Abstract
Iron influx increases the translation of the Alzheimer amyloid precursor protein (APP) via an iron-responsive element (IRE) RNA stem loop in its 5′-untranslated region. Equal modulated interaction of the iron regulatory proteins (IRP1 and IRP2) with canonical IREs controls iron-dependent translation of the ferritin subunits. However, our immunoprecipitation RT-PCR and RNA binding experiments demonstrated that IRP1, but not IRP2, selectively bound the APP IRE in human neural cells. This selective IRP1 interaction pattern was evident in human brain and blood tissue from normal and Alzheimer disease patients. We computer-predicted an optimal novel RNA stem loop structure for the human, rhesus monkey, and mouse APP IREs with reference to the canonical ferritin IREs but also the IREs encoded by erythroid heme biosynthetic aminolevulinate synthase and Hif-2α mRNAs, which preferentially bind IRP1. Selective 2′-hydroxyl acylation analyzed by primer extension analysis was consistent with a 13-base single-stranded terminal loop and a conserved GC-rich stem. Biotinylated RNA probes deleted of the conserved CAGA motif in the terminal loop did not bind to IRP1 relative to wild type probes and could no longer base pair to form a predicted AGA triloop. An AGU pseudo-triloop is key for IRP1 binding to the canonical ferritin IREs. RNA probes encoding the APP IRE stem loop exhibited the same high affinity binding to rhIRP1 as occurs for the H-ferritin IRE (35 pm). Intracellular iron chelation increased binding of IRP1 to the APP IRE, decreasing intracellular APP expression in SH-SY5Y cells. Functionally, shRNA knockdown of IRP1 caused increased expression of neural APP consistent with IRP1-APP IRE-driven translation.
Keywords: Amyloid, Blood, Brain, RNA-binding Protein, RNA Folding, Translation Control
Introduction
Alzheimer disease (AD)3 is a neurodegenerative process that is the leading cause of death worldwide for people over the age of 65 (1, 2). The pathological hallmarks of AD in the brain cortex are not only abundant extracellular amyloid plaques (3) and intracellular tangles of the neurofibrillary protein tau but also increased brain iron (in ferritin-associated plaques) (4).
The major component of these plaques is the 40–42-amino acid β-amyloid (Aβ) peptide cleaved from the amyloid precursor protein (APP) (5), which is the ubiquitously expressed trans-membrane metalloprotein (6). β-Secretase and γ-secretases generate the 40–42-amino acid Aβ peptide that is toxic to brain neurons, an event that is accelerated in the presence of iron (7–11). Intracellular iron levels control APP translation via iron-responsive 5′-untranslated region (APP 5′-UTR) sequences in the APP transcript (1, 12), likely mediated via internal ribosome entry mechanisms (13).
In mammalian cells, iron homeostasis is regulated by post-transcriptional events by iron regulatory proteins (IRP1 and IRP2) that bind at high affinity to iron-responsive element (IRE) RNA hairpins and repress the translation of the iron storage protein ferritin as well as transcripts of other iron-associated proteins (14–16). During conditions of iron influx, both IRP1 and IRP2 are released from the light (L) and heavy (H) ferritin IREs to increase the translational efficiency of the L- and H-chains of this intracellular iron storage multimer (17, 18). Transferrin receptor (TfR) mRNA stability is controlled by five IREs (19, 20) in its 3′-UTR, which binds IRP2 more avidly than IRP1 and thereby stabilizes TfR mRNA in a low intracellular iron milieu. Emerging evidence clearly supports iron directly interacting with the L- and H-ferritin IREs to weaken equal binding of IRP1-IRP2 (21).
The canonical IRE stem loops encoded by L- and H-ferritin and TfR mRNAs are highly conserved throughout evolution. Similar 5′-UTR-specific IREs control the translation of Hif-2α (22), ferroportin (IREG-1) (23), the erythroid heme biosynthetic aminolevulinate synthase (eALAS) (24), and mitochondrial aconitase (25), each of which bind avidly to IRP1 to a greater degree than IRP2. In contrast to TfR mRNA, which binds at high affinity to IRP2, the duodenal divalent metal ion transporter (DMT1) (26) encodes an IRE stem loop immediately downstream from its stop codon and preferentially binds IRP1 to mediate iron-dependent message stability. This event may be critical for setting rates of dietary iron uptake (26).
By comparison with the APP IRE in this report, the consensus IRE of all of these iron-associated transcripts, including L- and H-ferritin, is a 26–30-base-long conserved RNA stem loop that forms an RNA helix with a CAGUGN terminal loop (27). Biophysical studies revealed that CG base pairing within the terminal loop generates an AGU triloop and bulge nucleotide (25). An invariant to the canonical IRE RNA stem loop is the presence a bulge cytosine 6 bases upstream of the terminal loop (25). IRP1 was shown to interact with the H-ferritin IRE largely via hydrogen bonding between the phosphate bonds of the AGU trinucleotide loop and amino acids in the cleft between domains 2 and 3 of IRP1(28). The bulge C interacts with 10 amino acids in the domain 4 of IRP1 (28).
In conditions of iron influx, IRP1 no longer binds to IRE stem loops, but instead, an iron-sulfur cluster is assembled to convert IRP1 to an cytosolic aconitase enzyme (28, 29). In this state, IRP1 no longer represses ferritin, Hif-2α, and eALAS mRNA translation and stabilizes DMT-1 (TfR) mRNA stability. By modulating IRP2 stability by two cysteines (Cys-512 and Cys-516) that are predicted to lie in the RNA-binding cleft (30), iron is a significant regulator of IRE-mediated translation of the L- and H-ferritin chains, and IRP2 stabilizes TfR mRNA stability (31).
Supporting the clear physiological importance of IRP1, its expression is essential for growth, including a role in resistance to carcinogenesis (32). Cell growth is conferred by the cis-aconitase activity of IRP1 (33). The fact that many physiological and growth signals phosphorylate IRP1 (14, 34–36) is consistent with the fact that it is the predominant RNA-binding partner to the IRE encoded by the oxygen sensor HIF-2α (22), DMT1 and eALAS mRNAs.
This report supports a key role for IRP1 acting through the functional IRE that exists in the 5′-UTR of APP mRNA (1, 12). We determined the binding selectivity of the IRE in APP mRNA with respect to IRP1 and IRP2 as key RNA-binding proteins. Our biotin-labeled RNA (5′-UTR) pulldown assays ascertained that IRP1, and not IRP2, interacts with the APP IRE using both SH-SY5Y neuroblastoma cells and H4 neuroglioma cells. This selective IRP1 binding was found to functionally drive APP expression. The binding specificity of IRP1 > IRP2 in neural cells was reproduced in human brain and blood samples. In all cases, the pattern of H-ferritin IRE binding to IRP1 and IRP2 was employed as a positive control. Our results showed that the AD-specific APP transcript presents as an extreme example of an IRE-containing mRNA that selectively binds only to IRP1 as a translational repressor of APP expression during intracellular iron depletion with desferrioxamine.
EXPERIMENTAL PROCEDURES
Antibodies
Rabbit anti-human IRP1 antibody (Alpha Diagnostics International, San Antonio, TX) and anti-IRP1 (kind gift from Dr. Sharon Cooperman and Dr. Tracey Rouault, National Institutes of Health) each generated the same results in the assays shown; mouse anti-human IRP2 (Santa Cruz Biotechnology, Santa Cruz, CA) detected the H-ferritin IRE-IRP2 interaction, and a second antibody to IRP2 was also utilized to confirm the selectivity of IRP2 binding as detected with the Santa Cruz Biotechnology antibody. Anti-β-actin, anti-β-tubulin, rabbit anti-APP C-terminal antibody (A8717), and anti-biotin antibodies were from Sigma; and the APP N terminus-specific anti-22C11 antibody was from Chemicon, Temecula, CA.
Sequence Comparison
To generate evolutionary comparisons for an APP 5′-UTR alignment and to generate RNA secondary structure predictions, we downloaded the human, mouse, rhesus monkey APP family sequences from the National Center for Biotechnology Information (NCBI) Entrez data base. Genomic DNA sequences had specific exons and transcription start site (+1) explicitly identified. To ensure completeness, only verifiable 5′-UTR sequences were examined (37). Using ClustalW2, the central 57-base sequence of 5′-UTRs of the human APP 5′-UTR were aligned with the equivalent sequences in mouse and rhesus monkey APP genes (+47 from the 5′ cap site to −43 from the AUG codon) in addition to the IRE domain of the human L- and H-ferritin transcripts. The 5′-UTR-specific IREs in DMT-1, eALAS, and HIF-2α transcripts were compared at the same stringency settings in ClustalW2. All alignments used the same gap setting and were selected so that the central CAGUGC domain of the H-ferritin IRE was anchored in the center of the homology. Homology of the equivalent CAGAGC box in the APP IRE was sought between species where no less than 80% homology was considered notable only when they shared an aligned position with a 100% similarity in the vicinity of the CAGAGC loop motif.
RNA Secondary Structure Predictions
The same 57 APP 5′-UTR motifs used for sequence alignments were chosen to predict their most stably folded RNA secondary structures, as shown in Fig. 2B. Folding predictions were from the RNAshapes program (RNA Studio (38)). RNA secondary structure of the biotinylated APP IRE probes from wild type, CAGA-mutated, and APP IRE-like linear input sequences were each correlated for their relative capacity to bind to IRP1.
FIGURE 2.
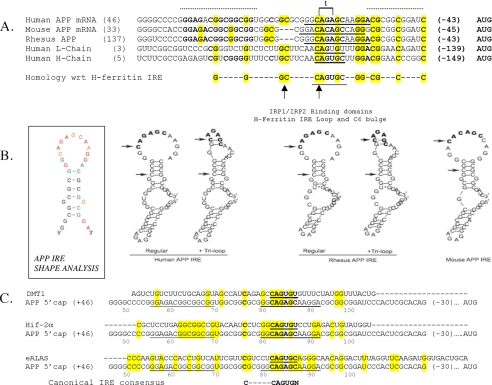
Sequence and RNA structural specificity of IRP1 binding to the IRE motif in the 5′-untranslated region of the APP transcript. Panel A, primary sequence homology among the human, rhesus monkey, and mouse APP IREs where 100% conservation among the three species is demarked by a dotted line above the alignment. Sequences encoding the canonical IRE RNA stem loops in the 5′-untranslated region of ferritin L- and H-chain are aligned to the APP IRE such that CAGUGC terminal loop of the H-ferritin IRE is bold and underlined. The arrows indicate the C-6 bulge and the start of the apical CAGUGN loop in the ferritin IRE. The homologous CAGAGC motif of the APP IRE (panel B) is bold and underlined as six nucleotides in the 13-base terminal loop predicted for the APP IRE (panel B below). The super-conserved homology among all three APP IREs and both L-ferritin and H-ferritin subunits is shown both in bold and highlighted lettering, whereas the evolutionary conservation is demarked only by bold lettering. The sequences matching the known L- and H-ferritin pseudo-triloops are indicated in brackets and by letter t. Panel B, left, RNA secondary structure of the APP IRE was assessed by the SHAPE technique (93). This measured the likelihood of whether any given nucleotide is base-paired to participate in a double-stranded RNA helix or is encoded by single-stranded RNA, where it is exposed to chemical modification (147 bases input APP 5′-UTR RNA). The normalized SHAPE reactivity was incorporated into the RNAstructure 4.6 software, which uses both nearest neighbor free energy parameters and SHAPE data as pseudo-energy parameters to develop a secondary structure prediction. Nucleotides are colored black (unreactive, SHAPE reactivity < 0.2), green (0.2 ≤ SHAPE reactivity < 0.4), orange (0.4 ≤ SHAPE reactivity < 0.6), and red (highly reactive, SHAPE reactivity ≥ 0.6), or gray (no data due to RNA degradation) (description of potential base-pairing within the terminal loop is provided in supplemental Fig. 1. Right, human, rhesus monkey, and mouse APP IRE secondary structures were predicted by the RNAshapes program. The human and rhesus monkey APP IREs were predicted to be capable of forming alternative RNA stem loops, either with no internal base pairing to generate a 13-base terminal loop or alternatively with two hydrogen bonds internal to the terminal loop that may generate an AGA triloop. The mouse APP IRE sequences do not permit this alternative base pairing within the terminal loop. Input nucleotide sequences were chosen to include the evolutionary hyper-conserved motifs (panel A, dotted line) including sequences from 35 and 16 bases on each side of the human APP-specific CAGAGC loop and 33 and 16 bases on each side of the rhesus monkey and mouse CAGAGC IRE loops (panel A). The arrows refer to the beginning position of the CAGAGC motif in the APP IRE, and the second arrow refers to the upstream cytosine (C-6). Panel C, pairwise homology between the APP IRE with corresponding sequences in the DMT-1 IRE, the Hif-2α IRE, and the eALAS IRE. The APP IRE exhibits 38% homology with the IREs of the transcripts for eALAS, Hif-2α, and L-ferritin with a lower 25% homology to the DMT-1 IRE (50% homology with the ferritin H-chain IRE over a 48-base region). A conserved C nucleotide is present +76 bases from the APP mRNA 5′ cap site; this C is similarly placed upstream of the APP terminal loop-specific CAGAGC motif as the conserved C-bulge that is 6 bases upstream of the H-ferritin CAGUGN terminal loop. Panel D, relative position of the functional APP IRE sequence (1) and the second APP IRE-like motif in of the Aβ region in the coding region of linear APP-695 mRNA (+1906 to +1931 with respect to APP-695). The specificity of interactions between IRP1 and biotinylated APP IRE probes relative to matched 37-base APP IRE-like probes was monitored by the pulldown assay as shown in Fig. 1D (n = 3). Row 1, Western blot detection of IRP1 that selectively bound to biotinylated APP IRE beads but not tRNA or biotinylated APP IRE-like beads (in triplicate). Row 2, biotin levels were monitored as loading controls to ensure equal bead recovery between lanes. Rows 3 and 4, Western blot confirmation that unchanged levels of IRP1 and β-actin were present in the lysate supernatants committed to the pulldown assay. CL, cell lysate. Panel E, RNA binding assays with the short and long biotinylated RNA probes encoding the wild type APP IRE when compared with the CAGA deletion. Lanes 1 and 2, 37-base wild type APP IRE (short form); Lanes 3 and 4, 57-base wild type APP IRE (long form); lanes 5 and 6, CAGA mutant version APP IRE (33 bases). IRP pulldown was registered by Western blots as described under “Experimental Procedures.” Panel F, the predicted secondary structures of the 37-nt RNA sequences encoding: 1 and 2, the canonical L-ferritin IRE (without and with the known AGU pseudo-triloop); 3 and 4, the APP IRE without and with the predicted AGA triloop in the GGCAGAGCAAGGA terminal loop; 5, the APP IRE ΔCAGA; 6, the APP Aβ region IRE-like domain. Predictions were by the RNAshapes program using the 37-base input sequences (shown as a positive control; the L-ferritin IRE CAGUGU terminal loop is 9 bases downstream from the 5′ end and 10 bases from the 3′ end).
SHAPE Analysis to Confirm Predicted RNA Structures for the APP IRE
The selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) technique was used to confirm RNA secondary structure predictions of APP 5′-UTR sequences. The human APP mRNA 5′-UTR transcripts were folded in a solution containing 6 mm Mg2+ and subjected to the SHAPE procedure. The normalized SHAPE reactivity was incorporated into the RNAstructure 4.6 software, which uses both nearest neighbor free energy parameters and SHAPE data as pseudo-energy parameters to develop a secondary structure prediction.
Cell Culture and Preparation of Lysates
Human SH-SY5Y neuroblastoma cells and H4 neuroglioblastoma cells were cultured in Dulbecco's modified essential medium supplemented with 10% FBS (Invitrogen) and penicillin/streptomycin (BioWhittaker, Walkersville, MD). Cells were exposed to desferrioxamine (0–100 μm, Calbiochem), iron (50 μm, National Institute of Standards and Technology (NIST), Gaithersburg, MD), bathocuproine (50 μm, Sigma) at times indicated. Iron was provided to cells as iron nitrate as indicated. Cytoplasmic protein lysates were prepared by homogenizing the cells in ribonucleoprotein immunoprecipitation buffer (25 mm Tris, pH 7.4, 1% Nonidet P-40, 0.5% sodium deoxycholate, 15 mm NaCl, protease inhibitors, RNase inhibitor, and 10 μm DTT).
Establishment of Stable IRP1 Knockdown Clones and Transfections with APP 5′-UTR Luciferase
SH-SY5Y cells were transfected with the pSilencer 2.1-U6-IRP1 siRNA (puro) and pSilencer 2.1-U6-Control (gift from Dr. Suzi Torti, Wake Forest University, Winston-Salem, NC). After transfection (24 h), 2 μg/ml puromycin was added to medium for selection (see Ref. 31). Stable clones were analyzed by real-time qPCR and Western blotting to confirm IRP knockdown. Neuroblastoma cells (SH-SY5Y), either those selected with puromycin for IRP1 knock-out or control knockdown shRNA cells, were transfected with 10 mg of DNA from the pGAL (APP 5′-UTR) construct and were co-transfected with 5 mg of DNA from a construct that expresses green fluorescent protein (GFP). Luciferase and GFP reporter genes were expressed from an SV40 promoter. Transfections were performed in the presence of Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). Typically neuroblastoma cells were grown in flasks (100 mm2) for each treatment. Each flask was transfected (12 h) and subsequently read for luciferase activity as reported previously (1).
IP-RT-PCR
Cell lysates (500 μg) were incubated for 30 min at 4 °C in the presence of 30 μl of protein A/G plus agarose (Santa Cruz Biotechnology). The lysates were then incubated for 3 h with agarose beads and 3 μg of either IRP1 (Alpha Diagnostics International) or IRP2 (Santa Cruz Biotechnology) antibodies at 4 °C. After washing (five times), RNA was extracted from both supernatants and protein A/G preparations using TRIzol reagent (Invitrogen). RT-PCR was performed using SuperScript III (Invitrogen and Integrated DNA Technologies, Coralville, Iowa) to detect the presence of mRNAs bound to IRP1 or IRP2 using gene-specific primer pairs of H-ferritin primers (forward, 5′-CTC TCC TTA GTC GCC GCC ATG-3′; reverse, 5′-AAT TCT TTG ATG GCT TTC ACC TGC-3′; 500 bp of product size; 55 °C of annealing temperature) and APP primers (forward, 5′-TGA GCG CAT GAA TCA GTC TC-3′; reverse, 5′-CCA GGC TGA ACT CTC CAT TC-3′; 250 bp of product size; 55 °C of annealing temperature).
Biotin Pulldown Assay
Biotinylated RNA oligonucleotides; H-ferritin IRE (biotin-5′-GGG UUU CCU GCU UCA ACA GUG CUU GGA CGG AAC CCG G-3′), APP IRE (5′-biotin-GC GGU GGC GGC GCG GGC AGA GCA AGG ACG CGG CGG AU-3′), APP IRE-ΔCAGA (5′-GCG GUG GCG GCG CGG GGC AAG GAC GCG GCG GAU-3′), and APP IRE-like (5′-biotin-G GUG UUG UCA UAG CGA CAG UGA UCG UCA UCA CCU UGG-3′) were purchased from Invitrogen. Cell lysates (100 μg) were incubated with 100 nm biotinylated oligonucleotide for each IREs for 3 h at room temperature. Paramagnetic streptavidin-conjugated Dynabeads (Invitrogen) were washed with ribonucleoprotein immunoprecipitation buffer and then added into lysates to bind IRP(1/2)-biotinylated-RNA complexes and incubated 1 h at room temperature. After five washes, the proteins bound to the beads were analyzed by Western blotting for IRP1, IRP2, and biotin. The blots were developed with chemiluminescence (Pierce) and visualized with a PhosphorImager, and IREs-bound IRPs were quantified using the Quantity One software (Bio-Rad).
RNA Gel Shift Analysis
For RNA electrophoretic mobility shift assays (REMSA), cytoplasmic extracts or recombinant IRP1 were incubated with radiolabeled transcript (2–10 × 104 cpm, 2.5–10 pg) for 30 min at 22 °C in a reaction volume of 10 μl made up with cytoplasmic extraction buffer. In some reactions, mouse polyclonal serum to IRP1 or mouse preimmune serum was included to generate supershifting complexes. Subsequently, RNase T1 (0–1 units) was added to the mixture for 10 min at 22 °C followed by heparin (0–10 mg/ml) (Sigma) for 10 min at 22 °C. RNA loading dye (0.1 volume of 90% glycerol, 10 mg/ml bromphenol blue, and xylene cyanol; 5× Tris-borate-EDTA) was added to each sample. The RNA-protein complexes were resolved at 4 °C for 15–20 min at 200 V in 0.5× Tris-borate-EDTA on a 1.5-mm 4–5% polyacrylamide mini gel (Bio-Rad) (acrylamide:bisacrylamide ratio of 36:1 or 70:1) after pre-electrophoresis (200 V for 20 min; 0.5× Tris-borate-EDTA running buffer). After electrophoresis, gels were fixed in 10% isopropyl alcohol, 7% acetic acid and vacuum-dried, and RNA-protein interactions were detected by use of a PhosphorImager (GE Healthcare).
Preparation of Human Brain Cytoplasmic Extracts
Human brain tissues were obtained from the Massachusetts Alzheimer Disease Research Center (ADRC) Tissue Resource Center. The brain tissues of three AD patients (70.3 years ± 12) and three age-matched controls (87 years ± 6.5) were homogenized in 3 volumes of ribonucleoprotein immunoprecipitation buffer. Nuclei and debris were removed by centrifugation at 3000 rpm for 30 min. The supernatant was further centrifuged at 14,000 rpm for 60 min. Biotin pulldown assay was performed on the resulting supernatant.
Real-time qPCR
Real-time qPCR was carried out on the ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA). The standard curve method was used for quantification. Total RNA was isolated using TRI Reagent (Sigma) reagent according to the manufacturer's instructions. After DNase I digestion, cDNAs were synthesized with SuperScript III first-strand qPCR supermix (Invitrogen) according to the manufacturer's instructions. Aliquots (1 μl) of cDNA were added to 24 μl of reaction mixture containing 2× SYBR Green PCR master mix (Applied Biosystems) and 300 nm primers. The IRP1, IRP2, β-actin, and TfR1 primers were designed by Dr. S. Torti (31) and ordered from Invitrogen. The APP primer set was purchased from Qiagen.
Competition Assay
Recombinant human IRP1 (rhIRP1) with an N-terminal His6 tag in Escherichia coli was expressed overnight at 37 °C in LB medium, and purified with nickel-nitrilotriacetic acid Fast Start kit (Qiagen, Valencia, CA) under native conditions. rhIRP1 (100 ng) was incubated for 3 h at room temperature with 25 nm of either biotinylated APP IRE or H-ferritin IRE in the presence of increasing concentrations of (25, 250, 625, 1250, 2500, 5000 nm) of the appropriate unlabeled competitor. The recombinant proteins bound to the IREs were precipitated using Dynabeads for 1 h at room temperature and analyzed by Western blotting. To measure IRP1-IREs binding affinities, we calculated the dissociation constant (Kd value) (26, 39).
Preparation of Human Blood Cell Lysates
Cell lysates of blood samples taken from six age-matched control subjects and six AD patients were analyzed for IRP1-APP IRE interactions by a biotin pulldown assay. Cytoplasmic extracts were prepared as was done for the brain.
Statistical Analysis
Values in the text and figures are presented as means ± standard deviations of experiments. Equal variance or separate variance from two sample two-tailed t tests were used to compare and evaluate significant differences between the groups. Data are means ± S.E., n = 7, p < 0.001, analyzed by two-tailed t tests, by analysis of variance + Dunnett's test.
RESULTS
Iron-responsive Element Sequences in the APP 5′-UTR Bind to IRP1, but Not IRP2, in SH-SY5Y and H4 Neural Cell Lines
Our published work identified an IRE-like sequence (APP IRE) (1) in the 5′-UTR of APP mRNA that was homologous to the well characterized canonical 5′ cap site IRE stem loops in the L- and H-chain ferritin mRNAs that bind equally to IRP1 and IRP2 (15) to control iron-dependent translation (40). To begin to investigate the specificity and mechanism of action of the APP IRE, we first aligned the sequences encoding 37 bases of the functional 5′-UTR-specific APP IRE with sequences encoding the IRE stem loops of L- and H-ferritin mRNAs (NCBI, BLAST). This alignment included comparison with an IRE-like sequence reported in the RNA encoding the Aβ domain specific to the coding sequences of the APP transcript (41).
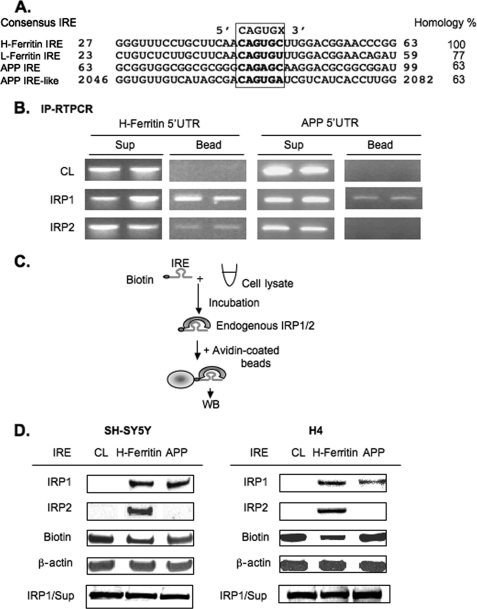
In the alignment of Fig. 1A, both the APP IRE and the APP IRE-like sequence in the downstream Aβ region of the APP transcript were each 63% homologous with the canonical L- and H-ferritin IREs. The H-ferritin IRE was 77% homologous with the L-ferritin IRE (in the vicinity surrounding the central IRP1-IRP2-binding CAGUGN IRE loop consensus motif). RNA gel-shift experiments were previously used to demonstrate that an IRP interacts with IRE sequence probes from the APP 5′-UTR and that a modulated interaction likely confers APP translational regulation (1). Here we investigated whether it is IRP1, IRP2, or both that interact with this RNA secondary structure by the use of both immunoprecipitation (IP)-linked RT-PCR experiments (IP-RT-PCR) and biotinylated RNA pulldown experiments (Fig. 1B).
FIGURE 1.
The iron-responsive element in the 5′-UTR of APP mRNA binds to IRP1, but not IRP2, in both SH-SY5Y and H4 neural cell lines. A, alignment of nucleotide sequence from H-ferritin, L-ferritin, and APP IREs showing homology to the H-ferritin IRE sequence. B, representative IP-RT-PCR experiment (n = 4) showing that APP mRNA binds IRP1, but not IRP2, whereas the H-ferritin mRNA binds both IRP1 and IRP2. SH-SY5Y cell lysates (500 μg) were incubated with 3 μg of either IRP1 or IRP2 antibody and 30 μl of A/G PLUS agarose bead slurry. Rabbit IgG, precleared with beads, was used as a negative control. Following IP, RT-PCR was performed from RNAs extracted from both beads and supernatant (Sup) where specific H-ferritin and APP primers were used to detect the presence/absence of each transcript. Data from duplicate samples are presented. C, a diagrammatic representation of the technique for the biotin pulldown assay. Briefly, biotin-labeled IRE probe was incubated with cell lysate (CL), and the binding protein-IRE complex was precipitated with streptavidin-coated beads followed by Western blotting (WB) of the attached proteins. D, SH-SY5Y (n = 8) and H4 (n = 3) lysates were incubated with 37-base-long biotinylated RNA probes encoding either H-ferritin or APP IRE sequences as shown. IRP1/2 IRE complexes were detected by WB using anti-IRP1/2 antibodies. Total lysate for each sample was probed for β-actin to ensure correctly balanced loading. IRP1 levels in the supernatants were unchanged across all pulldowns (bottom row).
IP-RT-PCR was employed to compare whether APP mRNAs might co-precipitate with IRP1 and/or IRP2 where both have been proven to bind equally to the canonical H-ferritin-TfR IREs (15). Using SH-SY5Y lysates and antibodies to IRP1 and IRP2, both the pulldown (Bead) and the supernatant (Sup) fractions were collected followed by RNA extraction. After reverse transcription (RT) and PCR (RT-PCR), the use of specific primers for either H-ferritin or APP transcripts generated correctly sized DNA amplicons specific for each cDNA (Fig. 1B). A representative IP-RT-PCR experiment with duplicate samples demonstrated that only IRP1 and not IRP2 bound to APP mRNA, whereas both IRP1 and IRP2 interacted with H-ferritin mRNA.
These results were consistent from six independently conducted IP-RT-PCR assays (see our Ref. 42) for a full method). Supernatants that maintained abundant endogenous mRNAs were used as a positive control for detecting the presence of both H-ferritin and APP RNA under all conditions (Fig. 1B). Immunoprecipitation using preimmune rabbit IgG instead of specific antibody was a negative control. We concluded that the H-ferritin mRNA bound to both IRP1 and IRP2 from SH-SY5Y cells, whereas APP mRNA bound only to IRP1.
To cross-reference this novel finding that IRP1 selectively (and not IRP2) binds to the APP mRNA and that this selective interaction takes place via the functional IRE sequences in the 5′-UTR of the transcript, we employed reverse technique by use of biotin pulldown assay (Fig. 1C). In this assay, we used biotinylated specific probes for core IRE sequences from the H-ferritin and APP mRNA 5′-UTRs (43) in the procedure shown in Fig. 1C. Confirming our IP-RT-PCR results in Fig. 1B, the data shown in Fig. 1D demonstrated that IRP1 and not IRP2 bound to the APP IRE (n = 8). From separately conducted experiments using two independent neural cell lines (SH-SY5Y and H4 cells), we observed a consistent pattern of selective IRP1, but not IRP2, binding to the APP IRE probes. Under identical conditions, the H-ferritin IRE sequences bound to both IRP1 and IRP2 (Fig. 1D), confirming established RNA gel shift assay data in previous reports (15, 26, 42). These biotinylated RNA pulldown results showed that APP IRE sequences selectively bound to IRP1 and confirmed the results of our IP-RT-PCR experiments (panel A), showing that that IRP2 did not bind to APP IRE sequences under conditions where IRP2 was readily detected to bind to the canonical H-ferritin IRE (n = 8). We concluded that the pattern of selective IRP1 binding to the APP 5′-UTR-specific IRE was consistently observed in two independent neural cell lines, that of H4 neuroglioblastoma cells (n = 3) and for SH-SY5Y neuroblastoma cells.
The canonical IRE encoded by the mRNAs for the ferritin L- and H-chains and the TfR mRNA 3′-UTR is a short conserved stem loop (∼30 nt). Using the Clustal2 DNA sequence search software (NCBI), we aligned 57 bases from APP IRE region, as encoded by 5′-UTR sequences +46 from the 5′ cap site to −43 bases from the start codon of the 146-nt human APP mRNA, with corresponding sequences from the rhesus monkey and mouse APP genes compared with the human L- and H-ferritin genes (Fig. 2A) (1, 44). This analysis revealed a strong mammalian homology anchored around the APP-specific CAGAGC motif (+82 to +87) similar to the canonical CAGUGC loop motif of the H-ferritin IRE stem loop (CAGUGN is the core IRE loop consensus). On each side of the APP CAGAGC motif, we identified conserved domains that were 100% conserved among the human, mouse, and rhesus monkey APP genes. In Fig. 2A, the core evolutionary conserved bases 5′ upstream of the APP CAGAGC motif are demarked by a dashed line, as are 11 bases 3′ downstream of this motif. The APP IRE exhibited a critical evolutionary conservation of the C nucleotide positioned 6 bases upstream of the APP CAGAGC motif equivalent to the spacing between the C bulge upstream of the CAGUGC terminal loop of the canonical H-ferritin IRE (Fig. 2). We extended this alignment of the APP IRE core domain to include IREs encoded by mRNAs that preferentially bind with IRP1 (>IRP2), as reported for 3′-UTR of DMT-1 mRNA (26) and the 5′-UTR of HIF-2α and eALAS mRNAs (22, 25). The relative positions of the terminal loops and upstream C-6 nucleotides of these three mRNAs also exhibited conserved sequence alignments as for the APP IREs as shown in Fig. 2C.
RNA Secondary Structure Determinants of IRP1 Binding to the IRE Stem Loop in the APP 5′-Untranslated Region
We employed further RNA binding assays to assess the impact of RNA secondary structure to influence the specificity by which IRP1 interacted with the 5′-UTR-specific APP IRE relative to related RNA probes of equal homology to the H-ferritin IRE (Fig. 1A) (41). For this purpose we used biotinylated RNA probes encoding either (i) the APP 5′-UTR-specific IRE or (ii) a second APP IRE-like RNA sequence in the APP coding region sequence (Fig. 1A). Using equal triplicate input loadings of SH-SY5Y cell lysates, we demonstrated that the 5′-UTR-specific biotinylated RNA probes encoding the core APP IRE interacted with IRP1, whereas under identical conditions of RNA binding assay, the matched RNA probe encoding the second APP IRE-like sequence did not bind to IRP1 (Fig. 2D). These data served as a selectivity control that IRP1 specifically binds only to the fully functional iron-responsive 5′-UTR sequences in the APP 5′-UTR (1). Control biotinylated tRNAs included in the these assays showed no binding to IRP1 or IRP2 (Fig. 2D).
Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) was used to identify those bases in the APP IRE likely to be in single-stranded and those participating in base-paired regions of the RNA secondary structure (Fig. 2B and supplemental Fig. 1). These results were found to be consistent with the computer-predicted RNA stem loop structure for the APP IRE (RNAshapes software as described in the legend for Fig. 2B). Local nucleotide flexibility of RNA was monitored by treating synthetic RNAs encoding the 147-nucleotide APP IRE with 2′-hydroxyl-reactive electrophiles, which selectively and covalently modify flexible nucleotides (i.e. single-stranded RNA nucleotides) at the 2′-ribose position. SHAPE data were found to be consistent with the presence of the predicted 13-base single-stranded terminal loop at the apex of six double-stranded GC nucleotides interrupted by two unpaired G nucleotides (Fig. 2, B, E, and F). It is possible for further GC base pairing to occur between GC dinucleotide at positions 2 and 3 of the terminal loop with CG at positions 7 and 8 in the human APP IRE (see alternative APP IREs, Fig. 2B). In the human APP IRE, this base pairing may generate a key AGA triloop that cannot form in the case of the APP IRE in the 5′-UTR of the mouse APP transcript (Fig. 2B).
To further investigate the sequence specificity of APP IRE binding to IRP1, the central CAGA quartet in the loop motif was deleted from the biotinylated wild type RNA probes (Fig. 2E). Shown is a representative biotinylated RNA pulldown assay in which we observed that the 33-nt RNA probe encoding the ΔCAGA-deleted version of the APP IRE no longer bound to lysate IRP1 from SH-SY5Y cells (10 μg of input lysate), whereas wild type biotinylated 37-nucleotide APP IRE RNA probes selectively bound to IRP1. IRP1 levels in the supernatants were unchanged (Fig. 2E), and both 37-nucleotide (short form) and 57-base-long input APP IRE probes avidly bound to IRP1 in SH-SY5Y lysates.
The RNA binding data shown in Fig. 2, D and E, was interpreted in the light of computer-based predictions of the base pairing of the RNA sequences for the APP IRE (predictions shown in Fig. 2F were conducted by RNAshapes as described under “Experimental Procedures”). In Fig. 2F, the most stable RNA folded from wild type APP IRE sequences was compared with (i) the L-ferritin IRE, (ii) the ΔCAGA-deleted version of the APP IRE, and (iii) the second APP IRE-like RNA stem loop.
Both the L-ferritin IRE and the APP IRE sequences were predicted to fold into two alternative equally stable RNA structures with similar ΔG values (Fig. 2F; stem loops 1 and 2 for L-ferritin IREs and stem loops 3 and 4 for APP IREs). Notably the L-ferritin IRE is known to coexist as an RNA stem loop with a CAGUGU terminal loop, but alternative internal loop G-C base pairing can generate an RNA structure with a terminal AGU pseudo-triloop (25). Critically, using the same folding parameters in RNAshapes, we predicted two alternating structures for the 37-base APP IRE such that the 13-base terminal loop could be refolded to form a AGA triloop resulting from two internal G-C base pairs (Fig. 2, B and F).
Using these same thermodynamic constraints, the predicted RNA structure of the APP IRE ΔCAGA deletion mutant was not capable of folding to form a AGT triloop (Fig. 2F, stem loop 5). Consistent with this prediction, RNA pulldown assays with biotinylated RNA probes encoding this deletion no longer exhibited the IRP1 binding of equivalent wild type biotinylated APP IRE probes (Fig. 2E). Also, the APP IRE-like sequence encoded by the Aβ domain of the APP coding region was also predicted to be capable of folding in the absence of a triloop in the CAGUGN loop motif (Fig. 2F, stem loop 6). In this configuration, this consensus was unavailable for IRP1/2 binding, which is consistent with the lack of IRP1 binding in Fig. 2D. As a control for these experiments, IRP1 was shown to be present in the supernatants after RNA pulldowns with all biotinylated mRNA probes (standardized by Western blotting with β-actin) (Fig. 2B).
IRP1 Binds to the APP IRE in a Dose- and Time-dependent Manner in Response to Intracellular Iron Chelation in SH-SY5Y Cells
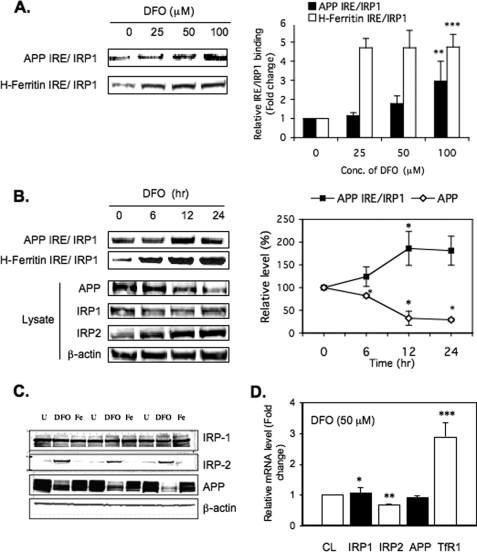
Many reports showed that iron chelation with DFO increased the binding of both IRP1 and IRP2 to H-ferritin mRNA and thereby repressed translation of its L- and H-subunits (14, 17, 45). Intracellular iron chelation was also shown to inhibit APP expression (1). To examine the mechanism by which DFO decreased APP expression in SH-SY5Y cells, potentially via translational repression, the experiments shown in Fig. 3, A and B, measured IRP1 binding to the APP IRE under dose-responsive and time-dependent conditions of iron chelation. Using the biotinylated RNA pulldown assay, as shown in Figs. 1 and 2, we found that IRP1 binding to the APP IRE was simultaneously increased by DFO treatment of SH-SY5Y cells similar to IRP1 binding and translational repression of the H-ferritin IRE.
FIGURE 3.
Increased IRP1 binding to APP IRE probes in response to dose- and time-dependent treatment of SH-SY5Y cells with the intracellular iron chelator, DFO. A, DFO dose response (n = 3). SH-SY5Y cells were treated with 0, 25, 50, or 100 μm DFO for 12 h followed by biotin pulldown assay of the IRP1-APP IRE and IRP1-H-ferrritin IRE interactions. Bead samples were Western blotted to detect the levels of bound IRP1. Quantitation of Western blots was performed using Quantity One software (Bio-Rad). The values plotted are means ± S.D. of n = 3 independent experiments for each time point. Conc., concentration. B, DFO time course (n = 3). Left panel, SH-SY5Y cells were treated with 50 μm DFO for 0, 6, 12, or 24 h followed by WB of IRP1 bound to either biotinylated APP-IRE or biotinylated H-ferritin IRE probes (top two rows) and WB of 5 μg of total lysate to measure APP, IRP1, IRP2, and β-actin levels. Right panel, densitometry and graphic measurement of the time course of DFO-induced binding of SH-SY5Y IRP1 to APP IRE probe correlated directly with total levels of APP in the same lysates. The values plotted are means ± S.D. of n = 3 independent experiments for each time point. C, second representative WB of an experiment showing DFO-dependent reduction of APP expression in SH-SY5Y cells (50 μm DFO (48 h) where β-actin and IRP1 levels were unchanged (IPR2 protein stabilization in response to DFO; experimental positive control)). D, real-time qPCR analysis and measurement of DFO-regulated gene expression (n = 3). SH-SY5Y cells were treated with 50 μm DFO for 12 h and then harvested for quantitative RT-PCR assays. The expression and steady-state mRNA levels of IRP1, IRP2, APP, and TfR1 genes were averaged relative to untreated control condition and corrected for the expression of the β-actin gene. TfR1 mRNA expression was used as a positive control because TfR levels are already known to be up-regulated by DFO via IRE-dependent stabilization of the receptor transcript. CL, cell lysate; U, untreated.
First, we performed dose-response experiments (n = 3) in which SH-SY5Y cells were treated with increasing amounts of DFO (Fig. 3A) to examine whether increased IRP1 binding to APP IRE sequences might correlate with inhibition of intracellular APP levels. Cells were harvested after a 12-h period, and the results reproducibly showed that increasing concentrations of DFO enhanced IRP1 binding to the 37-base biotinylated APP IRE probe. Maximum binding of IRP1 to APP IRE was achieved at the highest concentration used, 100 μm DFO representing almost 3-fold increases in IRP1-APP IRE binding. In comparison, IRP1-H-ferritin IRE interaction reached a plateau at the much lower dose of 25 μm DFO with a maintained maximal 4.74-fold increase in binding at 100 μm chelator. DFO treatment above 100 μm was determined to be toxic to SH-SY5Y cells. The statistical significance for increased APP IRE binding to SH-SY5Y IRP1 was p < 0.001, as for the H-ferritin IRE, each in response to 100 μm DFO treatment for 12 h.
Secondly, time course experiments (6, 12, and 24 h) were set up using the concentration of 50 μm DFO that induced 2-fold (12 h) increased binding of IRP1 to the APP IRE (100 nm biotinylated probe). As in the dose-response experiment, IRP1 binding to the APP IRE was increased 2-fold by 12 h with no further increase at 24 h of treatment with DFO in SH-SY5Y cells (Fig. 3B). Consistent with this result, 24 h of treatment with 100 μm DFO significantly decreased APP protein levels to 25% of original levels (n = 4).
Over several experiments, we observed a consistent average 2–3-fold difference in IRP1-APP IRE binding in cells treated with DFO (at 50–100 μm maximal concentrations) relative to controls. By contrast, the H-ferritin IRE binding to both IRP1 and IRP2 increased 5-fold in conditions of DFO treatment of SH-SY5Y cells (Fig. 3, A and B).
Thirdly, to ensure effective intracellular iron chelation with DFO, IRP2 levels increased by 8-fold in SH-SY5Y (positive control), whereas the levels of IRP1 were modestly influenced and showed no increase in the same cells (17, 34, 46) (Fig. 3, B and C). As expected, DFO treatment did not affect the intracellular steady-state levels of β-actin, and SH-SY5Y cells were shown to be fully viable. As a positive control, the binding interaction of IRP1 to biotinylated H-ferritin IRE probes was increased in response to intracellular iron chelation with DFO as was previously reported by several groups (17, 47, 48). In the representative experiment shown in Fig. 3 (n = 4), DFO treatment was observed to increase binding of biotinylated H-ferritin IRE probes to SH-Sy5Y-specific IRP2, whereas we continued to observe no IRP2 binding to the APP IRE (not shown).
Fourth, to determine whether DFO limited APP expression at the mRNA level in SH-SY5Y cells, we measured whether intracellular iron depletion changed the levels of the mRNAs for APP, TfR, IRP1, and IRP2, in DFO-treated cells (50 μm, 12 h) (Fig. 3D). The steady-state level of each mRNA was analyzed by qRT-PCR. As a positive control to confirm that DFO entered cells, the levels of TFR mRNA were shown to increase by 3-fold (positive control, see Refs. 26 and 49). We consistently observed (n = 5) that APP mRNA levels were unchanged in response to intracellular iron chelation (Fig. 3D), whereas at the same time, APP protein was decreased 4–5-fold in SH-SY5Y cells (p < 0.05) (Fig. 3C). During all conditions of changed iron levels, only IRP1 (not IRP2) remained the sole binding partner to biotinylated APP IRE probes, whereas matched H-ferritin IRE probes exhibited increased binding to both IRP1/2 during intracellular iron chelation with DFO.
We concluded from the representative data shown in Fig. 3 that intracellular iron chelation with DFO consistently decreased APP levels by up to 5-fold completely at the level of protein production and that IRP1 had a role in APP mRNA translational repression. For the purpose of ensuring specificity, each Western blot (n = 4) showed that β-actin and IRP-1 protein levels were unchanged in response to DFO treatment of SH-SY5Y cells. To confirm the metal specificity of DFO, we compared SH-SY5Y cells treated with either the copper chelator bathocuproine (50 μm) or DFO (50 μm) for 24 h (see supplemental Fig. 2). The results of standardized Western blots demonstrated that APP expression was reduced 2-fold by DFO when compared with a slight 30% increase in APP expression by the copper chelator. As controls, IRP2 expression was increased 5.4-fold after DFO treatment of SH-SY5Y cells, whereas the Wilson's ATPase, a copper-responsive protein, was induced 2.6-fold by bathocuproine and was slightly decreased after DFO treatment (supplemental Fig. 2).
IRP1 Knockdown in H4 Cells Increased APP Expression without Change to β-Actin mRNA
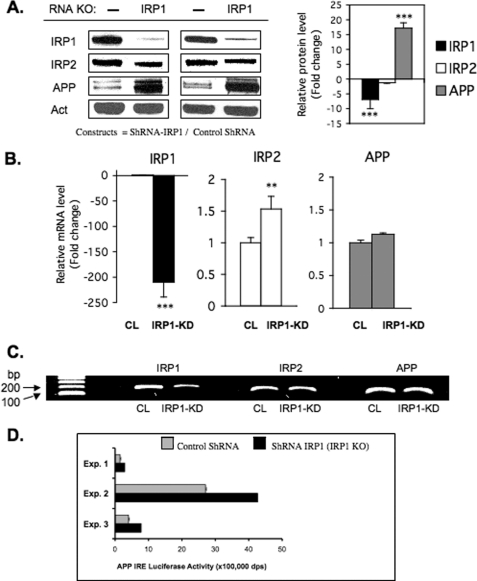
The experiments in Fig. 4A were conducted to determine the consequences of genetic IRP1 knockdown (KD) to specifically influence APP expression. H4 cells were stably transfected with shRNA plasmids encoding empty vector (Control) and IRP1-targeting siRNA sequences each under puromycin selection, as described by Torti and colleagues (31). IRP1 knockdown clones were isolated and grown, whereas IRP2 knockdown cells were refractory to growth and prohibited the separation of independent clones in these cells. We examined the expression of APP levels by Western blot analysis and found that APP expression was increased 17-fold in independent clones of shIRP knockdown cells (n = 4), whereas β-actin and IRP2 levels were unchanged.
FIGURE 4.
Stable knockdown of IRP1 increases the expression of APP in shRNA transfected H4 neural cells. A, total protein extracts were prepared from SH-SY5Y cells stably expressing shRNAs against human IRP1. A non-targeting siRNA sequence was transfected as a control. IRP1 and APP protein levels were determined by Western blot in control cells and IRP1 knockdown cells, and β-actin (Act) was used as a control (n = 4). The relative change of IRP1, IRP2, and APP protein levels was quantitated in two of the clones (left panel) and presented as -fold change (histogram; right panel). B, real-time qPCR (n = 6) was carried out on the ABI Prism 7000 sequence detection system (Applied Biosystems). Total RNA was isolated using TRIzol reagent (Sigma) according to the manufacturer's instructions. cDNA was synthesized with SuperScript III first-strand qPCR supermix (Invitrogen) according to the manufacturer's instructions. The primers (IRP1, IRP2, β-actin, TfR1) were designed as in Ref. 31 and ordered from Invitrogen. The APP primer set was purchased from Qiagen and has been benchmarked on several reports for accurate measurement of APP mRNA levels (62). CL, cell lysate. KD, knockdown. C, representative 2% agarose gel employed during qRT-PCR and analysis to generate the data shown in panel B. D, three separate reporter assays (Exp. 1–3) registering APP 5′-UTR-luciferase activity in IRP1 shRNA knockdown cells relative to normal IRP1 expression (empty shRNA control).
To determine whether increased APP expression was a consequence of transcription or at the protein translational level, we monitored APP mRNA levels by qRT-PCR analysis (Fig. 4B). For this purpose, we compared H4 cells transfected with the IRP1 knockdown plasmid with cells transfected with the control shRNA (also grown under conditions of puromycin selection). Indeed, although APP protein expression was increased 17-fold (panel A), we found that APP mRNA levels were unchanged. Thus, the consequence of genetically depleting H4 cells of IRP1 was a release of a translation block on APP expression by IRP1.
By correlating RT-PCR and Western blot analysis of IRP1 knockdown when compared with control knockdown cells, we concluded that the loss of IRP1 in the shRNA-IRP1 cells was likely the result both of IRP1 mRNA destabilization and inhibition of IRP1 translation. However, the lack of intracellular IRP1 caused no change to APP mRNA levels but a dramatic 17-fold increase in APP protein levels that was consistent with translational control of APP by IRP1 in H4 cells. These results are consistent with the model that IRP1 can repress APP mRNA translation by a similar mechanism to IRP1 block of ferritin mRNA expression at the level of mRNA translation.
In Fig. 4D, three independent transfection-based experiments showed that the reduced steady-state IRP1 levels activated expression of an APP IRE luciferase reporter gene (n = 3). We concluded that IRP1 regulates APP expression by repressing translation driven by APP 5′-UTR sequences (Fig. 4D). Thus, we provide functional data unequivocally demonstrating that expression of the Alzheimer APP is functionally driven by IRP1 via the APP IRE at the level of message translation.
Human rhIRP1 Interacted with the APP IRE with a Similar Binding Affinity in Vitro to That of the H-ferritin IRE Interaction
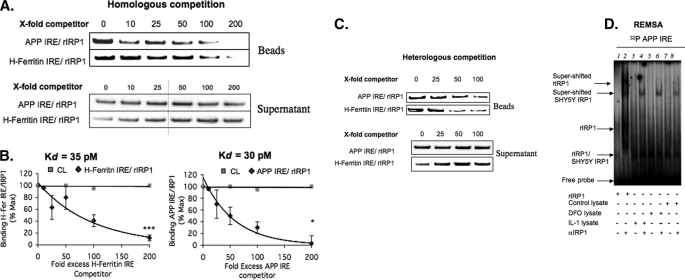
We measured the dissociation constant (Kd) for the RNA-protein interaction between human IRP1 (rhIRP1) (50) and biotinylated APP IRE probes. Using 25 nm biotinylated probes and 100 ng of purified rhIRP1 input, we demonstrated 90% inhibition of rhIRP1 binding at 200-fold excess unlabeled homologous probes for both APP IRE and H-ferritin IRE sequences (Fig. 5A). The addition of tRNA had no effect on the binding of the biotin APP IRE or H-ferritin IRE to rhIRP. The Kd value of the APP IRE-rhIRP1 complex was calculated by Scatchard analysis (39) at 30 pm, and the Kd value of the canonical H-ferritin-IRE complex was calculated at 35 pm (Fig. 5B). We then performed cross-competition experiments using the unlabeled H-ferritin or APP IRE to compete with the opposing biotin-labeled APP IRE (top row) and H-ferritin (bottom row) (heterologous competition shown in Fig. 5C). The same -fold excess unlabeled heterologous probe also competed biotinylated IREs for binding to rhIRP1 (Fig. 5C). Taken together these results confirmed published RNA gel-shift data (1) that the APP IRE binds to IRP1 at equivalent in vitro affinity (30 pm) to that of the classical interaction between the H-ferritin IRE and IRP1.
FIGURE 5.
rhIRP1 binds to APP IRE and H-ferritin IRE probes with similar binding affinity in vitro. Competition assays were employed (n = 4) with rhIRP1 and the use of incrementally increasing concentrations of APP IRE and H-ferritin-IRE RNAs in the presence of biotinylated RNA probes (“Experimental Procedures”). A, homologous competition (n = 3). rhIRP1 was incubated with either 25 nm biotinylated APP IRE or H-ferritin IRE in the presence of a 0-, 10-, 25-, 50-, 100-, or 200-fold (25, 250, 625, 1250, 2500, or 5000 nm) excess of unlabeled APP IREs or H-ferritin IRE. B, Scatchard plots from the quantitated data of panel A were used to calculate the dissociation constants (Kd) of rhIRP1 binding to H-ferritin IRE (left side) or APP IRE (right side). C, heterologous competition (n = 3). 25 nm biotinylated APP IRE was competed with 0-, 25-, 50-, or 100-fold unlabeled H-ferritin IRE, and likewise, 25 nm biotinylated H-ferritin IRE was competed with 0-, 25-, 50-, or 100-fold unlabeled APP IRE. D, RNA gel-shift analysis was performed with the 37-nt radiolabeled RNA probe encoding APP IRE (37 nt) sequences as used in the biotin pulldown assays (previously reported by Rogers et al. (1)), including a supershift assay demonstrating that the APP IRE probes employed for biotin pulldown assays detected rhIRP1- and SH-SY5Y-specific IRP1 binding with the same specificity. Lane 1, rhIRP1; lane 2, rhIRP + IRP-specific antibody; lane 3 SH-SY5Y/DFO, 100 μm; lane 4, SH-SY5Y/DFO 100 μm + IRP Ab; lane 5, SH-SY5Y/IL 1β; lane 6, SH-SY5Y/IL-1β + IRP Ab; lane 7 SH-SY5Y/Control; Lane 8, SH-SY5YControl + IRP1 Ab.
RNA gel-shift experiments (REMSA) were performed (1) to cross-reference that the same length probe used in biotin pulldowns would show specific binding between APP IRE probes and IRP1 (Fig. 5D). The results of a representative REMSA experiment are shown in panel D, confirming our previously published results that like the H-ferritin IRE, APP IRE probes bound selectively to rhIRP1 to form a co-migrating SH-SY5Y-specific IRP1-APP IRE REMSA complex that was specifically supershifted with antibody to IRP1. In this experiment, treatment with DFO increased IRP1 binding to the APP IRE as shown in Fig. 3. IL-1β also increased binding, consistent with our previous report (1).
Selective IRP1, but Not IRP2, Binding to the APP IRE Is Observed in Human Brain Temporal Cortex Tissues and in Human Blood
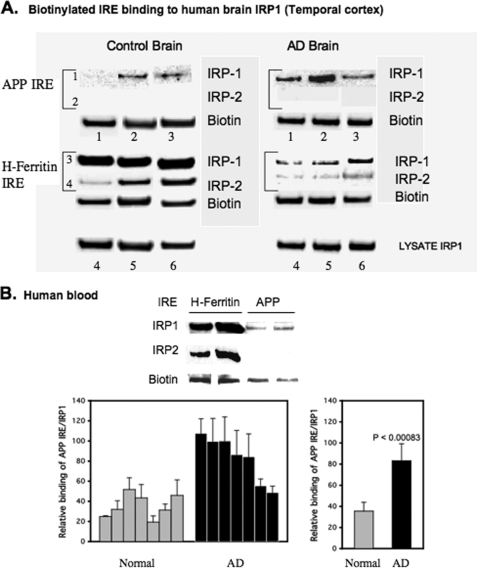
In AD brains, especially in regions such as the cortex and hippocampus that are particularly susceptible to the neuropathological changes of AD, increases in iron concentration have been measured by both MRI (51) and immunohistochemistry (52, 53) in neurons, astrocytes, and microglia (54). To examine the pattern of IRP1 relative to IRP2 binding to APP and H-ferritin IRE probes in human brain samples, we performed further biotin pulldown assays using temporal cortex tissue from control and AD patients (Alzheimer Disease Research Center, Massachusetts General Hospital). We assayed IRP1-IRP2 binding to both H-ferritin and APP IREs using the temporal cortex tissue lysates of three patients with AD as well as three age-matched control patients. Similar to SH-SY5Y and H4 neural cells, IRP1 derived from human brain tissues bound avidly to biotinylated APP IRE probes (row 1), whereas no IRP2 interaction was detected (row 2) (Fig. 6A).
FIGURE 6.
IRP1, but not IRP2, in human brain and blood tissue binds specifically to 5′-UTR-specific APP IRE sequences. A, biotin pulldown assay showing specificity of human brain IRP1-IRP2 binding to probes for the APP IRE relative to the H-ferritin IRE using lysates from postmortem temporal cortex from three AD subjects and three normal (age-matched) subjects. Lanes 1–6, pulldowns with biotinylated APP IRE (lanes 1–3) and H-ferritin IRE (lanes 4–6) RNA probes. Rows 1 and 2 are pulldowns with the 37-base biotinylated APP IRE probe. Rows 3 and 4 are pulldowns with the 37-base H-ferritin IRE, in each case visualized by Western blotting with IRP1 Ab (rows 1 and 3) or IRP2 Ab (rows 2 and 4). The presence of biotin was checked by Western blotting each bead sample to ensure equal efficiency of RNA probe recovery (IRE input). To ensure balanced loadings, the supernatants of lysates used in each pulldown were probed to determine unchanged levels of IRP1. B, human blood tissue samples were subjected to biotin pulldown assay using APP IREs, and associated IRP1 proteins were analyzed by WB. Data from duplicate samples were presented in the histogram shown. The values plotted are means ± S.E. of APP IRE-IRP1 binding from each of seven blood samples from age-matched controls (Normal) and AD patients.
Pulldown experiments performed with biotinylated H-ferritin IRE probe demonstrated specific binding to both human brain-derived IRP1 and human brain-derived IRP2 (Fig. 6, rows 3 and 4) (n = 3). To reconfirm the consistency of the APP IRE binding specificity toward IRP1 but not IRP2 in the human brain, the results were confirmed using hippocampal and frontal cortex (supplemental Fig. 3). We observed a trend toward increased binding of IRP1 to the APP IRE from data averaged for nine samples (three patients (supplemental data), which required further assessment.
We measured the specificity of APP-IRE interaction with IRP1 relative to IRP2 using human blood samples. Equal concentrations of biotinylated RNA probes, encoding either APP IRE or H-ferritin IRE sequences, were incubated with whole blood lysates prepared from AD patients versus age matched individuals (n = 7 per group). These biotin pulldown assays confirmed that IRP1, not IRP2, bound to biotinylated APP IRE RNA probes in lysates from packed red/white blood cells from AD patients and age-matched normal controls. As observed in human brain tissue and in SH-SY5Y and H4 cells, IRP1 and IRP2 bound equally to the H-ferritin IRE probe under matching assay conditions (positive control). Balanced amounts of protein were loaded between samples after standardization for protein content and hemoglobin levels (not shown). Densitometry of Western blot data from this biotin pulldown assay demonstrated that endogenous IRP1 binding to the biotinylated APP IRE probe was increased 2.5-fold in the blood of an AD patient when compared with control blood, suggesting a disease-specific increase in binding (Fig. 6B).
DISCUSSION
The amyloid precursor protein is key in the pathogenesis of AD, but it is also a copper/zinc metalloprotein and is a stress-inducible antioxidant, including its role to prevent oxidative damage to ribosomes (55). This study for the first time defines the specificity by which iron regulatory proteins (IRP1 relative to IRP2) interact with the functional iron-responsive element in the 5′-UTR of the APP transcript. We present key functional data for IRP1 as a central mediator of neural APP expression and in response to intracellular iron (Figs. 3 and 4).
There is considerable published biological, biophysical (28, 56), and bioinformatic (27) data that have defined the molecular bonds that are critical for IRP1 binding to the H-ferritin IRE. This information was used to shed light on the mechanism by which the APP IRE stem loop confers translation both at the basal level and in response to the levels of intracellular iron. This report supports citations that maintain a key function for IRP1 during iron homeostasis (57), including supporting that the IRP1 controls APP translation as a copper/zinc metalloprotein with ferroxidase activity (58). Structural variation exists in the family of iron-responsive RNA stem loops that mediate adaptive translational control responses when cellular iron and oxygen are changed, including that for the functional IRE-like sequences in α-hemoglobin transcript (59–61).
Here we employed previous knowledge from the L- and H-ferritin IRE interaction with IRP1 to extend the constraints of the known canonical IRE consensus required to account for full iron-dependent translational regulation of the APP transcript. Both NMR (56) and x-ray crystallography (28) have defined the key protein-RNA contacts that are important for IRP1-conferred binding to the ferritin IRE (reviewed in Refs. 25 and 27). Walden et al. (28) elegantly showed that the 2.8 Å resolution crystal structure of the IRP1-H-ferritin IRE complex was an open protein conformation when compared with that of cytosolic aconitase. The L-shaped IRP1 molecule (29), with four domains, specifically opens to embrace the H-ferritin IRE stem loop (28) through interactions with domains 1–3 and domain 4 separated by ∼30 Å, each involving protein-RNA bonds.
To understand and predict how APP IRE sequences might interact with IRP1, we conducted sequence alignments of the APP IRE from the human, rhesus monkey, and mouse genes (Fig. 2A). These revealed a strong mammalian homology anchored around the APP-specific CAGAGC motif that aligns to the canonical CAGUGC loop motif of the H-ferritin IRE (CAGUGN = core IRE loop consensus).
Consistent with SHAPE chemical modification results, use of the RNAshapes program predicted an APP IRE RNA stem loop encoding a 13-base 5′-GGCAGAGCAAGGA-3′ terminal loop and a 6-base stem with two unpaired G residues (Fig. 2B). Using human, rhesus monkey, and mouse transcripts as input, these analyses confirmed this conserved stem loop to be folded from the complete 57 bases encoding APP IRE sequences of each species (ΔG = −9.8 kcal/mol). SHAPE modifications were consistent with the human and rhesus monkey RNA stem loop prediction that permitted GC bases at positions 2 and 3 in the terminal loop to hydrogen-bond with the downstream GC dinucleotide at positions 7 and 8 (supplemental Fig. 1). The resulting RNA stem loop structure exhibits a key predicted AGA triloop within the context of a slightly more stable RNA stem loop (free energy: ΔG = −10.2 kcal mol). Confirming a functional role of these bases, deletion of the CAGA sequence in the central homology domain of the predicted APP IRE loop domain abrogated IRP1 binding to APP IRE probes as assessed by the biotin pulldown assay in Fig. 2, D and E. Also, the APP IRE does not confer baseline translation to luciferase reporter genes when this CAGAGC is no longer intact (not shown). In Fig. 2B, these alternate two most stable structures are shown for human and rhesus APP IREs although the open form is the sole predicted structure for the mouse APP transcript.
As a positive control, the same parameters in RNAshapes predicted the RNA secondary structure for the equivalent 57 bases of the L-ferritin IRE. The resulting RNA structure predictions confirmed known biophysical and chemical probing reports showing that the 6-base CAGUGN terminal loop forms a stable AGT trinucleotide because of single base pairing in the terminal CAGAGC (Fig. 2E). This is generated by CG base pairing at positions 1 and 5 of CAGUGU. Indeed, the ferritin L- and H-chain AGT pseudo-triloops account for the most molecular bonds with domains 2/3 of IRP1 (25).
We extended the alignment of the APP IRE core domain to include the IREs of transcripts known to preferentially interact with IRP1 (> IRP2). These included the IREs in the mRNAs encoding DMT-1, Hif-2α, and eALAS. In Fig. 2C, we identified which key terminal loop and bulge C positions were again conserved, revealing maintenance of the relative positioning of APP bases with the CAGUGN loop and upstream bulge C residues of these mRNAs. These may be critical for their IRP1 binding events (28) as for the H-ferritin IRE stem loop.
Several reports showed that the mRNAs for L- and H-ferritin and transferrin receptor bind to both IRP1 and IRP2 and indeed suggest redundancy for IRP1, emphasizing that IRP2 is a more critical controller of iron homeostasis (20, 31, 35, 63, 64). Translation of the ferritin L- and H-chains is preferentially regulated by IRP2 in response to cellular iron status (20, 35). The tissue distribution of IRP2 has been demonstrated to be expressed at relatively higher levels in brain neurons relative to IRP1 (20, 65, 66). Superoxide dismutase-1 (SOD1) knock-out mice demonstrated impaired IRP1 function with no misregulation of iron homeostasis (67).
Supporting its clear physiological role, IRP1 was, however, shown to be functionally significant as a regulator of carcinogenesis (32). Also, IRP1 accelerated the neurodegenerative pathology due to the ablation of the IRP2 gene in knock-out studies with one genetic strain of mice (57, 68, 69). IRP2 had no effect on brain iron metabolism in other genetic backgrounds (63). We report that IRP1 is the key post-transcriptional controller of APP gene expression, a known stress-induced antioxidant. Thus, IRP1 plays a critical role in the regulation of APP translation similar to that for Hif-2α mRNA where perhaps each are key to events related to their limiting hypoxic/ischemic damage and thus protecting tissue from oxidative stress (22).
Our IP-RT-PCR experiments cross-referenced these findings, demonstrating that APP mRNA was co-immunoprecipitated only with IRP1, and but not with IRP2, whereas H-ferritin transcripts interacted with both IRP1 and IRP2. Indeed, the APP IRE may well bind to IRP1 along its predicted “L” structure but may include additional sites to that of the ferritin IRE. We also included specificity controls to demonstrate that the 5′-UTR-specific APP IRE indeed bound to IRP1 at high affinity, where mutation and RNA binding competition assays demonstrated that IRP binding was a consequence of the shape of the APP IRE stem loop (RNA secondary structure) and not a function of the primary sequence (experiments in Fig. 2).
Metal chelation is one of the therapeutic strategies for Alzheimer disease (70, 71). Because APP mRNA was reported to encode a novel but fully functional IRE that controls APP translation (1, 44), this report focused on the pharmacological consequences of DFO to affect IRP1-dependent APP translation at the cellular level. We used conditions of treatment wherein this Fe3+ chelator increased both IRP1 and IRP2 binding to the canonical H-ferritin IRE associated with the known repression of L- and H-ferritin translation (18, 30, 34) (Fig. 3A). As a further positive control to ensure the correct conditions for intracellular iron chelation, TfR mRNA stability was increased 3-fold (Fig. 3D).
We identified a highly reproducible positive correlation that iron chelation with desferrioxamine selectively increased binding of IRP1 to APP IRE sequences, whereas APP protein levels were rapidly decreased in SH-SY5Y cells (Fig. 3B, right panel). At the same time, DFO significantly increased IRP2 expression by enhancing its protein stability (30) (Fig. 3C). Triplicate treatments of DFO treatment of SH-SY5Y cells reduced APP protein expression (>3-fold) in an exactly opposing expression profile to that of IRP2 (Fig. 3C).
The APP IRE exhibited a very close physiological interaction with IRP1 with a dissociation constant (Kd) of rhIRP1-APP IRE calculated to be 30 pm by Scatchard analysis. This Kd value is similar to that of IRP1-H-ferritin IRE (35 pm) (26) and clearly demonstrated that APP mRNA appears to exist as a tightly IRP1-bound ribonucleoprotein through its 5′-UTR, also associated with the other RNA-binding protein on other regions of the transcript, as described by Malter and colleagues (72). This close IRP1-APP IRE interaction was equally evident in SH-SY5Y cells, in H4 neuroglioma cells, and in human pathological brain lysates and in lysates from whole blood.
The degree of phosphorylation may differ among different tissues or between normal and Alzheimer patients, and this may not be limited to IRP1, but indeed to IRP2 (17, 73). However, each sample in this study of events in neural cells, without exception, demonstrated a selective interaction between human tissue IRP1 and the APP 5′-UTR-specific IRE probe.
The functional data with independent clonally derived H4 cells, depleted of IRP1, confirmed the physiological significance that the loss of human IRP1 caused dramatic consequences to result in a substantial 17-fold increase in APP protein production (Fig. 4). These data were highly reproducible under conditions of matched analysis using control, shRNA cells also grown under puromycin selection (no APP change), and where β-actin and compensatory IRP2 increases were not observed. Cell growth was accelerated in this line when compared with controls, perhaps consistent with the report of Ref. 32, which showed that the overexpression of IRP1 is associated with an apparent tumor suppressor phenotype and provides a direct regulatory link between the IRE-IRP system and cancer. Although APP protein levels were increased in the absence of IRP1, our qRT-PCR analysis confirmed that APP mRNA abundance was unchanged in these H4 cells relative to controls. These results are therefore consistent with the model that APP expression is regulated at the translational level, where IRP1 repressed APP mRNA access to ribosomes and thus its protein synthesis. To date, SH-SY5Y cells have been refractory to shRNA transfection and selection with puromycin to generate effective IRP1 knockdown.
Using the six brain frontal cortex tissues from three AD patients and three aged-matched subjects, we consistently observed that IRP1, not IRP2, specifically bound to our 5′-UTR-specific APP IRE probes (Fig. 6A), thus reproducing the same RNA-protein specificity in brain as we had observed for SH-SY5Y and H4 cells (Figs. 1 and 2). These in vivo findings suggest that APP may indeed be regulated post-transcriptionally in humans via radically different mechanisms from that of the well described H-ferritin translational regulation, which is controlled via both IRP1 and IRP2. Mounting opinion has favored a model in which IRP2 predominantly controls ferritin translation and TfR mRNA stability in response to iron or oxygen environment (31, 74).
Altered binding of IRP1 to the ferritin IRE has been reported to reflect an as yet uncharacterized pathological feature of the AD brain where Pinero et al. (45) demonstrated changed IRP1 binding to the H-ferritin IRE in 30% of AD samples relative to age-matched controls (45). Perhaps this reflected disease-specific iron mobilization out of the brain via the blood-brain barrier (75). In the experiment shown in Fig. 6, the binding of APP IRE to IRP1 in the temporal cortex collected from three AD and three age-matched controls was found to not be significantly changed (n = 3) where one normal individual showed very low levels of APP IRE binding to IRP1 in the temporal cortex samples (likely due to events associated with hypoxia during a recorded stroke). However, additional brain samples were used to measure IRP1 binding to the APP and H-ferritin IREs from control and AD tissue samples from the same six patients (supplemental Fig. 3). Using hippocampus and frontal cortex tissue in addition to temporal cortex from these individuals, we observed that there was a 2-fold average increased binding of biotinylated APP IRE probes to IRP1 in these AD brain samples relative to matched controls. As a positive control, on average, the levels of brain IRP1 binding to biotinylated H-ferritin IRE probes were decreased by a significant 20-fold when compared with control brains (n = 3, p < 0.008) (supplemental Fig. 3). The H-ferritin IRE bound to IRP2 (unlike the APP IRE), and this interaction was also diminished by the same margin in the AD brain (n = 3).
We concluded that disease-associated change in RNA binding pattern requires a larger sampling of both AD and normal brain tissue to measure potential disease-specific differences in IRP1 binding to APP 5′-UTR or indeed the interaction between the H-ferritin IRE and IRP1-IRP2. The selectivity of IRP1 binding to the APP 5′-UTR-specific IRE underscores that our results observed in tissue culture were reflected by RNA binding events in the human brain.
Interestingly, we consistently measured that the IRP1-APP IRE binding interaction was on average 3-fold increased in AD patient blood relative to age-matched controls (Fig. 6C) (n = 7) (p < 0.0001). There is no reported difference in serum iron (76) and ferritin (77) noted in AD blood and iron level. Thus, a simple blood-based biomarker for AD has yet to show promise to complement the use of clinical assessment with MRI imaging. Tests that involve the analysis of blood samples, which currently encompass assays of iron-binding protein p97 (78, 79), are in experimental stages; thus, our RNA binding assay may in the future be developed for the purpose of optimizing this assay as a biomarker for Alzheimer disease.
These findings have to be interpreted in light of strong evidence that high levels of metals (iron, copper, zinc) (80) accumulate in the vicinity of developing amyloid plaques in the cortex and hippocampus of the AD brain (81, 82), where APP expression is regulated by copper at the transcriptional level (83). Certainly, iron promotes Aβ neurotoxicity (11, 84, 85) by producing free radical damage and oxidative stress in the brain areas affected by neurodegeneration in AD (86, 87), and iron chelators showed clinical promise in human trials for pharmacological intervention in AD (70, 88–90).
To our knowledge, other reports show that the IRE first described in ferritin and TfR mRNAs may be more widespread, including recently discovered IRE in hemoglobin (60) and in the mitochondrial p75 protein (91). Our data, for the first time, showed that APP mRNA is a class of transcript that encodes a functional IRE in its 5′-UTR that binds physiologically with IRP1, and not IRP2. Recent reports strongly suggest that IRP2 dominates post-transcriptional regulation of iron metabolism in mammals (20, 92), whereas our data support that brain iron and/or oxidative control of APP translation appears to be different from systemic iron-dependent IRE controlled iron homeostasis (i.e. ferritin translation and transferrin receptor mRNA stability). To directly address this possibility, we plan to expand these studies to examine the brain-specific binding of IRP1/2 to other transcripts (for example, DMT-1 and ferroportin), including other neurodegenerative disease-associated mRNAs, which contain IRE sequences.
Acknowledgments
We are grateful to Dr. Suzi Torti at Wake Forest University, Winston-Salem, NC, who provided pSilencer constructs to knock down IRP1 expression. We thank Drs. Tracey Rouault and Sharon Cooperman at the National Institutes of Health, Dr. Betty Leibold (University of Utah), and Richard Eisenstein for the kind gifts of IRP1 and IRP2 antibodies. We appreciate the support of Dr. Ross Stein, Dr. Rudolph Tanzi, Dr. Jerrold Rosenbaum, and Dr. Marcie Glicksman. We are grateful to Avi Friedlich for making valuable suggestions on the first revision.
Note Added in Proof
While this article was in press, a complementary paper was published (Duce, J. A., Tsatsanis, A., Cater, M. A., James, S. A., Robb, E., Wikhe, K., Leong, S. L., Perez, K., Johanssen, T., Greenough, M. A., Cho, H. H., Galatis, D., Moir, R. D., Masters, C. L., McLean, C., Tanzi, R. E., Cappai, R., Barnham, K. J., Ciccotosto, G. D., Rogers, J. T., and Bush, A. I. (2010) Cell, in press).
This work was supported, in whole or in part, by National Institutes of Health Grant AG20181 (to J. T. R.). This work was also supported by a Zenith award from the Alzheimer's Association (to J. T. R.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- AD
- Alzheimer disease
- IRE
- iron-responsive element
- Aβ
- β-amyloid
- APP
- amyloid precursor protein
- IRP
- iron regulatory protein
- H- and L-ferritin
- heavy and light ferritin
- TfR
- transferrin receptor
- DFO
- desferrioxamine
- eALAS
- erythroid heme biosynthetic aminolevulinate synthase
- nt
- nucleotide
- SHAPE
- selective 2′-hydroxyl acylation analyzed by primer extension
- REMSA
- RNA electrophoretic mobility shift assay
- IP
- immunoprecipitation
- WB
- Western blotting
- qPCR
- quantitative PCR
- rh
- recombinant human
- Ab
- antibody.
REFERENCES
- 1.Rogers J. T., Randall J. D., Cahill C. M., Eder P. S., Huang X., Gunshin H., Leiter L., McPhee J., Sarang S. S., Utsuki T., Greig N. H., Lahiri D. K., Tanzi R. E., Bush A. I., Giordano T., Gullans S. R. (2002) J. Biol. Chem. 277, 45518–45528 [DOI] [PubMed] [Google Scholar]
- 2.Wolfe M. S. (2006) Sci. Am. 294, 72–79 [DOI] [PubMed] [Google Scholar]
- 3.Walsh D. M., Klyubin I., Fadeeva J. V., Cullen W. K., Anwyl R., Wolfe M. S., Rowan M. J., Selkoe D. J. (2002) Nature 416, 535–539 [DOI] [PubMed] [Google Scholar]
- 4.Lee V. M., Giasson B. I., Trojanowski J. Q. (2004) Trends Neurosci. 27, 129–134 [DOI] [PubMed] [Google Scholar]
- 5.Wolfe M. S., Xia W., Ostaszewski B. L., Diehl T. S., Kimberly W. T., Selkoe D. J. (1999) Nature 398, 513–517 [DOI] [PubMed] [Google Scholar]
- 6.Multhaup G., Schlicksupp A., Hesse L., Beher D., Ruppert T., Masters C. L., Beyreuther K. (1996) Science 271, 1406–1409 [DOI] [PubMed] [Google Scholar]
- 7.Huang X., Atwood C. S., Hartshorn M. A., Multhaup G., Goldstein L. E., Scarpa R. C., Cuajungco M. P., Gray D. N., Lim J., Moir R. D., Tanzi R. E., Bush A. I. (1999) Biochemistry 38, 7609–7616 [DOI] [PubMed] [Google Scholar]
- 8.Fraering P. C., Ye W., LaVoie M. J., Ostaszewski B. L., Selkoe D. J., Wolfe M. S. (2005) J. Biol. Chem. 280, 41987–41996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H., Wolfe M. S., Selkoe D. J. (2009) Structure 17, 326–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osenkowski P., Li H., Ye W., Li D., Aeschbach L., Fraering P. C., Wolfe M. S., Selkoe D. J., Li H. (2009) J. Mol. Biol. 385, 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schubert D., Chevion M. (1995) Biochem. Biophys. Res. Commun. 216, 702–707 [DOI] [PubMed] [Google Scholar]
- 12.Bandyopadhyay S., Huang X., Cho H., Greig N. H., Youdim M. B., Rogers J. T. (2006) J. Neural. Transm. Suppl. 237–247 [DOI] [PubMed] [Google Scholar]
- 13.Beaudoin M. E., Poirel V. J., Krushel L. A. (2008) Nucleic Acids Res. 36, 6835–6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomson A. M., Rogers J. T., Leedman P. J. (2000) J. Biol. Chem. 275, 31609–31615 [DOI] [PubMed] [Google Scholar]
- 15.Kim H. Y., Klausner R. D., Rouault T. A. (1995) J. Biol. Chem. 270, 4983–4986 [DOI] [PubMed] [Google Scholar]
- 16.Cox T. C., Bawden M. J., Martin A., May B. K. (1991) EMBO J. 10, 1891–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke S. L., Vasanthakumar A., Anderson S. A., Pondarré C., Koh C. M., Deck K. M., Pitula J. S., Epstein C. J., Fleming M. D., Eisenstein R. S. (2006) EMBO J. 25, 544–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ke Y., Wu J., Leibold E. A., Walden W. E., Theil E. C. (1998) J. Biol. Chem. 273, 23637–23640 [DOI] [PubMed] [Google Scholar]
- 19.Rouault T. A. (2006) Nat. Chem. Biol. 2, 406–414 [DOI] [PubMed] [Google Scholar]
- 20.Meyron-Holtz E. G., Ghosh M. C., Iwai K., LaVaute T., Brazzolotto X., Berger U. V., Land W., Ollivierre-Wilson H., Grinberg A., Love P., Rouault T. A. (2004) EMBO J. 23, 386–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan M. A., Walden W. E., Goss D. J., Theil E. C. (2009) J. Biol. Chem. 284, 30122–30128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmer M., Ebert B. L., Neil C., Brenner K., Papaioannou I., Melas A., Tolliday N., Lamb J., Pantopoulos K., Golub T., Iliopoulos O. (2008) Mol. Cell 32, 838–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKie A. T., Barrow D., Latunde-Dada G. O., Rolfs A., Sager G., Mudaly E., Mudaly M., Richardson C., Barlow D., Bomford A., Peters T. J., Raja K. B., Shirali S., Hediger M. A., Farzaneh F., Simpson R. J. (2001) Science 291, 1755–1759 [DOI] [PubMed] [Google Scholar]
- 24.Wingert R. A., Galloway J. L., Barut B., Foott H., Fraenkel P., Axe J. L., Weber G. J., Dooley K., Davidson A. J., Schmid B., Paw B. H., Shaw G. C., Kingsley P., Palis J., Schubert H., Chen O., Kaplan J., Zon L. I. (2005) Nature 436, 1035–1039 [DOI] [PubMed] [Google Scholar]
- 25.Goforth J. B., Anderson S. A., Nizzi C. P., Eisenstein R. S. (2010) RNA 16, 154–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gunshin H., Allerson C. R., Polycarpou-Schwarz M., Rofts A., Rogers J. T., Kishi F., Hentze M. W., Rouault T. A., Andrews N. C., Hediger M. A. (2001) FEBS Lett. 509, 309–316 [DOI] [PubMed] [Google Scholar]
- 27.Piccinelli P., Samuelsson T. (2007) RNA 13, 952–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walden W. E., Selezneva A. I., Dupuy J., Volbeda A., Fontecilla-Camps J. C., Theil E. C., Volz K. (2006) Science 314, 1903–1908 [DOI] [PubMed] [Google Scholar]
- 29.Dupuy J., Volbeda A., Carpentier P., Darnault C., Moulis J. M., Fontecilla-Camps J. C. (2006) Structure 14, 129–139 [DOI] [PubMed] [Google Scholar]
- 30.Zumbrennen K. B., Wallander M. L., Romney S. J., Leibold E. A. (2009) Mol. Cell Biol. 29, 2219–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang W., Di X., D'Agostino R. B., Jr., Torti S. V., Torti F. M. (2007) J. Biol. Chem. 282, 24650–24659 [DOI] [PubMed] [Google Scholar]
- 32.Chen G., Fillebeen C., Wang J., Pantopoulos K. (2007) Carcinogenesis 28, 785–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pitula J. S., Deck K. M., Clarke S. L., Anderson S. A., Vasanthakumar A., Eisenstein R. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10907–10912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fillebeen C., Chahine D., Caltagirone A., Segal P., Pantopoulos K. (2003) Mol. Cell Biol. 23, 6973–6981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schalinske K. L., Blemings K. P., Steffen D. W., Chen O. S., Eisenstein R. S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 10681–10686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eisenstein R. S., Tuazon P. T., Schalinske K. L., Anderson S. A., Traugh J. A. (1993) J. Biol. Chem. 268, 27363–27370 [PubMed] [Google Scholar]
- 37.Maloney B., Ge Y. W., Greig N., Lahiri D. K. (2004) FASEB J. 18, 1288–1290 [DOI] [PubMed] [Google Scholar]
- 38.Janssen S., Giegerich R. (2010) Bioinformatics 26, 632–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allerson C. R., Cazzola M., Rouault T. A. (1999) J. Biol. Chem. 274, 26439–26447 [DOI] [PubMed] [Google Scholar]
- 40.Rogers J. T., Bush A. I., Cho H. H., Smith D. H., Thomson A. M., Friedlich A. L., Lahiri D. K., Leedman P. J., Huang X., Cahill C. M. (2008) Biochem. Soc. Trans. 36, 1282–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanzi R. E., Hyman B. T. (1991) Nature 350, 564. [DOI] [PubMed] [Google Scholar]
- 42.Thomson A. M., Cahill C. M., Cho H. H., Kassachau K. D., Epis M. R., Bridges K. R., Leedman P. J., Rogers J. T. (2005) J. Biol. Chem. 280, 30032–30045 [DOI] [PubMed] [Google Scholar]
- 43.López de Silanes I., Zhan M., Lal A., Yang X., Gorospe M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2987–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bandyopadhyay S., Huang X., Cho H., Greig N. H., Youdim M. B., Rogers J. T. (2006) J. Neural Transm. Suppl. 2006, 237–247 [DOI] [PubMed] [Google Scholar]
- 45.Piñero D. J., Hu J., Connor J. R. (2000) Cell Mol. Biol. 46, 761–776 [PubMed] [Google Scholar]
- 46.Fillebeen C., Caltagirone A., Martelli A., Moulis J. M., Pantopoulos K. (2005) Biochem. J. 388, 143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muckenthaler M., Gray N. K., Hentze M. W. (1998) Mol. Cell 2, 383–388 [DOI] [PubMed] [Google Scholar]
- 48.Cooperman S. S., Meyron-Holtz E. G., Olivierre-Wilson H., Ghosh M. C., McConnell J. P., Rouault T. A. (2005) Blood 106, 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morse L. J., Payton S. M., Cuny G. D., Rogers J. T. (2004) J. Mol. Neurosci. 24, 129–136 [DOI] [PubMed] [Google Scholar]
- 50.Gray N. K., Quick S., Goossen B., Constable A., Hirling H., Kühn L. C., Hentze M. W. (1993) Eur. J. Biochem. 218, 657–667 [DOI] [PubMed] [Google Scholar]
- 51.Bartzokis G., Tishler T. A., Shin I. S., Lu P. H., Cummings J. L. (2004) Ann. N.Y. Acad. Sci. 1012, 224–236 [DOI] [PubMed] [Google Scholar]
- 52.Jellinger K., Paulus W., Grundke-Iqbal I., Riederer P., Youdim M. B. (1990) J. Neural. Transm. Park Dis. Dement. Sect. 2, 327–340 [DOI] [PubMed] [Google Scholar]
- 53.Zecca L., Youdim M. B., Riederer P., Connor J. R., Crichton R. R. (2004) Nat. Rev. Neurosci. 5, 863–873 [DOI] [PubMed] [Google Scholar]
- 54.Connor J. R., Snyder B. S., Arosio P., Loeffler D. A., LeWitt P. (1995) J. Neurochem. 65, 717–724 [DOI] [PubMed] [Google Scholar]
- 55.Honda K., Smith M. A., Zhu X., Baus D., Merrick W. C., Tartakoff A. M., Hattier T., Harris P. L., Siedlak S. L., Fujioka H., Liu Q., Moreira P. I., Miller F. P., Nunomura A., Shimohama S., Perry G. (2005) J. Biol. Chem. 280, 20978–20986 [DOI] [PubMed] [Google Scholar]
- 56.Addess K. J., Basilion J. P., Klausner R. D., Rouault T. A., Pardi A. (1997) J. Mol. Biol. 274, 72–83 [DOI] [PubMed] [Google Scholar]
- 57.Ghosh M. C., Tong W. H., Zhang D., Ollivierre-Wilson H., Singh A., Krishna M. C., Mitchell J. B., Rouault T. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12028–12033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rogers J. (May2, 2002) U. S. Patent WO/2002/034766
- 59.Leipuviene R., Theil E. C. (2007) Cell. Mol. Life Sci. 64, 2945–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.dos Santos C. O., Dore L. C., Valentine E., Shelat S. G., Hardison R. C., Ghosh M., Wang W., Eisenstein R. S., Costa F. F., Weiss M. J. (2008) J. Biol. Chem. 283, 26956–26964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wallander M. L., Leibold E. A., Eisenstein R. S. (2006) Biochim. Biophys. Acta 1763, 668–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie Z., Dong Y., Maeda U., Xia W., Tanzi R. E. (2007) J. Biol. Chem. 282, 4318–4325 [DOI] [PubMed] [Google Scholar]
- 63.Galy B., Ferring D., Minana B., Bell O., Janser H. G., Muckenthaler M., Schümann K., Hentze M. W. (2005) Blood 106, 2580–2589 [DOI] [PubMed] [Google Scholar]
- 64.Galy B., Hölter S. M., Klopstock T., Ferring D., Becker L., Kaden S., Wurst W., Gröne H. J., Hentze M. W. (2006) Nat. Genet. 38, 967–970 [DOI] [PubMed] [Google Scholar]
- 65.Ke Y., Sierzputowska-Gracz H., Gdaniec Z., Theil E. C. (2000) Biochemistry 39, 6235–6242 [DOI] [PubMed] [Google Scholar]
- 66.Leibold E. A., Gahring L. C., Rogers S. W. (2001) Histochem. Cell Biol. 115, 195–203 [DOI] [PubMed] [Google Scholar]
- 67.Starzynski R. R., Lipinski P., Drapier J. C., Diet A., Smuda E., Bartlomiejczyk T., Gralak M. A., Kruszewski M. (2005) J. Biol. Chem. 280, 4207–4212 [DOI] [PubMed] [Google Scholar]
- 68.LaVaute T., Smith S., Cooperman S., Iwai K., Land W., Meyron-Holtz E., Drake S. K., Miller G., Abu-Asab M., Tsokos M., Switzer R., 3rd, Grinberg A., Love P., Tresser N., Rouault T. A. (2001) Nat. Genet. 27, 209–214 [DOI] [PubMed] [Google Scholar]
- 69.Smith S. R., Cooperman S., Lavaute T., Tresser N., Ghosh M., Meyron-Holtz E., Land W., Ollivierre H., Jortner B., Switzer R., 3rd, Messing A., Rouault T. A. (2004) Ann. N.Y. Acad. Sci. 1012, 65–83 [DOI] [PubMed] [Google Scholar]
- 70.Crapper McLachlan D. R., Dalton A. J., Kruck T. P., Bell M. Y., Smith W. L., Kalow W., Andrews D. F. (1991) Lancet 337, 1304–1308 [DOI] [PubMed] [Google Scholar]
- 71.Adlard P. A., Cherny R. A., Finkelstein D. I., Gautier E., Robb E., Cortes M., Volitakis I., Liu X., Smith J. P., Perez K., Laughton K., Li Q. X., Charman S. A., Nicolazzo J. A., Wilkins S., Deleva K., Lynch T., Kok G., Ritchie C. W., Tanzi R. E., Cappai R., Masters C. L., Barnham K. J., Bush A. I. (2008) Neuron 59, 43–55 [DOI] [PubMed] [Google Scholar]
- 72.Westmark C. J., Gourronc F. A., Bartleson V. B., Sayin U., Bhattacharya S., Sutula T., Malter J. S. (2005) J. Neuropathol. Exp. Neurol. 64, 1037–1045 [DOI] [PubMed] [Google Scholar]
- 73.Schalinske K. L., Eisenstein R. S. (1996) J. Biol. Chem. 271, 7168–7176 [DOI] [PubMed] [Google Scholar]
- 74.Muckenthaler M. U., Galy B., Hentze M. W. (2008) Annu. Rev. Nutr. 28, 197–213 [DOI] [PubMed] [Google Scholar]
- 75.Burdo J. R., Connor J. R. (2003) Biometals 16, 63–75 [DOI] [PubMed] [Google Scholar]
- 76.Squitti R., Pasqualetti P., Cassetta E., Dal Forno G., Cesaretti S., Pedace F., Finazzi-Agrò A., Rossini P. M. (2003) Neurology 60, 2013–2014 [DOI] [PubMed] [Google Scholar]
- 77.Fischer P., Götz M. E., Danielczyk W., Gsell W., Riederer P. (1997) Life Sci. 60, 2273–2278 [DOI] [PubMed] [Google Scholar]
- 78.Kim D. K., Seo M. Y., Lim S. W., Kim S., Kim J. W., Carroll B. J., Kwon D. Y., Kwon T., Kang S. S. (2001) Neuropsychopharmacology 25, 84–90 [DOI] [PubMed] [Google Scholar]
- 79.Jefferies W. A., Dickstein D. L., Ujiie M. (2001) J. Alzheimers Dis. 3, 339–344 [DOI] [PubMed] [Google Scholar]
- 80.Connor J. R., Menzies S. L., St Martin S. M., Mufson E. J. (1992) J. Neurosci. Res. 31, 75–83 [DOI] [PubMed] [Google Scholar]
- 81.Lovell M. A., Robertson J. D., Teesdale W. J., Campbell J. L., Markesbery W. R. (1998) J. Neurol. Sci. 158, 47–52 [DOI] [PubMed] [Google Scholar]
- 82.Liu G., Huang W., Moir R. D., Vanderburg C. R., Lai B., Peng Z., Tanzi R. E., Rogers J. T., Huang X. (2006) J. Struct. Biol. 155, 45–51 [DOI] [PubMed] [Google Scholar]
- 83.Bellingham S. A., Lahiri D. K., Maloney B., La Fontaine S., Multhaup G., Camakaris J. (2004) J. Biol. Chem. 279, 20378–20386 [DOI] [PubMed] [Google Scholar]
- 84.Rottkamp C. A., Raina A. K., Zhu X., Gaier E., Bush A. I., Atwood C. S., Chevion M., Perry G., Smith M. A. (2001) Free Radic Biol. Med. 30, 447–450 [DOI] [PubMed] [Google Scholar]
- 85.Cahill C. M., Lahiri D. K., Huang X., Rogers J. T. (2009) Biochim. Biophys. Acta 1790, 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smith M. A., Richey Harris P. L., Sayre L. M., Beckman J. S., Perry G. (1997) J. Neurosci. 17, 2653–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smith C. D., Carney J. M., Starke-Reed P. E., Oliver C. N., Stadtman E. R., Floyd R. A., Markesbery W. R. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 10540–10543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reznichenko L., Amit T., Zheng H., Avramovich-Tirosh Y., Youdim M. B., Weinreb O., Mandel S. (2006) J. Neurochem. 97, 527–536 [DOI] [PubMed] [Google Scholar]
- 89.Avramovich-Tirosh Y., Amit T., Bar-Am O., Zheng H., Fridkin M., Youdim M. B. (2007) J. Neurochem. 100, 490–502 [DOI] [PubMed] [Google Scholar]
- 90.Ritchie C. W., Bush A. I., Mackinnon A., Macfarlane S., Mastwyk M., MacGregor L., Kiers L., Cherny R., Li Q. X., Tammer A., Carrington D., Mavros C., Volitakis I., Xilinas M., Ames D., Davis S., Beyreuther K., Tanzi R. E., Masters C. L. (2003) Arch. Neurol. 60, 1685–1691 [DOI] [PubMed] [Google Scholar]
- 91.Lin E., Graziano J. H., Freyer G. A. (2001) J. Biol. Chem. 276, 27685–27692 [DOI] [PubMed] [Google Scholar]
- 92.Meyron-Holtz E. G., Ghosh M. C., Rouault T. A. (2004) Science 306, 2087–2090 [DOI] [PubMed] [Google Scholar]
- 93.Wilkinson K. A., Merino E. J., Weeks K. M. (2006) Nat. Protoc. 1, 1610–1616 [DOI] [PubMed] [Google Scholar]