Abstract
Vitamin D3 hydroxylase (Vdh) isolated from actinomycete Pseudonocardia autotrophica is a cytochrome P450 (CYP) responsible for the biocatalytic conversion of vitamin D3 (VD3) to 1α,25-dihydroxyvitamin D3 (1α,25(OH)2VD3) by P. autotrophica. Although its biological function is unclear, Vdh is capable of catalyzing the two-step hydroxylation of VD3, i.e. the conversion of VD3 to 25-hydroxyvitamin D3 (25(OH)VD3) and then of 25(OH)VD3 to 1α,25(OH)2VD3, a hormonal form of VD3. Here we describe the crystal structures of wild-type Vdh (Vdh-WT) in the substrate-free form and of the highly active quadruple mutant (Vdh-K1) generated by directed evolution in the substrate-free, VD3-bound, and 25(OH)VD3-bound forms. Vdh-WT exhibits an open conformation with the distal heme pocket exposed to the solvent both in the presence and absence of a substrate, whereas Vdh-K1 exhibits a closed conformation in both the substrate-free and substrate-bound forms. The results suggest that the conformational equilibrium was largely shifted toward the closed conformation by four amino acid substitutions scattered throughout the molecule. The substrate-bound structure of Vdh-K1 accommodates both VD3 and 25(OH)VD3 but in an anti-parallel orientation. The occurrence of the two secosteroid binding modes accounts for the regioselective sequential VD3 hydroxylation activities. Moreover, these structures determined before and after directed evolution, together with biochemical and spectroscopic data, provide insights into how directed evolution has worked for significant enhancement of both the VD3 25-hydroxylase and 25(OH)VD3 1α-hydroxylase activities.
Keywords: Crystal Structure, Cytochrome P450, Heme, Hydroxylase, Molecular Dynamics, Vitamin D, Biocatalysis, Biotransformation, Directed Evolution
Introduction
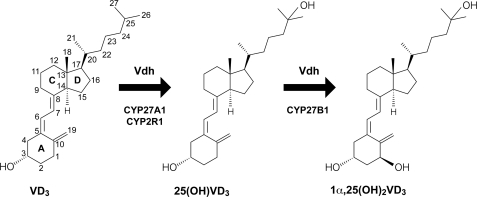
Vitamin D3 (VD3)3 is a B-ring opening secosteroid involved in a wide variety of biological functions in mammals (1). In humans, VD3 is converted into its physiologically active form, 1α,25-dihydroxyvitamin D3 (1α,25(OH)2VD3), via hydroxylation reactions that are catalyzed by several cytochrome P450s (CYPs) (1, 2). The first hydroxylation is done at the C25 position of VD3 by CYP27A1 (2, 3) and CYP2R1 (2, 4) in the liver to produce 25-hydroxyvitamin D3 (25(OH)VD3). The second proceeds at the C1α position of 25(OH)VD3 by CYP27B1 in the kidney (5) (Fig. 1). The final product, 1α,25(OH)2VD3, functions as a hormone with a critical role in maintaining calcium and phosphate homeostasis as well as in controlling the differentiation and proliferation of multiple cell types (1, 2, 6). Indeed, the many symptoms associated with VD3 deficiency and the VD metabolic disorder, which include psoriasis, osteoporosis, rickets, and hypoparathyroidism, are treated using 1α,25(OH)2VD3 and its derivatives (1).
FIGURE 1.
Scheme of catalytic conversion of VD3 to 1α,25(OH)2VD3. In humans, VD3 is physiologically activated mainly by CYP27A1, CYP2R1, and CYP27B1 in the liver and kidney. Vdh from P. autotrophica also catalyzes the conversion of VD3 to 1α,25(OH)2VD3 by regio- and stereoselective sequential hydroxylations.
Although the chemical synthesis of 1α,25(OH)2VD3 from cholesterol is an established method, it is inefficient, the maximum yield is no more than 1% (7). Alternatively, biocatalytic conversion by the actinomycete Pseudonocardia autotrophica is currently in practical use for the industrial production of 1α,25(OH)2VD3 (8, 9). We have recently cloned the gene encoding the VD3 hydroxylating enzyme (Vdh) of P. autotrophica, showing that Vdh is a CYP with ∼43% amino acid sequence similarities with the members of the CYP107 family (10). There were two more interesting findings of this study. First, we found that Vdh is capable of catalyzing regio- and stereoselective sequential hydroxylations at the C25 and C1α positions of VD3. An in vitro assay using a recombinant enzyme revealed that Vdh clearly exhibits VD3 25-hydroxylase and 25(OH)VD3 1α-hydroxylase activities (Fig. 1). These enzymatic properties are consistent with phenomena in the course of biotransformation: the 25(OH)VD3 level initially increases, followed by a gradual increase in the 1α,25(OH)2VD3 level in culture media. No 1α-hydroxyvitamin D3 was detected in either in vivo or in vitro experiments. Second, the VD3 25-hydroxylase activity of Vdh is comparable with that of physiologically VD3-metabolizing CYP27A1 (2, 3) and CYP2R1 (2, 4) despite the fact that VD3 is probably a nonnative substrate for Vdh.
CYPs are hemoproteins found across almost all of the Trees of Life and have extremely diverse biological functions, including biosynthesis of steroid hormones, lipids, and complex antibiotics, such as polyketides, detoxification of xenobiotics, drug metabolism, and bioconversion of recalcitrant molecules into usable carbon sources (11). A typical reaction catalyzed by CYPs is hydroxylation (monooxygenation), although several CYPs catalyze even more complex reactions such as epoxidation, C-C bond coupling and cleavage, and dehalogenation. This intriguing functional diversity of CYPs has led to many studies on their three-dimensional structures, which attempt to understand the structural mechanism of substrate recognition, interactions with redox partner proteins, electron and proton transfer, and catalytic reactions (12). Additionally, the versatility of CYPs is highly attractive for their potential uses as biocatalysts in chemical and pharmaceutical applications. Furthermore, protein engineering has been applied to improve and specialize the function of CYPs from bacteria and mammals (13–18). To realize more effective bioconversion of VD3, biotransformation activity from VD3 to 25(OH)VD3 was improved by a combination of random and site-saturated mutagenesis, and finally, a highly active Vdh mutant (Vdh-K1) was obtained (10). Vdh-K1 is a quadruple mutant (T70R/V156L/E216M/E384R) that is ∼22 times more active in the hydroxylation of VD3 to 25(OH)VD3 than wild-type Vdh (Vdh-WT) in in vivo bioconversion using Escherichia coli cells (10). Intriguingly, the four mutational points do not lie in the substrate-binding pocket, but are scattered throughout the molecule. Thus, it is of considerable interest as to how these mutations effect the significant enhancement in Vdh activity and how the enzyme recognizes each substrate.
Here we report the crystal structures of Vdh-WT and Vdh-K1 in the presence and absence of VD3/25(OH)VD3. The directed evolution and the x-ray structures show the importance of the conformational transition from open to closed states rather than the local active-site structure for activity enhancement. Furthermore, the VD3- and 25(OH)VD3-bound Vdh-K1 structures shed light on the occurrence of two anti-parallel substrate-binding modes, reliably enabling regioselective sequential hydroxylation of VD3.
EXPERIMENTAL PROCEDURES
Materials
All enzymes used for genetic manipulation were purchased from New England Biolabs, Takara, Promega, and Stratagene. Spinach ferredoxin and ferredoxin reductase used for enzyme assay were purchased from Sigma. 25(OH)VD3 was kindly provided by Mercian Co. Ltd. Partially methylated β-cyclodextrin (PMβCD) was purchased from Junsei Chemical Inc. All other chemicals were purchased from Sigma or WAKO Pure Chemicals Inc.
Protein Expression and Purification
The recombinant proteins used in this work contained a His6 tag at the C terminus. Overproduction of Vdh-WT was carried out using a Rhodococcus erythropolis expression system (19) or E. coli, and purified by Ni-affinity chromatography and anion exchange chromatography, as previously described (10, 20). The highly active Vdh-K1 mutant (T70R/V156L/E216M/E384R) was generated by directed evolution (10). Recombinant Vdh-K1 was produced using pET22b expression vector (Novagen) in E. coli strain BL21(DE3). Overexpression was induced by the addition of 0.1 mm isopropyl β-d-thiogalactoside for 20 h at 22 °C in Luria-Bertani (LB) medium supplemented with 100 μm FeSO4 and 80 μg/ml of 5-aminolevurinic acid. The cells were harvested and resuspended in buffer A (50 mm Tris-HCl, pH 7.5, and 10% glycerol) supplemented with 2 mm dithiothreitol (DTT). The cells were lysed by sonication, and the homogenate was clarified by centrifugation. The supernatant was dialyzed against buffer A, and then applied to a Ni-affinity column (HIS-SELECT resin; Sigma) pre-equilibrated with buffer A. The column was washed with buffer A, and the enzyme was eluted with a linear gradient of 0–400 mm imidazole in buffer A. The red fractions were collected, dialyzed against buffer B (20 mm Tris-HCl, pH 7.5, 10% glycerol, 1 mm DTT, and 0.5 mm EDTA), and subsequently applied to a DEAE-Sephacel anion-exchange column (GE Healthcare) pre-equilibrated with buffer B. The enzyme was eluted with a linear gradient of 0–400 mm NaCl in buffer B. The individual fractions were analyzed by SDS-PAGE. A carbon monoxide difference spectral assay was performed to verify the Vdh concentration (21). Vdh single mutants (T70R, V156L, E216M, and E384R) were also prepared using inverse PCR (22) with appropriate synthetic oligonucleotides as primers and pET29-Vdh-WT (10) as the template. Each mutant was expressed by E. coli BL21(DE3) and purified with the same procedure as for Vdh-K1.
UV-visible Spectral Measurements
The substrate-induced spectral shift was monitored using a JASCO V-630 biospectrophotometer with 1-cm path length quartz cells. VD3 and 25(OH)VD3 were dissolved in dimethyl sulfoxide at concentrations ranging from 50 μm to 2 mm, and 10 μl of the substrate solution was added to 990 μl of reaction mixture containing 2.5 μm Vdh-WT or Vdh mutants, 20 mm Tris-HCl, pH 7.5, and 0.05% PMβCD or 0.02% γ-cyclodextrin (γCD). PMβCD and γCD were used to increase the water solubility of VD3 and 25(OH)VD3, respectively (9, 23). CD concentrations referred to those adopted in actual VD3 bioconversion by P. autotrophica. The correct Kd values cannot be determined due to the low substrate solubility and the presence of inclusion complex of substrate and CD.
Enzyme Activity Measurements
VD3 25-hydroxylase and 25(OH)VD3 1α-hydroxylase activities were measured by reconstitution experiments. One milliliter of reaction mixture containing 500 nm Vdh-WT or 100 nm Vdh-K1, 81.4 mm Tris-HCl, pH 7.5, 100 mm NaCl, 96 μg/ml of spinach ferredoxin, 0.1 units/ml of spinach ferredoxin reductase, 3 units/ml of glucose dehydrogenase, 2 mm NADH, 2 mm NADPH, 60 mm d-glucose, 1% dimethyl sulfoxide, and 30 μm VD3 or 25(OH)VD3 was used for enzyme activity measurements. Enzymatic reaction was initiated by the addition of NAD(P)H, followed by incubation at 30 °C. Aliquots of the reaction mixture were collected after 20 (Vdh-WT) or 5 min (Vdh-K1), and extracted once with 1.5 ml and once with 0.75 ml of ethyl acetate. The combined ethyl acetate phase was allowed to dry, and the resulting residue was dissolved in 200 μl of methanol. The methanol solution was analyzed by a high-performance liquid chromatography (HPLC) apparatus equipped with a J'sphere ODS-H80 (I.D. 4.6 × 75 mm; YMC, Japan) controlled at a column temperature of 40 °C. Samples were resolved on the column using a linear gradient of 50 to 100% acetonitrile for 12 min, followed by elution with 100% acetonitrile for 13 min at a flow rate of 1.5 ml/min. The metabolites were detected by UV at 265 nm.
Crystallization and X-ray Diffraction Studies
Purified Vdh-WT was concentrated to 20 mg/ml in 20 mm Tris-HCl, pH 7.5, and 0–0.2% PMβCD (for VD3 complex) or 0–0.2% γCD (for 25(OH)VD3 complex), using a centrifugal ultrafiltration device with a 30-kDa cutoff membrane (Millipore). Vdh-K1 was also concentrated to 20 mg/ml under the same conditions as Vdh-WT, but supplemented with 20 mm NaCl. For substrate complex formation, saturated VD3/25(OH)VD3 dissolved in dimethyl sulfoxide (∼100 mm) was added to each sample at the volume ratio 1:99, and incubated overnight at 4 °C. The crystallization conditions were screened with 96-well sitting-drop plates at 20 °C by the sparse matrix approach. Optimization of the hit conditions were carried out by the hanging-drop vapor diffusion technique in 24-well plates at 20 °C. Vdh-WT crystals were grown using reservoir solution containing 0.1 m BisTris, pH 7.5, 50 mm CaCl2, 40–120 mm NaCl or KCl, and 32–40% PEG 400 or PEG 550 monomethyl ether or 20% PEG 1000. Vdh-K1 was crystallized using reservoir solution containing 0.1 m calcium acetate and 10–14% PEG 3350. Crystals were flash-cooled under a nitrogen gas stream at 100 K for x-ray diffraction studies. Prior to flash cooling, crystals of Vdh-K1 were short-soaked with cryoprotectant, which is the crystallization mother liquor supplemented with 20% glycerol. All diffraction data were collected at the Photon Factory (PF, Tsukuba, Japan), using the charge-coupled device detector (ADSC). The data were indexed, integrated, and merged using the HKL2000 program package (24). Vdh-WT was crystallized in two crystal forms: the trigonal crystals were obtained in space group P31 with unit cell parameters a = b = 62, c = 99 Å, both in the presence and absence of VD3/25(OH)VD3, and the orthorhombic crystals were grown in space group P212121 with unit cell parameters a = 63.6, b = 65.8, c = 102.3 Å in the presence of saturated 25(OH)VD3. Vdh-K1 was crystallized in orthorhombic space group P212121 with unit cell dimensions of a = 77, b = 172, c = 189 Å, both in the presence and absence of VD3/25(OH)VD3.
Structure Determination and Model Analysis
The structure of Vdh-WT in trigonal crystal form was determined by the molecular replacement method with the program AMoRe (25), using P450 EryF model fragments (PDB code 1JIN) (26) as search probes. The structures of Vdh-WT and Vdh-K1 in orthorhombic crystal forms were solved by molecular replacement with the program MOLREP (27), using the trigonal Vdh-WT structure as a search model. All models were refined using the program REFMAC5 (28), and were inspected and corrected in the graphic program COOT (29). A randomly selected 5% of the observed reflections were set aside for cross-validation analysis and used to monitor various refinement stage. VD3 and 25(OH)VD3 models in Vdh-K1 were built based on a Fo − Fc difference Fourier map in COOT. Electron density for 25(OH)VD3 is clear, whereas that for VD3 is partly ambiguous due to the high mobility and high temperature factors. The validities of all substrate models were verified by calculating the simulated annealing 2Fo − Fc composite omit map with the program CNS 1.1 (30) (supplemental Fig. S4). The stereochemical qualities of the final refined models were assessed by the program PROCHECK (31). The x-ray data collection and refinement statistics are summarized in Table 1. Structure superposition and assessment of conformational changes with difference distance matrices were performed with the program SuperPose (32). Molecular drawings were prepared using the program PyMOL (33).
TABLE 1.
Crystallographic parameters and refinement statistics
| PDB code |
|||||
|---|---|---|---|---|---|
| 3A4G | 3A4H | 3A4Z | 3A50 | 3A51 | |
| Model | Vdh-WT | Vdh-WT | Vdh-K1 | Vdh-K1 | Vdh-K1 |
| Conformation | Open | Open | Closed | Closed | Closed |
| No. monomers in the AU | 1 | 1 | 5 | 5 | 5 |
| Substrates | Substrate-free | Substrate-free | Substrate-free | VD3 | 25(OH)VD3 |
| Data collection | |||||
| Beamline | PF-BL5A | PF-BL17A | AR-NE3A | AR-NE3A | AR-NW12A |
| Wavelength (Å) | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Space group | P31 | P212121 | P212121 | P212121 | P212121 |
| Unit cell parameters (Å, deg) | 61.7, 61.7, 98.8, 90, 90, 120 | 63.6, 65.8, 102.3, 90, 90, 90 | 77.4, 172.5, 189.9, 90, 90, 90 | 77.4, 172.3, 189.1, 90, 90, 90 | 77.2, 171.8, 189.1, 90, 90, 90 |
| Resolution range (Å)a | 50–1.75 (1.81–1.75) | 50–3.05 (3.16–3.05) | 50–2.20 (2.24–2.20) | 50–2.05 (2.09–2.05) | 50–2.00 (2.07–2.0) |
| Unique reflections | 42,012 | 8,366 | 129,009 | 158,815 | 170,383 |
| Redundancya | 5.3 (3.2) | 3.6 (3.8) | 7.3 (6.0) | 7.5 (6.3) | 7.4 (7.0) |
| Completeness (%)a | 99.3 (93.2) | 97.6 (97.9) | 100.0 (99.8) | 100.0 (99.8) | 100.0 (100.0) |
| I/σ (I)a | 21.9 (3.7) | 18.9 (6.0) | 26.0 (2.9) | 26.9 (2.9) | 32.5 (3.3) |
| Rmergea,b | 0.075 (0.316) | 0.090 (0.317) | 0.101 (0.500) | 0.107 (0.540) | 0.077 (0.594) |
| Wilson B-factor (Å2) | 23.2 | 45.0 | 27.9 | 23.3 | 29.5 |
| Refinement | |||||
| Rwork/Rfreec | 0.185/0.236 | 0.217/0.270 | 0.201/0.244 | 0.201/0.240 | 0.197/0.234 |
| R.m.s.deviation bond lengths (Å) | 0.015 | 0.006 | 0.015 | 0.013 | 0.014 |
| R.m.s.deviation bond angles (deg) | 1.53 | 0.96 | 1.56 | 1.50 | 1.55 |
| Total atoms | 3,451 | 3,138 | 16,621 | 16,977 | 17,293 |
| Average B-factors (Å2) | |||||
| Overall | 28.7 | 44.1 | 34.1 | 28.9 | 30.4 |
| Ligands | 35.3 (PEG) | 69.0 (VD3) | 41.3 (25(OH)VD3) | ||
| Ramachandran plot (%) | 89.6/10.4/0.0/0.0 | 89.5/10.5/0.0/0.0 | 91.1/8.9/0.0/0.0 | 90.6/9.4/0.0/0.0 | 90.6/9.4/0.0/0.0 |
a Data for the highest resolution shell are provided in parentheses.
b Rmerge = Σh Σi|Ih,i − 〈Ih〉|/ΣhΣi Ih,i, where 〈Ih〉 is the mean intensity of a set of equivalent reflections.
c Rwork = Σ|Fobs − Fcalc|/Σ Fobs for the 95% of the reflection data used in the refinement. Fobs and Fcalc are observed and calculated structure factor amplitudes, respectively. Rfree is the equivalent of Rwork, except that it was calculated for a randomly chosen 5% test set excluded from the refinement.
RESULTS
Spectroscopic Characterization of Vdh
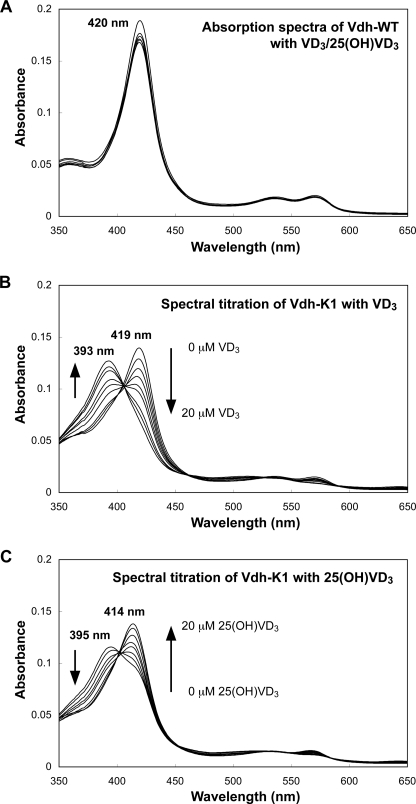
The binding affinities of VD3/25(OH)VD3 to Vdh-WT and Vdh-K1 were examined by UV-visible spectroscopy (Fig. 2). A small amount of PMβCD or γCD were added to the assay mixture to dissolve VD3 or 25(OH)VD3, respectively (see “Experimental Procedures”). The absorption spectra of Vdh-WT showed a Soret peak at 420 nm both in the presence and absence of VD3 and 25(OH)VD3 (Fig. 2A). This indicates that Vdh-WT is not affected by substrate addition and the heme iron is present consistently in a water-coordinated low-spin state. In contrast to Vdh-WT, significant spectral changes were detected for Vdh-K1. Purified Vdh-K1 is present as a mixture of high-spin (Soret peak at ≈390 nm) and low-spin (Soret peak at ≈420 nm) states, and the ratio of high/low-spin states is very sensitive to the addition of CDs. Typically, Vdh-K1 is present primarily as a low-spin state in the presence of PMβCD, whereas it prefers a high-spin state in the presence of γCD. Spectral titration of Vdh-K1 with VD3 showed a spectral shift from the low-spin to high-spin state, which is a typical substrate-binding spectral change (type I spectral change; Fig. 2B). Unexpectedly, spectral titration of Vdh-K1 with 25(OH)VD3 produced a spectral shift from the high-spin (389 nm) to low-spin (414 nm) state (reverse type I spectral change), suggesting that an unidentified ligand is coordinated to the heme iron in the presence of 25(OH)VD3 (Fig. 2C). The ligand exhibiting the Soret peak at 414 nm was identified as the C3 OH group (3β-OH) of 25(OH)VD3 by x-ray structure analysis, as described later.
FIGURE 2.
Spectral changes of Vdh-WT and Vdh-K1 with VD3/25(OH)VD3. UV-visible spectra for Vdh-WT on addition of 0, 10, and 20 μm VD3/25(OH)VD3 (panel A), and for Vdh-K1 on addition of 0–20 μm VD3 (panel B) and 0–20 μm 25(OH)VD3 (panel C). Arrows indicate the directions of spectral changes in response to the increase in VD3/25(OH)VD3 concentrations. Wavelength (nm) at maximum height of Soret band observed in each spectral measurement is also indicated.
General Description of Vdh Structure
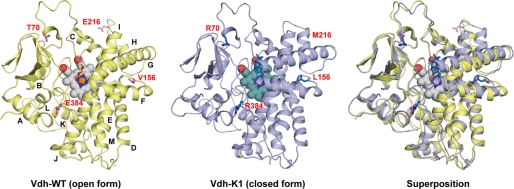
The crystal structures of Vdh-WT in the substrate-free form and Vdh-K1 in both the substrate-free and substrate-bound forms were determined by molecular replacement using P450 EryF (CYP107A1) as a search model (Fig. 3 and Table 1). Molecules in all crystal forms have a well defined continuous electron density, except for several N-terminal residues and C-terminal His tag regions. The overall structure of Vdh exhibits a typical triangular-shaped P450-fold consisting of 13 α-helices (A-helix to M-helix) and 8 β-strands (a-strand to h-strand). The heme cofactor is located at the center of the molecule and is sandwiched between distal I-helix and proximal L-helix. The heme propionate groups are tightly bound to side chains Arg289, His95, and His345. The thiol group of Cys347 is coordinated to the heme iron, as shown in previously reported CYP structures. A sequence similarity search indicated that Vdh is a member of the family CYP107. The structures of three molecular species of CYP107 (EryF (26, 35, 36), PikC (37, 38), and BioI (39)) have been solved to date; a pairwise structure comparison revealed that Vdh is the most related to PikC (CYP107L1).
FIGURE 3.
Ribbon diagram of overall structure of Vdh-WT (open form) and Vdh-K1 (closed form). Overlay of these two structures is also shown. The heme cofactor is in the sphere, and the bound-substrate (25(OH)VD3) and the side chains of mutational residues at positions 70, 156, 216, and 384 are in sticks. The α-helices are labeled alphabetically from the N to C terminus.
Vdh-WT Forms an Open Conformation
The crystal structure of Vdh-WT was determined in the substrate-free form at a resolution of 1.75 Å (Fig. 3). The asymmetric unit comprises one Vdh-WT monomer. Vdh-WT is most structurally related to substrate-free PikC in the open conformation (PDB code 2BVJ, chain B) (37) with a root mean square deviation (r.m.s. deviation) of 2.6 Å for 376 Cα atoms (supplemental Fig. S3). Differences in the main chain conformation are observed in two loop regions: an N-terminal loop (residues 10–17) and a loop between B-helix and C-helix commonly referred to as a “BC-loop” (residues 71–85). The BC-loop is known to be flexible and is partially disordered in the substrate-free PikC structure. The BC-loop in Vdh-WT protrudes from the globular body of the P450-fold, and the substrate-binding pocket on the distal side of the heme is widely open and exposed to the solvent (Fig. 3 and supplemental Fig. S5A). The water ligand is bound to the distal side of the heme iron with a Fe-O distance of 2.4 Å. Extra electron density is also observed in the large pocket mainly surrounded by hydrophobic residues, in which a model of polyethylene glycol (PEG) was reasonably fitted. The bound PEG is likely a crystallization artifact.
We also obtained crystals of Vdh-WT in the presence of substrates in two different forms. The same trigonal crystals were grown with saturated VD3 or 25(OH)VD3, but the structures were completely similar to that in the substrate-free open conformation. Vdh-WT also crystallized in the orthorhombic space group P212121 with saturated 25(OH)VD3, and the structure was determined at 3.05-Å resolution. The overall structure in the orthorhombic crystal also superposed very well onto that in trigonal form, including the BC-loop, with an r.m.s. deviation of 1.0 Å for 395 Cα atoms (supplemental Fig. S2A). In both structures, no electron density for VD3/25(OH)VD3 bound to the active-site pocket was observed. These results indicate that the open conformation of Vdh-WT is stable and not affected by crystal lattice forces, even in the presence of saturated VD3/25(OH)VD3.
Closed Structure of Vdh-K1 in Substrate-free and Substrate-bound Forms
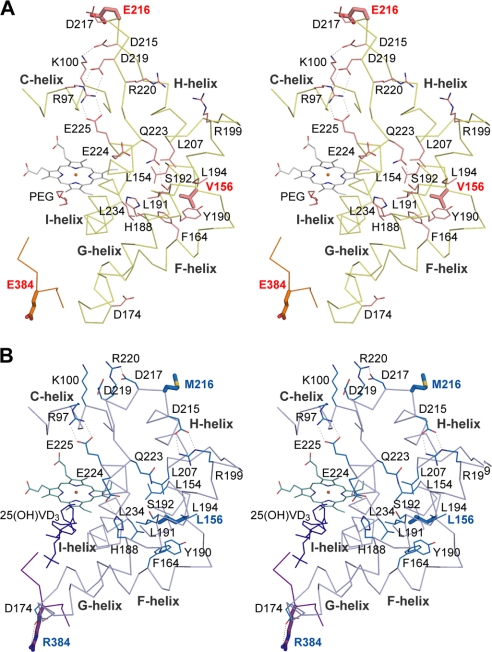
The structures of Vdh-K1 in the substrate-free, VD3-bound, and 25(OH)VD3-bound forms were determined at 2.3-, 2.05-, and 2.0-Å resolutions, respectively (Fig. 3). Crystallization screening of Vdh-K1 yielded only one orthorhombic form with relatively large unit-cell dimensions compared with Vdh-WT crystals. The asymmetric unit contains 5 Vdh-K1 monomers that are not related by point-group symmetry. The structures of all the refined models of Vdh-K1 (15 monomers) are almost identical (r.m.s. deviation, < 0.7 Å), and no relatively large structural differences were observed, suggesting that the conformation of Vdh-K1 is also stable even in the substrate-free, VD3-bound, and 25(OH)VD3-bound states. Notably, local structural differences were found between Vdh-WT and Vdh-K1 (Figs. 3, 4, supplemental Fig. S2B, and Movies S1 and S2). The differences were most evident in the repositioning of the F- and G-helices, which are shifted by a distance of up to 8 Å, and the loop connecting the F- and G-helix (FG-loop) contacts the opposite loop (C-terminal loop) via two hydrogen bonds. This motion leads to a closed conformation where the substrate-binding pocket is covered by FG-helices (Figs. 3 and 4). An additional conformational change occurs in a loop connecting H- and I-helix (HI-loop), and induces the extension of H-helix by 4 residues (Fig. 4). Similar conformational changes were also observed in the PikC structure, and Vdh-K1 is more structurally related to the closed form of PikC (PDB code 2BVJ, chain A; r.m.s. deviation of 3.0 Å for 378 Cα atoms) than to the open form of PikC (PDB code 2BVJ, chain B; r.m.s. deviation of 4.0 Å for 378 Cα atoms; supplemental Fig. S3) (37). The BC-loop is known to often move toward the substrate-binding pocket in response to substrate binding, whereas it is motionless in Vdh (Fig. 3). Therefore, the substrate-binding pocket is not fully closed and is partially accessible by the solvent. The four mutational positions in Vdh-K1 (T70R, V156L, E216M, and E384R) are not located at the substrate-binding site but are scattered throughout the molecule. We discuss later how these mutations are involved in the open/closed conformational change (see “Discussion”).
FIGURE 4.
Stereo view Cα trace of the Vdh-WT (panel A) and Vdh-K1 (panel B) in the region composing C-, FG-, and HI-helices and the C-terminal loop. Mutational residues at positions 156, 216, and 384 are depicted in sticks. Salt bridges and hydrogen bonds broken and reconstituted during open/close motion are shown as dashed lines. Side chains involving these hydrophilic interactions or hydrophobic residues nearby Leu156/Val156 creating hydrophobic core are in thin sticks and labeled.
VD3 and 25(OH)VD3 Are Bound to Vdh-K1 in an Anti-parallel Orientation
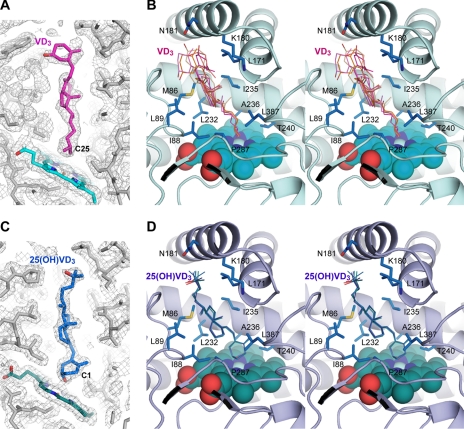
Vdh catalyzes the two-step hydroxylation of VD3 to 1α,25(OH)2VD3 via 25(OH)VD3. Because the hydroxylation positions are discrete, VD3 and 25(OH)VD3 must be bound in different orientations. The electron density shows that VD3 is bound in an extended conformation at the active-site pocket, and the aliphatic side chain including C25 is located in the vicinity of the heme iron (Fig. 5, A and B). The mean distance between the C25 of VD3 and the heme iron of each molecule is 4.6 ± 0.1 Å. No solvent is coordinated to the distal site of the heme iron probably due to VD3 binding. These observations support the high-spin state of the heme iron as shown by spectroscopic data. The mean refined B-factor value for VD3 atoms is 69 Å2, suggesting that VD3 is not tightly bound to the enzyme. Actually, the conformation of the five refined VD3 models in each monomer is varied, especially in the A-ring moiety for which the electron density is even lower and unclear (Fig. 5, A and B, and supplemental Fig. S4). The aliphatic side chain and the CD-rings of VD3 are surrounded by hydrophobic side chain atoms of residues Met86, Ile88, Leu89, Leu171, Ile235, Leu232, Pro287, and Leu387 with distances ranging from 3.6 to 4.5 Å, whereas the A-ring moiety is considerably exposed to the solvent (Fig. 5B). Although Trp67, Pro83, Lys180, Asn181, and Met184 derived from BC-loop, FG-loop, and G-helix lie near the A-ring, there are no specific interatomic interactions with a unique hydrophilic 3β-OH of A-ring. In contrast, the electron density for 25(OH)VD3 is very clear, and it is evidently bound in an anti-parallel orientation (Fig. 5, C and D, and supplemental Fig. S4). The mean B-factor value for 25(OH)VD3 atoms is 41 Å2, which is lower than that for VD3. The clear electron density also revealed that A-ring forms in a boat conformation with an axial 3β-OH occupying the sixth coordinate position of the heme iron with a mean distance of 2.5 ± 0.1 Å. The hydroxylating site C1 is located at a distance of 5.0 ± 0.0 Å from the heme iron. The 3β-OH coordination to the heme iron explains the Soret peak at 414 nm in the spectroscopic data. Unlike the A-ring of VD3, the aliphatic side chain of 25(OH)VD3 does not fluctuate even though it is partly exposed to the solvent and can be immobilized alongside of the FG-loop via van der Waals forces. In addition, in 4 of 5 molecules, hydrogen bonds form between 25-OH and one of the solvent molecules clustered at the entrance of the substrate-binding pocket; these also contribute to the attachment of the aliphatic side chain.
FIGURE 5.
VD3 and 25(OH)VD3 binding to Vdh-K1. Final 2Fo − Fc electron density map for VD3 (panel A) and 25(OH)VD3 (panel C) contoured at 0.8σ level (chain ID, A). Unbiased simulated annealing composite omit map for all chains in the asymmetric unit are shown in supplemental Fig. S4. B, stereo view active-site structure with bound VD3 in chain A. All five refined models of VD3 in each chain in the asymmetric unit are superimposed and shown as thin sticks. Residues creating the substrate-binding pocket are shown in sticks and are labeled. The heme cofactor is the sphere. D, stereo view active site structure with bound 25(OH)VD3 prepared in the same way and in the same orientation to panel B.
DISCUSSION
Interpretation of Spectroscopic Data
The absorption spectra of Vdh-WT show that the heme iron is present in a water-liganded low-spin state both in the presence and absence of a substrate (Fig. 2A). The crystal structures of Vdh-WT also reveals that no substrate is bound to Vdh-WT even in the presence of saturated VD3/25(OH)VD3. These results clearly show a very low substrate binding affinity despite the fact that the catalytic activity of Vdh-WT is comparable with those of physiologically VD3-metabolizing CYP27A1 and CYP2R1 (2–4). One possible explanation is that substrate binding to Vdh-WT may occur depending on the association with a redox partner protein. Several cases have been reported to date where a redox protein functions as an effector that promotes substrate binding to CYPs (40, 41). In contrast to Vdh-WT, Vdh-K1 exhibits spectral changes in response to the addition of VD3/25(OH)VD3 (Fig. 2, B and C). The absorption peak in the Soret band is shown at 393 nm with VD3 and at 414 nm with 25(OH)VD3. The former clearly corresponds to a five-coordinate high-spin state of the heme iron caused by VD3 binding, whereas the latter indicates a six-coordinate low-spin state. The structure of Vdh-K1 with bound 25(OH)VD3 revealed that the 3β-OH group of 25(OH)VD3 is a sixth coordination ligand that is positioned very close to the heme iron. Ligation of the OH group to the heme iron was previously reported in the structure of P450cam complexed with the product 5-exo-hydroxycamphor (42). In addition, several mammalian CYPs show the reverse type I spectral change in response to the addition of a substrate or inhibitor carrying OH groups (Ref. 43 and references therein). Obviously, the OH-bound heme iron reduces the catalytic activity of CYP, because the low-spin ferric state with very low redox potential precludes its reduction of the heme iron that is needed for the subsequent electron transfer and dioxygen binding. Therefore, further repositioning of the A-ring of 25(OH)VD3 would be required for promoting the reduction and monooxygenation at the C1α site.
The spectral data suggest that the binding affinities of both substrates to Vdh increase by directed evolution. The specific activity measurements also showed that Vdh-K1 is ∼12 times more active for VD3 25-hydroxylation and 25 times more active for 25(OH)VD3 1α-hydroxylation than Vdh-WT (supplemental Table S1). The high binding affinity and catalytic activity of Vdh-K1 for 25(OH)VD3 are totally unexpected, because we monitored only the VD3 25-hydroxylase activity in the directed evolution experiments. These results strongly suggest that the four mutations do not improve the substrate specificity but affect the underlying enzyme mechanisms that are common to both hydroxylation activities.
Structural Insights into Activity Enhancement by Directed Evolution
Current structural studies reveal that the conformational change from the open to closed state results from the four amino acid substitutions generated by directed evolution. Then the question arises as to how these individual mutations contribute to the conformational change. Val156 lies at the loop connecting E- and F-helix, and its side chain is buried inside the hydrophobic cluster formed by the side chains of Phe164, Leu191, Leu194, Leu154, Leu207, and Tyr190 (Fig. 4A). The substitution from Val to Leu gives rise to steric hindrance with the side chains Phe164 and Tyr190 in the open state, thus requiring the spatial repositioning of these hydrophobic side chains. In the Vdh-K1 structure, the hydrophobic cluster is actually rearranged by changing the torsion geometries of these side chains together with the approximately ∼8-Å shift of FG-helices, and consequently, the interatomic distances are reoptimized (Fig. 4B). Glu216 is located at the protrusive HI-loop, and its side chain is fully exposed to the solvent with a relatively high B-factor value (about 55 Å2) in the Vdh-WT structure. The solvent-exposed methionine is unfavorable for protein stability, and the E216M mutation possibly functions as a driving force for conformational change of the HI-loop and extension of H-helix involving Met216. It is of interest to note that several salt bridges and hydrogen bonds are broken and reconstructed between residues derived from the C-, FG-, and HI-helices region during the conformational change (Fig. 4). These rearrangements of interatomic interactions imply that motions in the FG- and HI-helices regions are mutually cooperative. In contrast, the two other mutations (E384R and T70R) lie in the structurally rigid region. Arg384 is located at the C-terminal loop that interacts with the FG-loop in the closed Vdh-K1 structure. The main chain O of Arg384 and side chain OH of Ser385 form hydrogen bonds to the main chain N of Asp174 and main chain O of Val172 at the FG-loop, respectively. In addition, electrostatic interactions occur between the side chain guanidyl of Arg384 and carboxyl of Asp174 with distances of 3.5–5 Å (Fig. 4B). It is thus likely that substitution from acidic to basic residues at position 384 somewhat increases the stability of the closed conformation. It is unclear whether Arg70 contributes to the conformational change and increase in substrate binding affinity. In several CYPs, arginine residues at the entrance of the substrate access channel are suggested to play a crucial role in substrate recognition and/or guiding substrate entry and egress (44, 45). Likewise, Arg70 in Vdh-K1 is located at the entrance of the substrate-binding pocket and may provide a weak primary binding site that improves the efficiency of substrate entry and egress. To investigate which mutations contribute most to the conformational change, the absorption spectra of each single mutant (T70R, V156L, E216M, and E384R) were also measured on addition of 0–20 μm substrates. As shown in supplemental Fig. S1, only the V156L mutant showed an obvious but relatively small substrate-induced spectral change, whereas no spectral changes were observed for three other mutants similar to Vdh-WT. The results suggest that the combination of these mutations is essential to elicit high substrate binding affinities observed in Vdh-K1.
Elaborate rearrangements of interatomic interactions around CFGHI helices imply that the closed structure observed in Vdh-K1 is not a novel conformational state that resulted from directed evolution but rather a naturally pre-existing state. The original residues at mutational positions seemingly cause no serious problems even in the closed conformation. Furthermore, open Vdh-WT and closed Vdh-K1 are structurally related to the naturally existing open and closed states of wild-type PikC (supplemental Fig. S3). Several CYPs are known to undergo conformational changes even in the absence of a substrate, and the substrate complex formation likely arises from a pre-existing conformational equilibrium rather than an induced fit mechanism (37, 46–48). In the present work, it is likely that the directed evolution was responsible for the shift in conformational equilibrium toward the closed conformation and facilitation of VD3/25(OH)VD3 binding. Vdh-WT may surmount an energy barrier and transform from the open to closed state in response to the native substrate binding, and the current Vdh-K1 structure may mimic the putative closed form of Vdh-WT when bound to the native substrate. Many CYPs show extremely broad substrate specificity, presumably involving an open/close motion (49). Typically, the open and closed structures correspond to the substrate-free and substrate-bound forms, respectively, and our results thus introduce the possibility that identification of mutational positions inducing a shift in conformational equilibrium toward the closed form would unselectively increase the substrate binding affinity and catalytic activities.
We observe a remarkable parallel between our directed evolution studies of Vdh and natural selection of CYP51 mutants identified in Candida albicans azole-resistant isolates that emerged under selective pressure of antifungal drug fluconazole (50). In both cases, substrate/inhibitor binding affinities are greatly influenced by mutations in remote loops. These mutations presumably play a role in orchestrating the conformational transition and enhancing the enzyme performance. The parallel suggests that the mechanism could be widely used in nature to adapt CYPs to ever-emerging new conditions and substrates.
Anti-parallel Substrate Binding Modes Enabling Sequential Hydroxylation
On the basis of spectroscopic and crystallographic data, it is strongly suggested that Vdh-K1 has an exactly specified binding orientation for each substrate. Intriguingly, the backbone and side chain conformations at the active-site pocket are identical among the substrate-free, VD3-bound, and 25(OH)VD3-bound forms of Vdh-K1, and no induced-fit mechanisms are observed, although VD3 is asymmetric (Fig. 5). In light of these structural findings, it is likely that the binding orientation depends on simply whether the C25 site is hydroxylated. VD3 is bound to the enzyme in an orientation that positions the aliphatic side chain containing C25 toward the heme iron and the A-ring containing the hydrophilic 3β-OH group toward the entrance of the substrate-binding pocket exposed to the solvent (Fig. 5B). This binding orientation is possibly a consequence of the need to prevent exposure of the hydrophobic aliphatic side chain to the solvent, and is also consistent with the fact that Vdh exhibits only 25-hydroxylase activity against VD3. Once C25 is hydroxylated, 25(OH)VD3 is bound to the enzyme in an anti-parallel orientation with 3β-OH of A-ring anchored to the heme iron, thereby enabling the subsequent 1α-hydroxylation. The existence of two substrate binding modes in different orientations implies that Vdh-K1 is not functionally specialized.
In mammals, VD3 is hydroxylated by mainly CYP27A1, CYP2R1, and CYP27B1. Of these CYPs, the crystal structure of CYP2R1 has been determined in complex with VD3 (4). In CYP2R1, VD3 is fully buried in the hydrophobic active site pocket, and the A-ring of VD3 facing the protein surface is also surrounded and stabilized by hydrophobic interactions with several of the aromatic side chains (supplemental Fig. S5D). The well fitted substrate-bound form of CYP2R1 exemplifies the functionally specialized structure of a VD3 25-hydroxylase. Recently, the structures of P450 SU-1 (CYP105A1) from Streptomyces griseolus have been reported, which is another example of bacterial CYP that is capable of hydroxylating VD3 (34, 45). The product 1α,25(OH)2VD3 lies sideways within the large active site pocket of CYP105A1, and the hydroxylating sites, C25 and C1, are located at a distance of about 11 Å from the heme iron (supplemental Fig. S5C). The authors suggested that the 1α,25(OH)2VD3 binding site far from the heme iron is presumably a primary binding site, and the substrate would be required to move closer to the heme iron for a catalytic reaction (34, 45). These different VD3 binding conditions give a glimpse into the impressive versatility of CYPs.
Supplementary Material
Acknowledgments
We thank the entire beamline staff at Photon Factory for technical support in the collection of x-ray diffraction data.
This work was supported in part by the Development of Basic Technologies for Advanced Production Methods Using Microorganism Functions project of the New Energy and Industrial Technology Development Organization (NEDO) of Japan. Synchrotron radiation experiments were conducted under the approval of 2007G650 at Photon Factory.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1, Figs. S1–S5, and Movies S1 and S2.
- VD3
- vitamin D3
- CYP
- cytochrome P450
- Vdh
- vitamin D3 hydroxylase
- 25(OH)VD3
- 25-hydroxyvitamin D3
- CD
- cyclodextrin
- PMβCD
- partially methylated β-cyclodextrin
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- r.m.s.
- root mean square.
REFERENCES
- 1.Jones G., Strugnell S. A., DeLuca H. F. (1998) Physiol. Rev. 78, 1193–1231 [DOI] [PubMed] [Google Scholar]
- 2.Prosser D. E., Jones G. (2004) Trends Biochem. Sci. 29, 664–673 [DOI] [PubMed] [Google Scholar]
- 3.Sawada N., Sakaki T., Ohta M., Inouye K. (2000) Biochem. Biophys. Res. Commun. 273, 977–984 [DOI] [PubMed] [Google Scholar]
- 4.Strushkevich N., Usanov S. A., Plotnikov A. N., Jones G., Park H. W. (2008) J. Mol. Biol. 380, 95–106 [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto K., Uchida E., Urushino N., Sakaki T., Kagawa N., Sawada N., Kamakura M., Kato S., Inouye K., Yamada S. (2005) J. Biol. Chem. 280, 30511–30516 [DOI] [PubMed] [Google Scholar]
- 6.Deeb K. K., Trump D. L., Johnson C. S. (2007) Nat. Rev. Cancer 7, 684–700 [DOI] [PubMed] [Google Scholar]
- 7.Zhu G. D., Okamura W. H. (1995) Chem. Rev. 95, 1877–1952 [Google Scholar]
- 8.Sasaki J., Miyazaki A., Saito M., Adachi T., Mizoue K., Hanada K., Omura S. (1992) Appl. Microbiol. Biotechnol. 38, 152–157 [DOI] [PubMed] [Google Scholar]
- 9.Takeda K., Asou T., Matsuda A., Kimura K., Okamura K., Okamoto R., Sasaki J., Adachi T., Omura S. (1994) J. Ferment. Bioeng. 78, 380–382 [Google Scholar]
- 10.Fujii Y., Kabumoto H., Nishimura K., Fujii T., Yanai S., Takeda K., Tamura N., Arisawa A., Tamura T. (2009) Biochem. Biophys. Res. Commun. 385, 170–175 [DOI] [PubMed] [Google Scholar]
- 11.Coon M. J., Ding X. X., Pernecky S. J., Vaz A. D. (1992) FASEB J. 6, 669–673 [DOI] [PubMed] [Google Scholar]
- 12.Denisov I. G., Makris T. M., Sligar S. G., Schlichting I. (2005) Chem. Rev. 105, 2253–2277 [DOI] [PubMed] [Google Scholar]
- 13.Julsing M. K., Cornelissen S., Buhler B., Schmid A. (2008) Curr. Opin. Chem. Biol. 12, 177–186 [DOI] [PubMed] [Google Scholar]
- 14.Gillam E. M. J. (2008) Chem. Res. Toxicol. 21, 220–231 [DOI] [PubMed] [Google Scholar]
- 15.Gillam E. M. J. (2007) Arch. Biochem. Biophys. 464, 176–186 [DOI] [PubMed] [Google Scholar]
- 16.Fasan R., Meharenna Y. T., Snow C. D., Poulos T. L., Arnold F. H. (2008) J. Mol. Biol. 383, 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S., Chaulagain M. R., Knauff A. R., Podust L. M., Montgomery J., Sherman D. H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18463–18468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis J. C., Bastian S., Bennett C. S., Fu Y., Mitsuda Y., Chen M. M., Greenberg W. A., Wong C. H., Arnold F. H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16550–16555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitani Y., Meng X., Kamagata Y., Tamura T. (2005) J. Bacteriol. 187, 2582–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yasutake Y., Fujii Y., Cheon W. K., Arisawa A., Tamura T. (2009) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 372–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Omura T., Sato R. (1964) J. Biol. Chem. 239, 2370–2378 [PubMed] [Google Scholar]
- 22.Hemsley A., Arnheim N., Toney M. D., Cortopassi G., Galas D. J. (1989) Nucleic Acids Res. 17, 6545–6551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villalonga R., Cao R., Fragoso A. (2007) Chem. Rev. 107, 3088–3116 [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 25.Navaza J. (1994) Acta Crystallogr. Sect. A 50, 157–163 [Google Scholar]
- 26.Cupp-Vickery J. R., Garcia C., Hofacre A., McGee-Estrada K. (2001) J. Mol. Biol. 311, 101–110 [DOI] [PubMed] [Google Scholar]
- 27.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 28.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 29.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 31.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 32.Maiti R., Van Domselaar G. H., Zhang H., Wishart D. S. (2004) Nucleic Acids Res. 32, W590–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 34.Sugimoto H., Shinkyo R., Hayashi K., Yoneda S., Yamada M., Kamakura M., Ikushiro S., Shiro Y., Sakaki T. (2008) Biochemistry 47, 4017–4027 [DOI] [PubMed] [Google Scholar]
- 35.Cupp-Vickery J. R., Poulos T. L. (1995) Nat. Struct. Biol. 2, 144–153 [DOI] [PubMed] [Google Scholar]
- 36.Cupp-Vickery J., Anderson R., Hatziris Z. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 3050–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherman D. H., Li S., Yermalitskaya L. V., Kim Y., Smith J. A., Waterman M. R., Podust L. M. (2006) J. Biol. Chem. 281, 26289–26297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li S., Ouellet H., Sherman D. H., Podust L. M. (2009) J. Biol. Chem. 284, 5723–5730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cryle M. J., Schlichting I. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15696–15701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schenkman J. B., Jansson I. (2003) Pharmacol. Ther. 97, 139–152 [DOI] [PubMed] [Google Scholar]
- 41.Tosha T., Yoshioka S., Takahashi S., Ishimori K., Shimada H., Morishima I. (2003) J. Biol. Chem. 278, 39809–39821 [DOI] [PubMed] [Google Scholar]
- 42.Li H. Y., Narasimhulu S., Havran L. M., Winkler J. D., Poulos T. L. (1995) J. Am. Chem. Soc. 117, 6297–6299 [Google Scholar]
- 43.Shimada T., Tanaka K., Takenaka S., Foroozesh M. K., Murayama N., Yamazaki H., Guengerich F. P., Komori M. (2009) Chem. Res. Toxicol. 22, 1325–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winn P. J., Lüdemann S. K., Gauges R., Lounnas V., Wade R. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5361–5366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayashi K., Sugimoto H., Shinkyo R., Yamada M., Ikeda S., Ikushiro S., Kamakura M., Shiro Y., Sakaki T. (2008) Biochemistry 47, 11964–11972 [DOI] [PubMed] [Google Scholar]
- 46.Savino C., Montemiglio L. C., Sciara G., Miele A. E., Kendrew S. G., Jemth P., Gianni S., Vallone B. (2009) J. Biol. Chem. 284, 29170–29179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott E. E., He Y. A., Wester M. R., White M. A., Chin C. C., Halpert J. R., Johnson E. F., Stout C. D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13196–13201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang W. C., Westlake A. C., Maréchal J. D., Joyce M. G., Moody P. C., Roberts G. C. (2007) J. Mol. Biol. 373, 633–651 [DOI] [PubMed] [Google Scholar]
- 49.Poulos T. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13121–13122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Podust L. M., Poulos T. L., Waterman M. R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3068–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.