β-catenin and γ-catenin interact with E-cadherin in a mutually exclusive manner, their association with hypoglycosylated E-cadherin drives the interaction of AJs with distinct cytoskeletal components and with different functional outcomes (49). We have shown before that the DPAGT1-dependent remodeling of AJs involves changes in the association of γ-catenin with E-cadherin scaffolds, with mature hypoglycosylated AJs recruiting more γ-catenin where it acts as a junctional stabilizer (7, 10, 15). The abundance of γ-catenin is prominently diminished in immature, highly N-glycosylated AJs from a subset of oral squamous cell carcinoma tissues overexpressing DPAGT1 (10). In light of our present results with A253 cells, which also overexpress DPAGT1 and form weak AJs, it is possible that in cancer tissues, γ-catenin is not sequestered by immature AJs, which allows it to accumulate in the cytoplasm and to translocate to the nucleus where it up-regulates DPAGT1. Interestingly, in A253 cells, γ-catenin appeared to accumulate in the perinuclear region, suggesting that its entry to the nucleus may be a regulated, multistep process (Fig. 6C). Collectively, our studies provide evidence that both β-catenin and γ-catenin may serve as links coordinating N-glycosylation with Wnt and intercellular adhesion, although in different cellular contexts.
In addition to E-cadherin, the Wnt/β-catenin pathway interacts with a number of other signaling networks, including Notch, Hedgehog, transforming growth factor β (TGFβ), and bone morphogenetic protein signaling (16). To date, at least 80 target genes of Wnt/β-catenin signaling have been identified. Some of the prominent transcriptional targets include the genes that encode cyclin D1, c-Myc, c-Jun, mitochondrial cytochrome oxidase II, E-cadherin, fibronectin, and matrix metalloproteinases (46). Notably absent among these transcriptional targets are the genes involved in cellular metabolism. Our studies now add DPAGT1, a key gene in the metabolic pathway of protein N-glycosylation, to the list of target genes of the canonical Wnt/β-catenin pathway.
This work was supported by National Institutes of Health Grants RO1 DE010183 and RO1 DE015304 from NIDCR (to M. A. K.).
P. K. Sengupta and M. A. Kukuruzinska, unpublished observations.
- Tcf
- T-cell factor
- AJ
- adherens junctions
- ChIP
- chromatin immunoprecipitation
- CHO
- Chinese hamster ovary
- CM
- conditioned medium
- LEF
- lymphoid enhancer-binding factor
- MDCK
- Madin-Darby canine kidney
- N-glycans
- asparagine-linked oligosaccharides
- EndoH
- endoglycosidase H
- PNGase
- peptide:N-glycosidase.
REFERENCES
- 1.Helenius A., Aebi M. (2001) Science 291, 2364–2369 [DOI] [PubMed] [Google Scholar]
- 2.Kophengnavong T., Michnowicz J. E., Blackwell T. K. (2000) Mol. Cell. Biol. 20, 261–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marek K. W., Vijay I. K., Marth J. D. (1999) Glycobiology 9, 1263–1271 [DOI] [PubMed] [Google Scholar]
- 4.Ioffe E., Stanley P. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 728–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Y. Y., Takahashi M., Gu J. G., Miyoshi E., Matsumoto A., Kitazume S., Taniguchi N. (2008) Cancer Sci. 99, 1304–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rademacher T. W., Parekh R. B., Dwek R. A. (1988) Annu. Rev. Biochem. 57, 785–838 [DOI] [PubMed] [Google Scholar]
- 7.Liwosz A., Lei T., Kukuruzinska M. A. (2006) J. Biol. Chem. 281, 23138–23149 [DOI] [PubMed] [Google Scholar]
- 8.Lau K. S., Partridge E. A., Grigorian A., Silvescu C. I., Reinhold V. N., Demetriou M., Dennis J. W. (2007) Cell 129, 123–134 [DOI] [PubMed] [Google Scholar]
- 9.Guo H. B., Lee I., Kamar M., Pierce M. (2003) J. Biol. Chem. 278, 52412–52424 [DOI] [PubMed] [Google Scholar]
- 10.Nita-Lazar M., Noonan V., Rebustini I., Walker J., Menko A. S., Kukuruzinska M. A. (2009) Cancer Res. 69, 5673–5680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dennis J. W., Granovsky M., Warren C. E. (1999) Biochim. Biophys. Acta 1473, 21–34 [DOI] [PubMed] [Google Scholar]
- 12.Mendelsohn R. D., Helmerhorst E. J., Cipollo J. F., Kukuruzinska M. A. (2005) Biochim. Biophys. Acta 1723, 33–44 [DOI] [PubMed] [Google Scholar]
- 13.Wu X., Rush J. S., Karaoglu D., Krasnewich D., Lubinsky M. S., Waechter C. J., Gilmore R., Freeze H. H. (2003) Hum. Mutat. 22, 144–150 [DOI] [PubMed] [Google Scholar]
- 14.Fernandes R. P., Cotanche D. A., Lennon-Hopkins K., Erkan F., Menko A. S., Kukuruzinska M. A. (1999) Histochem. Cell Biol. 111, 153–162 [DOI] [PubMed] [Google Scholar]
- 15.Nita-Lazar M., Rebustini I., Walker J., Kukuruzinska M. A. (2010) Exp. Cell Res. 316, 1871–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Amerongen R., Nusse R. (2009) Development 136, 3205–3214 [DOI] [PubMed] [Google Scholar]
- 17.Logan C. Y., Nusse R. (2004) Annu. Rev. Cell Dev. Biol. 20, 781–810 [DOI] [PubMed] [Google Scholar]
- 18.MacDonald B. T., Tamai K., He X. (2009) Dev. Cell 17, 9–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Angers S., Moon R. T. (2009) Nat. Rev. Mol. Cell Biol. 10, 468–477 [DOI] [PubMed] [Google Scholar]
- 20.Tamai K., Zeng X., Liu C., Zhang X., Harada Y., Chang Z., He X. (2004) Mol. Cell 13, 149–156 [DOI] [PubMed] [Google Scholar]
- 21.Nusse R. (2005) Cell Res. 15, 28–32 [DOI] [PubMed] [Google Scholar]
- 22.Gordon M. D., Nusse R. (2006) J. Biol. Chem. 281, 22429–22433 [DOI] [PubMed] [Google Scholar]
- 23.He T. C., Sparks A. B., Rago C., Hermeking H., Zawel L., da Costa L. T., Morin P. J., Vogelstein B., Kinzler K. W. (1998) Science 281, 1509–1512 [DOI] [PubMed] [Google Scholar]
- 24.Tetsu O., McCormick F. (1999) Nature 398, 422–426 [DOI] [PubMed] [Google Scholar]
- 25.Shimizu M., Fukunaga Y., Ikenouchi J., Nagafuchi A. (2008) Mol. Cell. Biol. 28, 825–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shim J. S., Kim D. H., Kwon H. J. (2004) Oncogene 23, 1704–1711 [DOI] [PubMed] [Google Scholar]
- 27.Brembeck F. H., Rosário M., Birchmeier W. (2006) Curr. Opin. Genet. Dev. 16, 51–59 [DOI] [PubMed] [Google Scholar]
- 28.Jamora C., Fuchs E. (2002) Nat. Cell Biol. 4, 101–108 [DOI] [PubMed] [Google Scholar]
- 29.Chen K. H., Tung P. Y., Wu J. C., Chen Y., Chen P. C., Huang S. H., Wang S. M. (2008) Cancer Lett. 267, 37–48 [DOI] [PubMed] [Google Scholar]
- 30.Gottardi C. J., Gumbiner B. M. (2004) J. Cell Biol. 167, 339–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheon S. S., Cheah A. Y., Turley S., Nadesan P., Poon R., Clevers H., Alman B. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 6973–6978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soler C., Grangeasse C., Baggetto L. G., Damour O. (1999) FEBS Lett. 442, 178–182 [DOI] [PubMed] [Google Scholar]
- 33.Green K. J., Simpson C. L. (2007) J. Invest. Dermatol. 127, 2499–2515 [DOI] [PubMed] [Google Scholar]
- 34.Chidgey M., Dawson C. (2007) Br. J. Cancer 96, 1783–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Komekado H., Yamamoto H., Chiba T., Kikuchi A. (2007) Genes Cells 12, 521–534 [DOI] [PubMed] [Google Scholar]
- 36.Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kukuruzinska M. A., Lennon-Hopkins K. (1999) Biochim. Biophys. Acta 1426, 359–372 [DOI] [PubMed] [Google Scholar]
- 38.Klein P. S., Melton D. A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 8455–8459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyons J. P., Mueller U. W., Ji H., Everett C., Fang X., Hsieh J. C., Barth A. M., McCrea P. D. (2004) Exp. Cell Res. 298, 369–387 [DOI] [PubMed] [Google Scholar]
- 40.Zhurinsky J., Shtutman M., Ben-Ze'ev A. (2000) Mol. Cell. Biol. 20, 4238–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhurinsky J., Shtutman M., Ben-Ze'ev A. (2000) J. Cell Sci. 113, 3127–3139 [DOI] [PubMed] [Google Scholar]
- 42.Heuberger J., Birchmeier W. (2010) Cold Spring Harbor Perspect. Biol. 2, a002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo H. B., Randolph M., Pierce M. (2007) J. Biol. Chem. 282, 22150–22162 [DOI] [PubMed] [Google Scholar]
- 44.Dennis J. W., Laferté S., Waghorne C., Breitman M. L., Kerbel R. S. (1987) Science 236, 582–585 [DOI] [PubMed] [Google Scholar]
- 45.Tice D. A., Soloviev I., Polakis P. (2002) J. Biol. Chem. 277, 6118–6123 [DOI] [PubMed] [Google Scholar]
- 46.Sethi J. K., Vidal-Puig A. (2010) Biochem. J. 427, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anagnostou S. H., Shepherd P. R. (2008) Biochem. J. 416, 211–218 [DOI] [PubMed] [Google Scholar]
- 48.Nelson W. J., Nusse R. (2004) Science 303, 1483–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jamal B. T., Nita-Lazar M., Gao Z., Amin B., Walker J., Kukuruzinska M. A. (2009) Cell Health Cytoskeleton 1, 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vagin O., Tokhtaeva E., Yakubov I., Shevchenko E., Sachs G. (2008) J. Biol. Chem. 283, 2192–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yook J. I., Li X. Y., Ota I., Fearon E. R., Weiss S. J. (2005) J. Biol. Chem. 280, 11740–11748 [DOI] [PubMed] [Google Scholar]
- 52.Bolós V., Peinado H., Pérez-Moreno M. A., Fraga M. F., Esteller M., Cano A. (2003) J. Cell Sci. 116, 499–511 [DOI] [PubMed] [Google Scholar]