Abstract
Cholera toxin (CT) is an AB5 toxin that moves from the cell surface to the endoplasmic reticulum (ER) by retrograde vesicular transport. In the ER, the catalytic A1 subunit dissociates from the rest of the toxin and enters the cytosol by exploiting the quality control system of ER-associated degradation (ERAD). The driving force for CTA1 dislocation into the cytosol is unknown. Here, we demonstrate that the cytosolic chaperone Hsp90 is required for CTA1 passage into the cytosol. Hsp90 bound to CTA1 in an ATP-dependent manner that was blocked by geldanamycin (GA), an established Hsp90 inhibitor. CT activity against cultured cells and ileal loops was also blocked by GA, as was the ER-to-cytosol export of CTA1. Experiments using RNA interference or N-ethylcarboxamidoadenosine, a drug that inhibits ER-localized GRP94 but not cytosolic Hsp90, confirmed that the inhibitory effects of GA resulted specifically from the loss of Hsp90 activity. This work establishes a functional role for Hsp90 in the ERAD-mediated dislocation of CTA1.
Keywords: Bacterial Toxins, Chaperone Chaperonin, Cholera Toxin, Endoplasmic Reticulum(ER), Protein Translocation, Protein-Protein Interactions, RNA Interference (RNAi), Surface Plasmon Resonance (SPR), ER-associated Degradation, Hsp90
Introduction
Cholera toxin (CT)4 is one of the main virulence factors produced by Vibrio cholerae (1, 2). It is an AB-type protein toxin that contains separate catalytic and cell-binding subunits. The catalytic A subunit is initially synthesized as a 27 kDa protein, which undergoes proteolytic nicking to generate a disulfide-linked CTA1/CTA2 heterodimer. The ADP-ribosyltransferase activity of CT resides in the 22 kDa CTA1 polypeptide, while the 5 kDa CTA2 polypeptide maintains numerous non-covalent interactions with the B subunit and thereby links the enzymatic A1 moiety to the cell-binding B moiety. The CTB subunit, built from 11 kDa monomers, is a homopentameric ring-like structure that binds to GM1 gangliosides on the plasma membrane of a target cell.
CT travels as an intact holotoxin from the cell surface to the ER (3). Environmental conditions in the ER facilitate reduction of the CTA1/CTA2 disulfide bond and dissociation of CTA1 from CTA2/CTB5. This process occurs at the resident redox state of the ER and involves the action of protein-disulfide isomerase (PDI), an ER-localized oxidoreductase (4–8). Unfolding of the dissociated CTA1 subunit allows it to move into the cytosol through one or more protein-conducting channels in the ER membrane (9–11). Cytosolic CTA1 then refolds into an active conformation and modifies its Gsα target.
ER-associated degradation (ERAD), a host quality control mechanism, is responsible for the ER-to-cytosol dislocation of CTA1 (12–14). A variety of ER-localized chaperones, lectins, and oxidoreductases function in ERAD (15–17). These proteins recognize features that are present in misfolded proteins such as surface-exposed hydrophobic residues or improper patterns of N-linked glycosylation. When a misfolded protein is identified by the ERAD system, it is exported to the cytosol through Sec61 and/or Derlin-1 protein-conducting channels. Dislocated ERAD substrates are usually appended with polyubiquitin chains that serve as a molecular tag for degradation by the 26 S proteasome. However, CTA1 avoids the standard ERAD route of ubiquitin-dependent proteasomal degradation because it has a paucity of lysine residues for ubiquitin conjugation (18, 19).
ERAD substrates are extracted from the ER through a mechanism that often involves the AAA ATPase Cdc48/p97 (16). However, p97 appears to play a minimal role in CTA1 dislocation (20, 21). An alternative model has proposed a ratchet mechanism, which involves the spontaneous refolding of CTA1 as it enters the cytosol (19). Recent CTA1 structural studies do not support this model, as it has been shown that the isolated CTA1 subunit is a partially disordered, thermally unstable protein (22–24). Because CTA1 is in an unfolded conformation at 37 °C, an as yet unidentified host protein must provide the driving force for CTA1 extraction from the ER.
In this report we demonstrate that Hsp90 is required for CTA1 dislocation to the cytosol. The loss of Hsp90 function did not prevent CT transport to the ER but did inhibit the ER-to-cytosol export of CTA1. This, in turn, blocked CT activity against both cultured cells and ileal loops. Hsp90 has been shown to maintain membrane-embedded ERAD substrates in a soluble state and to help determine the fate of misfolded proteins (25, 26). This work establishes a new role for Hsp90 in the extraction of a soluble ERAD substrate from the ER.
EXPERIMENTAL PROCEDURES
Materials
CT was purchased from List Biological Laboratories (Campbell, CA); His-tagged CTA1 was purified in the laboratory as previously described (24). Cell culture and transfection reagents were purchased from Invitrogen (Carlsbad, CA). Geldanamycin (GA) was purchased from Stressgen (Ann Arbor, MI), while N-ethylcarboxamidoadenosine (NECA), MG132, ALLN, and Brefeldin A (BfA) were purchased from Sigma-Aldrich.
Surface Plasmon Resonance (SPR)
Experiments with the Reichert (Depew, NY) SR7000 SPR Refractometer were performed as previously described (22, 24). To detect toxin-chaperone interactions, the SPR sensor slide was coated with a His-tagged CTA1 construct (22). Binding affinities and stoichiometries were calculated using the BioLogic (Campbell, Australia) Scrubber 2 software. To detect CTA1 in the cytosol or medium of toxin-treated cells, the SPR sensor slide was coated with an anti-CTA1 antibody (24). CTA standards (100, 10, 1, and 0.1 ng/ml) from Calbiochem (La Jolla, CA) were used for these experiments; only the 1 and 0.1 ng/ml standards are shown on the SPR traces for scaling purposes. The cytosolic samples used for this assay were brought to a final volume of 1 ml, thus allowing for a direct comparison between the standards and experimental samples.
Toxicity Assays
Cells continually exposed to various concentrations of CT for 2 h in the absence or presence of drug treatment were analyzed for cAMP content using an [125I]cAMP competition assay from Perkin-Elmer (Boston, MA) as previously described (24). Cells continually exposed to various concentrations of ricin for 4 h in the absence or presence of drug treatment were analyzed for protein synthesis levels by the incorporation of [35S]methionine (Perkin-Elmer) into newly synthesized proteins (13). Cells grown in 24-well plates were used for these experiments.
A rat ligated ileal loop model was used to evaluate the in vivo effect of GA on CT enterotoxicity. Sprague-Dawley rats (n = 4) between 90–120 g in weight were used. Rats were starved for 16 h before initiating the assays. All animal experiments were done after approval from the CINVESTAV-IPN Animal Ethical Committee (CICUAL). To perform laparotomy and exposing the small intestines, animals were anesthetized with xylacin (6.5 mg/kg) and ketamin (34.5 mg/kg). Once the intestines were exposed, the ileocecal valve was localized, and, 20 cm upstream on the small intestine, four ileal loops of 3 cm with 2–4 cm between them were ligated. The ileal loops were inoculated with 200 μl of CT, at 2 μg/ml in phosphate-buffered saline (PBS), alone or with 15 μm of GA. Two other loops were injected with 200 μl of 15 μm GA alone or with 200 μl of PBS. Preliminary experiments indicated that 9 μm GA was not an effective inhibitory concentration, and that 25 μm GA produced results similar to those obtained with 15 μm GA. After inoculums, intestines were returned to the abdominal cavity and the incision was sutured. Inoculated rats were kept alive during 7 h and then sacrificed by cervical dislocation. Ileal loops were photographed and dissected; the intestinal contents were collected by gentle pressure. The volume of fluid in each loop was measured and expressed as a ratio of the amount (μl) of fluid per unit length (cm) of loop.
Dislocation and Secretion Assays
Experiments were performed as previously described (24). In brief, HeLa cells were incubated at 4 °C for 30 min with 1 μg/ml of CT. The cells were then chased at 37 °C for 2 h in toxin-free medium lacking or containing the specified drugs. Media samples were collected, and the cells were lifted from the plate with EDTA in PBS. The collected cell pellet was resuspended in 1 ml (SPR assay) or 0.1 ml (Western blot analysis) HCN buffer (50 mm Hepes pH 7.5, 150 mm NaCl, 2 mm CaCl2, 10 mm N-ethylmaleimide, and a protease inhibitor mixture) containing 0.04% digitonin (Calbiochem). After 10 min at 4 °C, cytosolic (i.e. supernatant) and organelle (i.e. pellet) fractions were collected by centrifugation. For Western blot analysis, 120 μl of 1× sample buffer was added to the pellet and 20 μl of 4× sample buffer was added to the supernatant. Cells were grown in 6-well plates for these experiments; three wells were used for each condition.
Western Blot Analysis
As previously described (24), antibodies used in Western blot analysis were rabbit anti-Hsp90 (Stressgen) at 1:20,000 dilution, rabbit anti-PDI (Stressgen) at 1:5,000 dilution, rabbit anti-CTA (Sigma-Aldrich) at 1:20,000 dilution, rabbit anti-actin (Abcam, Cambridge, MA) at 1:10,000 dilution, and horseradish-peroxidase-conjugated goat anti-rabbit IgG (Jackson Immunoresearch Laboratories, West Grove, PA) at 1:20,000 dilution. Separate blots were run for each protein, except for Hsp90 and actin which were probed on the same blot. Samples were resolved by reducing or non-reducing SDS-PAGE with 15% polyacrylamide gels. When determining the fidelity of our fractionation procedure, anti-Hsp90 and anti-PDI blots each detected only a single band of the expected molecular mass on the entire gel.
Transfection and Immunoprecipitation
CHO cells seeded to 80% confluency in 6-well plates were transfected with pcDNA3.1/ssCTA1 (27) using Lipofectamine (Invitrogen) according to the manufacturer's instructions. At 24 h post-transfection, cells were incubated in methionine-free medium for 1 h before [35S]methionine was added for another hour. Where indicated, 0.1 μm NECA or 0.1 μm GA was present during both the methionine starvation and the radiolabeling. Digitonin was then used to generate membrane and cytosolic fractions as described above. Both fractions were immunoprecipitated with an anti-CTA antibody. SDS-PAGE with PhosphorImager analysis was used to visualize and quantify the immunoisolated material. Control experiments with mock transfected cells did not detect the 21 kDa protein immunoisolated from cells transfected with pcDNA3.1/ssCTA1. The extent of CTA1 dislocation for each experimental condition was calculated with the following equation: % cytosolic CTA1 = CTA1 supernatant signal/[CTA1 supernatant signal + CTA1 pellet signal]. The percentage obtained from untreated control cells was then set as a 100% value, and the percentages obtained from the experimental conditions were expressed as a fraction of the 100% control value.
RNA Interference (RNAi)
1.5 × 104 HeLa cells were seeded to 24-well plates in DMEM supplemented with 10% fetal bovine serum (Atlanta Biologicals) the day before transfection. Cells were incubated at 37 °C with 5% CO2 overnight and allowed to reach 60–80% confluency. On the day of transfection, cells were washed twice with PBS before 0.4 ml of DMEM was added to each well. Transfection reagents were prepared in Optimem-reduced serum media. Lipofectamine 2000 was diluted 1:50 in Optimem and allowed to incubate for 10 min at room temperature. In separate tubes, Hsp90α/β siRNA and control siRNA-A (Santa Cruz Biotechnology, Santa Cruz, CA) were diluted to 10 nm in Optimem and allowed to incubate for 10 min at room temperature. The diluted siRNAs and Lipofectamine 2000 were then combined and allowed to incubate at room temperature for 20 min. 100 μl of the appropriate mixture was then added to the cells. After an overnight incubation at 37 °C and 5% CO2, 50 μl of 100% fetal bovine serum was added to each well. The cells were incubated for another 24 h before experiments were performed.
RESULTS AND DISCUSSION
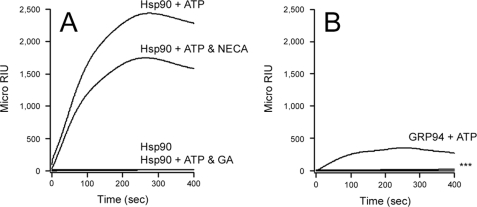
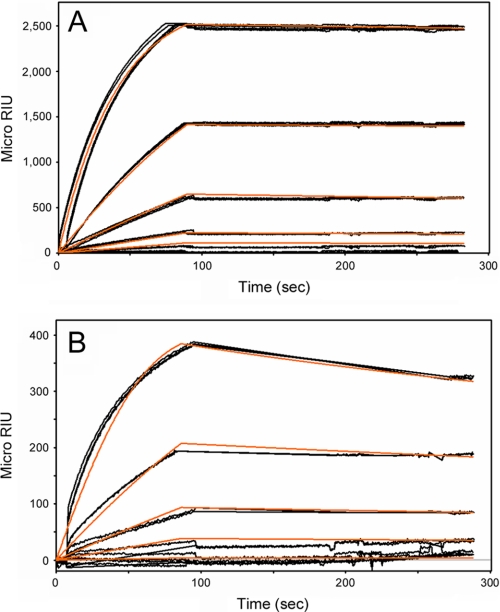
Hsp90 functions in toxin translocation across the endosomal membrane (28–30), but its potential role in ERAD-mediated toxin dislocation has not been examined. To address this issue, SPR was used to determine whether Hsp90 could directly interact with the isolated CTA1 subunit at physiological temperature (Fig. 1A). Hsp90 binding to CTA1 occurred in an ATP-dependent manner that was blocked by GA. This demonstrated that Hsp90 could bind to CTA1 at 37 °C. Because CTA1 is in an unfolded conformation at 37 °C (22, 24), this also suggested that Hsp90 recognizes an unfolded conformation of CTA1 during the dislocation event. The interaction between CTA1 and Hsp90-ATP was not substantially affected by NECA, a drug that inhibits GRP94 but not Hsp90. GRP94, an ER-localized Hsp90, also bound to CTA1 in an ATP-dependent process that was blocked by both GA and NECA (Fig. 1B). The interaction between Hsp90 and CTA1 was much stronger than the interaction between GRP94 and CTA1: Hsp90 bound to CTA1 with a KD of 7 nm, whereas GRP94 bound to CTA1 with a KD of 292 nm (Fig. 2 and Table 1). Consistent with the dimeric natures of Hsp90 and GRP94 (31), both chaperones bound to CTA1 in a 2:1 ratio of chaperone:toxin (Fig. 2). The specific, high-affinity interaction between Hsp90-ATP and CTA1 indicated that Hsp90 could be involved with the CT intoxication process.
FIGURE 1.
GA but not NECA inhibits the interaction between CTA1 and Hsp90. Either Hsp90 (A) or GRP94 (B) was perfused over a CTA1-coated SPR sensor slide at 37 °C under the following conditions: no ATP in the perfusion buffer, ATP in the perfusion buffer, ATP and GA in the perfusion buffer, or ATP and NECA in the perfusion buffer. Ligand was removed from the perfusion buffer 240 s into the experiment. One of four representative experiments is shown. In panel B, *** indicates all experimental conditions other than GRP94 + ATP.
FIGURE 2.
Binding of CTA1 to Hsp90 or GRP94. A, Hsp90 was perfused over a CTA1-coated SPR sensor slide at 100, 400, 800, 1,600, and 3,200 nm concentrations. The ligand was removed from the perfusion buffer after 115 s. B, GRP94 was perfused over a CTA1-coated SPR sensor slide at 100, 200, 400, 800, and 1,600 nm concentrations. The ligand was removed from the perfusion buffer after 118 s. Note that the results for Hsp90 and GRP94 are plotted on different scales. For all conditions, ATP was present in the 37 °C perfusion buffer. Measurements collected from three independent experiments are shown. The orange lines represent best fit curves derived from the raw data using 1:2 (CTA1:Hsp90 or CTA1:GRP94) binding models. The SPR traces display, from top to bottom of the graph, results for decreasing concentrations of the ligand.
TABLE 1.
CTA1 exhibits a high affinity interaction with Hsp90 and a lower affinity interaction with GRP94
On rates (ka), off rates (kd), and equilibrium dissociation constants (KD) between CTA1 and either Hsp90 or GRP94 were calculated from the data presented in Fig. 2.
| CTA1 binding partner | ka | kd | KD |
|---|---|---|---|
| 1/Ms | 1/s | nm | |
| Hsp90 | 9,929 | 7.8 × 10−5 | 7 |
| GRP94 | 1,537 | 4.5 × 10−4 | 292 |
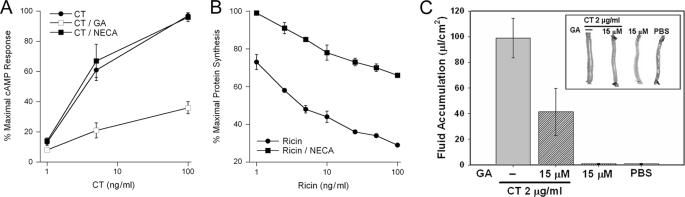
To detect a functional role for Hsp90 in CT intoxication, CT toxicity assays were performed in the presence or absence of GA (Fig. 3A). The elevated levels of cAMP resulting from CT intoxication were strongly inhibited in GA-treated cells. Whereas CT activity against untreated control cells exhibited a half-maximal effective concentration at 4 ng CT/ml, GA-treated cells only produced 36% of the maximal cAMP response when exposed to 100 ng CT/ml. Thus, at least 25-fold higher concentrations of CT would be required to elicit the same degree of intoxication in GA-treated cells as compared with the untreated control cells. In contrast, NECA-treated cells exhibited the same level of sensitivity to CT as the untreated control cells (Fig. 3A). GA-induced toxin resistance could not, therefore, be attributed to the inactivation of GRP94. NECA-treated cells were resistant to ricin, another AB toxin that uses the ERAD system for A chain dislocation to the cytosol (Fig. 3B). This observation was consistent with published results (32) and demonstrated that NECA was functional at the concentration used in our CT assay. Additional control experiments demonstrated that GA did not block the cytopathic activity of a CTA1 construct that was expressed directly in the cytosol of cells transfected with a CTA1-encoding plasmid (27) (supplemental Fig. S1). The inhibitory effect of GA thus appeared to involve an event upstream of toxin-target interactions. Furthermore, GA did not inhibit the forskolin-induced elevation of intracellular cAMP: cells treated with GA and forskolin produced 100 ± 2% of the cAMP levels recorded for cells treated with forskolin alone (n = 3). Forskolin activates adenylate cyclase without the input of Gsα, so this observation demonstrated that GA did not directly inhibit the production of cAMP by adenylate cyclase.
FIGURE 3.
GA but not NECA inhibits CT intoxication. A, CHO cells were incubated with varying concentrations of CT in the absence of additional treatment, in the presence of 0.1 μm GA, or in the presence of 0.1 μm NECA. After 2 h of continual toxin exposure, toxicity was assessed from the elevated levels of intracellular cAMP. The means ± S.E. of at least four independent experiments with triplicate samples are shown. B, CHO cells were incubated for 4 h with varying concentrations of ricin in the absence or presence of 0.1 μm NECA. Toxicity was then determined from the incorporation of [35S]methionine into newly synthesized proteins. The averages ± ranges of two independent experiments with triplicate samples are shown. C, surgically sealed sections of rat intestine were injected with 2 μg/ml of CT in the absence or presence of 15 μm GA. Two other loops were injected with only 15 μm GA or with PBS. Morphological examination (inset) and the calculation of fluid accumulation were performed 7 h post-injection. Results presented in the graph represent the averages ± S.D. of data obtained from 4 rats.
GA also inhibited CT activity in the ileal loop model of intoxication (Fig. 3C). Surgically sealed sections of intestine were injected with 2 μg/ml of CT in the absence or presence of GA. Seven hours later, the CT-injected loop displayed the distended morphology indicative of water accumulation resulting from productive intoxication. In contrast, loops that were injected with both CT and 15 μm GA exhibited substantially attenuated fluid accumulation and intestinal distension. GA derivatives have been evaluated as anti-cancer agents in clinical trials (33, 34). The protective effect of GA in a physiological model of intoxication suggests that it could also be used as a therapeutic to prevent or possibly treat cholera.
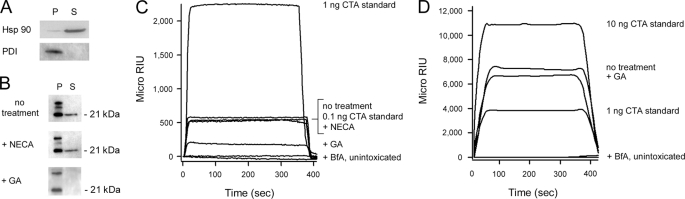
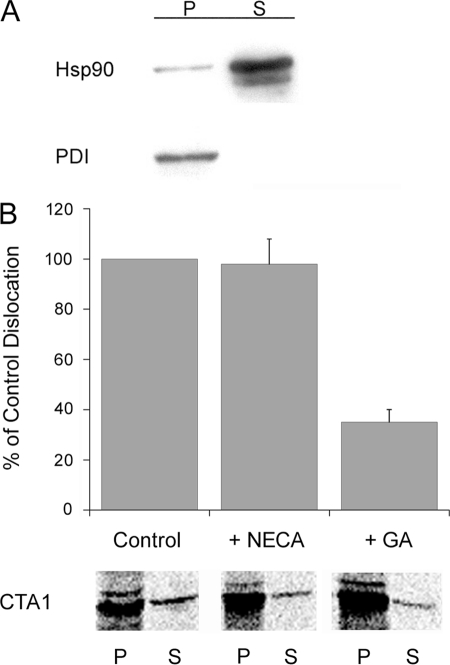
Toxin resistance can result from an inhibition of toxin dislocation from the ER to the cytosol (9, 11, 13, 24). To determine if GA blocked toxin export to the cytosol, we used an established dislocation assay (24, 35) to monitor the appearance of CTA1 in the cytosol of intoxicated HeLa cells (Fig. 4). Like CHO cells, GA-treated HeLa cells were protected from CT as assessed by cAMP production (data not shown). After a 30 min exposure to CT at 4 °C, HeLa cells were chased for 2 h at 37 °C in the absence of additional toxin. Separate organelle and cytosol fractions were then collected from digitonin-permeabilized cells. Control experiments demonstrated the fidelity of our fractionation protocol: Western blot analysis detected the majority of Hsp90 in the supernatant (i.e. cytosolic) fraction, while PDI, a soluble ER protein, was found exclusively in the pellet fraction, which contained the intact ER and other membranes (Fig. 4A). After background subtraction, semi-quantitative analysis of the PDI Western blot detected a negligible amount of PDI (0.5%) in the supernatant fraction (n = 3). Additional Western blot analysis with non-reducing SDS-PAGE was performed in order to track the intracellular localization of CTA1 (Fig. 4B). The 21 kDa CTA1 subunit is initially synthesized as part of a larger, 26 kDa CTA precursor (1, 2). Proteolytic nicking of CTA generates a disulfide-linked CTA1/CTA2 heterodimer, which is reductively cleaved in the ER; only the reduced CTA1 subunit enters the cytosol. Consistent with these observations, we detected both disulfide-linked CTA1/CTA2 and reduced CTA1 in the organelle fractions but only detected reduced CTA1 in the cytosol fractions. In comparison to the untreated control cells, the distribution of cytosolic CTA1 was unaffected by NECA treatment. However, GA-treated cells contained less cytosolic CTA1 than either untreated or NECA-treated cells. It thus appeared that Hsp90 function was required for efficient passage of CTA1 into the cytosol.
FIGURE 4.
GA but not NECA inhibits CTA1 dislocation to the cytosol. HeLa cells were pulse-labeled at 4 °C for 30 min with 1 μg/ml of CT. The cells were then chased for 2 h at 37 °C in toxin-free medium containing no additions, 0.1 μm GA, 0.1 μm NECA, or 5 μg/ml of BfA. Permeabilization of the plasma membrane with digitonin was used to partition cell extracts into separate organelle (pellet; P) and cytosolic (supernatant; S) fractions. A, both fractions from untreated cells were probed by Western blot to establish the distributions of cytosolic marker Hsp90 and ER marker PDI. B, both fractions from untreated (no treatment) or drug-treated cells were probed for the presence of CTA1 by Western blot analysis of non-reducing SDS-PAGE gels. The upper band represents the disulfide-linked CTA1/CTA2 heterodimer; the middle band results from nonspecific cross-reactivity; and the lower band represents reduced CTA1. C, an SPR sensor slide coated with an anti-CTA antibody was used to detect the cytosolic pool of CTA1 from untreated or drug-treated cells. D, media samples taken from cells at the end of the chase were perfused over an SPR sensor slide coated with an anti-CTA antibody. For both C and D, CTA standards were perfused over the sensor slide as positive controls. The cytosolic fraction and medium from unintoxicated cells were also perfused over the sensor slides as negative controls for panels C and D, respectively. At the end of each experiment, bound sample was stripped from the sensor slide. One of four representative experiments is shown for each SPR experiment.
To strengthen our Western blot analysis with an alternative and quantitative detection method, we employed the technique of SPR. HeLa cells exposed to CT at 4 °C were again fractionated into membrane and cytosolic components after a 2 h chase at 37 °C. In this experiment, the cytosolic fractions were perfused over a SPR sensor slide that had been coated with an anti-CTA antibody (Fig. 4C). A negligible background signal was obtained from unintoxicated cells, whereas control cells intoxicated in the absence of drug treatment produced a signal that was comparable to the response obtained from the 0.1-ng CTA standard (n = 4). The low level of cytosolic CTA1 in control cells was consistent with the known inefficiency of CT trafficking from the cell surface to the ER dislocation site (4, 7, 36). NECA-treated cells produced an SPR signal similar to the response obtained from the untreated control cells. In contrast, GA-treated cells produced an SPR signal that was substantially less than the response obtained from the untreated control cells. Exposure of untreated or GA-treated cells to proteasome inhibitors did not alter these results, which indicated the data were not affected by potential toxin degradation in the cytosol (supplemental Fig. S2). The detection of a minor pool of cytosolic CTA1 in GA-treated cells by SPR but not by Western blot reflects the greater level of sensitivity provided by SPR-based assays. CTA1 was not detected in the cytosol of cells treated BfA, a drug that prevents CT transport from the cell surface to the ER dislocation site (4, 7, 37).
Protein concentration is directly proportional to the SPR-derived association rate constant (ka) (38), so we calculated the levels of cytosolic CTA1 from a plot of the ka values for the CTA standards. With this method, we estimated that 2.4-fold less CTA1 was in the cytosol of GA-treated cells than in the cytosol of untreated or NECA-treated cells (supplemental Table S1). A substantial inhibition of intoxication resulting from reduced, but not absent, levels of cytosolic CTA1 has previously been reported (13, 14, 24). The degree of intoxication appears to be influenced by numerous factors, including (i) the extent of CTA1 dislocation to the cytosol; (ii) the rate of CTA1 degradation in the cytosol; (iii) CTA1 activity against Gsα; and (iv) the de-activation of ADP-ribosylated Gsα by proteolysis or by the action of ADP-ribosyl(arginine)protein hydrolase (9, 13, 14, 19, 24, 39–41). The balance between these factors usually favors CTA1 activity against Gsα and, thus, productive intoxication. However, as shown here and in previous work, this balance can be shifted to protect cells from intoxication without completely eliminating the cytosolic pool of CTA1.
To ensure that GA did not inhibit the intracellular trafficking of CT, we screened media samples from toxin-treated cells for the presence of free CTA1. The CT holotoxin travels by vesicle carriers from the cell surface to the ER (3). Reduction and chaperone-assisted dissociation of CTA1 from the rest of the toxin then allows the free A1 subunit to move from the ER to the cytosol. In addition, a fraction of dissociated CTA1 enters the secretory pathway and is released into the extracellular medium (24, 36). Consistent with these observations, our SPR assay detected the secretion of CTA1 from intoxicated HeLa cells (Fig. 4D). Nearly equivalent amounts of CTA1 were secreted into the medium of untreated cells and GA-treated cells. No signal was obtained from the medium of BfA-treated cells, thus demonstrating that toxin trafficking to the ER was a prerequisite for CTA1 secretion into the medium. The aforementioned results were obtained with a SPR sensor slide that had been coated with an anti-CTA antibody. When the media samples were perfused over an SPR sensor slide that had been coated with an antibody against the cell-binding CTB subunit, no positive signals were obtained (data not shown). Thus, the secreted toxin was free CTA1 and not the CT holotoxin. Because untreated and GA-treated cells released equivalent amounts of free CTA1, GA did not appear to block holotoxin trafficking to the ER, CTA1 dissociation from the rest of the toxin in the ER, or the secretion of free CTA1.
To further demonstrate that GA inhibited the CTA1 dislocation event rather than an upstream CT trafficking step, we combined our dislocation assay with a plasmid-based system to express CTA1 directly in the ER of transfected CHO cells (12, 27). With this system, CTA1 is co-translationally inserted into the ER and then dislocated back to the cytosol. Similar strategies have been used by other labs to monitor toxin dislocation (10, 35, 42–45). Control experiments again demonstrated the fidelity of our fractionation protocol in CHO cells: Western blot analysis detected Hsp90 mainly in the cytosolic fraction, while PDI was only found in the organelle fraction (Fig. 5A). Consistent with the SPR-based dislocation assay which used exogenously applied toxin, the plasmid-based expression system detected a 2.9-fold block of CTA1 dislocation in GA-treated cells (Fig. 5B). No inhibitory effect on CTA1 dislocation was observed for NECA-treated cells, thus demonstrating that the GA-induced block of CTA1 dislocation did not result from an effect on GRP94 (Fig. 5B).
FIGURE 5.
GA inhibits dislocation of an ER-localized CTA1 construct. CHO cells were transfected with a plasmid encoding a CTA1 subunit appended with an ER-targeting signal sequence. Permeabilization of the plasma membrane with digitonin was used to partition cell extracts into separate organelle (pellet; P) and cytosolic (supernatant; S) fractions. A, both fractions from transfected cells were probed by Western blot to establish the distributions of Hsp90 and PDI. B, distribution of CTA1 immunoprecipitated from transfected cells after a 1 h radiolabeling was visualized and quantified by SDS-PAGE with PhosphorImager analysis. Experiments were performed with untreated control cells, cells exposed to 0.1 μm NECA, or cells exposed to 0.1 μm GA. The upper band in the gel represents the population of CTA1 with an intact, uncleaved signal sequence. The means ± S.E. of three independent experiments are presented in the graph.
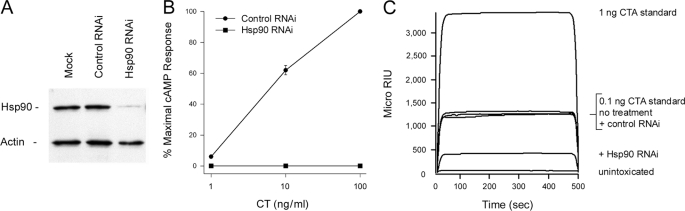
To confirm the specific role of Hsp90 in CTA1 dislocation, we used RNAi to deplete the cellular pool of Hsp90 (Fig. 6). Western blot analysis confirmed the near-complete elimination of Hsp90 protein after transfection with the RNAi construct targeting Hsp90 but not after transfection with the control RNAi construct (Fig. 6A). Cells with reduced levels of Hsp90 were highly resistant to CT intoxication (Fig. 6B) and did not effectively deliver CTA1 to the cytosol as assessed by our SPR-based dislocation assay (Fig. 6C). The nearly identical results obtained by drug treatment and RNAi indicated that our data were not influenced by off-target effects from a single experimental technique.
FIGURE 6.
Hsp90 depletion prevents CT intoxication and CTA1 dislocation. HeLa cells were mock transfected, transfected with control siRNA, or transfected with Hsp90 siRNA. A, cell extracts were probed by Western blot analysis with an anti-Hsp90 antibody or an anti-actin antibody. After standardization to the actin-loading control, transfection with the Hsp90 siRNA was estimated to reduce Hsp90 protein levels to 10 ± 6% (n = 3) of control levels from the mock-transfected cells. B, transfected cells were exposed to varying concentrations of CT for 2 h before intracellular cAMP levels were determined. The means ± S.E. of four independent experiments with triplicate samples are shown. C, SPR sensor slide coated with an anti-CTA antibody was used to detect the cytosolic pool of CTA1 from transfected cells. CTA standards were perfused over the sensor slide as positive controls, while a cytosolic fraction generated from unintoxicated cells was used as a negative control. One of three representative experiments is shown.
A complete inhibition of CT intoxication was recorded for the RNAi experiment but not for the GA experiment, which suggested that RNAi was more effective at abrogating Hsp90 function than drug treatment. Consistent with this interpretation, we found a greater reduction of cytosolic CTA1 in the RNAi experiment (3.2-fold) than in the GA experiment (2.4-fold) (supplemental Table S1). The residual pool of CTA1 that appeared in the cytosol of cells lacking Hsp90 was not sufficient to produce a cAMP response, possibly because of compensating cellular processes to prevent or reverse CT activity against Gsα (14, 39–41). The complete inhibition of CT-induced cAMP production upon RNAi knockdown of Hsp90 was remarkable, and it highlighted the importance of Hsp90 for the CT intoxication process.
Recently, Hsp90 was shown to be involved with the renaturation of an unfolded protein that had passed from the cell surface to the ER and from the ER to the cytosol (46). The Hsp90-assisted refolding of denatured proteins may be linked to its dislocation activity: by coupling dislocation with refolding, Hsp90 would prevent the (re)folded CTA1 protein from sliding back into the dislocation pore. This ratchet mechanism would thus provide the driving force for CTA1 dislocation. Although a previous report suggested the ATP-dependent dislocation of CTA1 does not require cytosolic factors (10), we and others (35, 47) have detected a membrane-associated pool of Hsp90 which may have been purified with the microsomal preparation used in that study. Our work clearly shows that Hsp90 exhibits a high affinity interaction with CTA1, and that the disruption of this interaction inhibits both CTA1 dislocation and CT intoxication in vivo.
Hsp90 is also involved with ricin intoxication, although in this case Hsp90 apparently prepares the catalytic A chain of ricin for proteasomal degradation (32). The GA-induced inactivation of Hsp90 thus allows ricin A chain to accumulate in the cytosol and thereby generates cellular sensitization to this AB-type plant toxin. In contrast, we have shown that GA-treated cells are resistant to CT. The dislocation of ricin A chain also involves GRP94 and p97 (32, 48), whereas these proteins do not appear to be active in CTA1 dislocation (20, 21). Thus, while both CTA1 and ricin A chain exploit ERAD for passage into the cytosol, distinct molecular events are involved with the dislocation of the two toxins.
ERAD substrates, including ricin A chain, are usually exported to the cytosol through the action of p97 (16, 48). However, there is also a p97-independent dislocation route for some ERAD substrates such as CTA1 (20, 21, 49, 50). This report demonstrates that the p97-independent dislocation of CTA1 requires Hsp90 function. We have thus established a new role for Hsp90 in the extraction of a soluble ERAD substrate from the ER.
Supplementary Material
Acknowledgment
We thank Mansfield Burlingame for technical assistance with the CT/GA toxicity assays in HeLa cells.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AI073783 (to K. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Table S1.
- CT
- cholera toxin
- BfA
- brefeldin A
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- GA
- geldanamycin
- NECA
- N-ethylcarboxamidoadenosine
- PDI
- protein-disulfide isomerase
- SPR
- surface plasmon resonance.
REFERENCES
- 1.De Haan L., Hirst T. R. (2004) Mol. Membr. Biol. 21, 77–92 [DOI] [PubMed] [Google Scholar]
- 2.Sánchez J., Holmgren J. (2008) Cell Mol. Life Sci. 65, 1347–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wernick N. L. B., Chinnapen D. J.-F., Cho J. A., Lencer W. I. (2010) Toxins 2, 310–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lencer W. I., de Almeida J. B., Moe S., Stow J. L., Ausiello D. A., Madara J. L. (1993) J. Clin. Invest. 92, 2941–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Majoul I., Ferrari D., Söling H. D. (1997) FEBS Lett. 401, 104–108 [DOI] [PubMed] [Google Scholar]
- 6.Orlandi P. A. (1997) J. Biol. Chem. 272, 4591–4599 [PubMed] [Google Scholar]
- 7.Orlandi P. A., Curran P. K., Fishman P. H. (1993) J. Biol. Chem. 268, 12010–12016 [PubMed] [Google Scholar]
- 8.Tsai B., Rodighiero C., Lencer W. I., Rapoport T. A. (2001) Cell 104, 937–948 [DOI] [PubMed] [Google Scholar]
- 9.Bernardi K. M., Forster M. L., Lencer W. I., Tsai B. (2008) Mol. Biol. Cell 19, 877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmitz A., Herrgen H., Winkeler A., Herzog V. (2000) J. Cell Biol. 148, 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dixit G., Mikoryak C., Hayslett T., Bhat A., Draper R. K. (2008) Exp Biol Med. 233, 163–175 [DOI] [PubMed] [Google Scholar]
- 12.Teter K., Allyn R. L., Jobling M. G., Holmes R. K. (2002) Infect Immun. 70, 6166–6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teter K., Holmes R. K. (2002) Infect Immun. 70, 6172–6179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teter K., Jobling M. G., Holmes R. K. (2003) Traffic 4, 232–242 [DOI] [PubMed] [Google Scholar]
- 15.Tsai B., Ye Y., Rapoport T. A. (2002) Nat. Rev. Mol. Cell Biol. 3, 246–255 [DOI] [PubMed] [Google Scholar]
- 16.Bar-Nun S. (2005) Curr. Top Microbiol. Immunol. 300, 95–125 [DOI] [PubMed] [Google Scholar]
- 17.Nakatsukasa K., Brodsky J. L. (2008) Traffic 9, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hazes B., Read R. J. (1997) Biochemistry 36, 11051–11054 [DOI] [PubMed] [Google Scholar]
- 19.Rodighiero C., Tsai B., Rapoport T. A., Lencer W. I. (2002) EMBO Rep 3, 1222–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kothe M., Ye Y., Wagner J. S., De Luca H. E., Kern E., Rapoport T. A., Lencer W. I. (2005) J. Biol. Chem. 280, 28127–28132 [DOI] [PubMed] [Google Scholar]
- 21.McConnell E., Lass A., Wójcik C. (2007) Biochem. Biophys. Res. Commun. 355, 1087–1090 [DOI] [PubMed] [Google Scholar]
- 22.Pande A. H., Scaglione P., Taylor M., Nemec K. N., Tuthill S., Moe D., Holmes R. K., Tatulian S. A., Teter K. (2007) J. Mol. Biol. 374, 1114–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ampapathi R. S., Creath A. L., Lou D. I., Craft J. W., Jr., Blanke S. R., Legge G. B. (2008) J. Mol. Biol. 377, 748–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massey S., Banerjee T., Pande A. H., Taylor M., Tatulian S. A., Teter K. (2009) J. Mol. Biol. 393, 1083–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Youker R. T., Walsh P., Beilharz T., Lithgow T., Brodsky J. L. (2004) Mol. Biol. Cell 15, 4787–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X., Venable J., LaPointe P., Hutt D. M., Koulov A. V., Coppinger J., Gurkan C., Kellner W., Matteson J., Plutner H., Riordan J. R., Kelly J. W., Yates J. R., 3rd, Balch W. E. (2006) Cell 127, 803–815 [DOI] [PubMed] [Google Scholar]
- 27.Teter K., Jobling M. G., Holmes R. K. (2004) Infect Immun. 72, 6826–6835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ratts R., Zeng H., Berg E. A., Blue C., McComb M. E., Costello C. E., vanderSpek J. C., Murphy J. R. (2003) J. Cell Biol. 160, 1139–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haug G., Aktories K., Barth H. (2004) Infect Immun. 72, 3066–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haug G., Leemhuis J., Tiemann D., Meyer D. K., Aktories K., Barth H. (2003) J. Biol. Chem. 278, 32266–32274 [DOI] [PubMed] [Google Scholar]
- 31.Zuehlke A., Johnson J. L. (2010) Biopolymers 93, 211–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spooner R. A., Hart P. J., Cook J. P., Pietroni P., Rogon C., Höhfeld J., Roberts L. M., Lord J. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17408–17413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powers M. V., Workman P. (2006) Endocr. Relat. Cancer 13, Suppl. 1, S125–S135 [DOI] [PubMed] [Google Scholar]
- 34.Blagosklonny M. V. (2002) Leukemia 16, 455–462 [DOI] [PubMed] [Google Scholar]
- 35.Forster M. L., Sivick K., Park Y. N., Arvan P., Lencer W. I., Tsai B. (2006) J. Cell Biol. 173, 853–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fishman P. H. (1982) J. Cell Biol. 93, 860–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nambiar M. P., Oda T., Chen C., Kuwazuru Y., Wu H. C. (1993) J. Cell. Physiol. 154, 222–228 [DOI] [PubMed] [Google Scholar]
- 38.Homola J. (2003) Anal. Bioanal. Chem. 377, 528–539 [DOI] [PubMed] [Google Scholar]
- 39.Chang F. H., Bourne H. R. (1989) J. Biol. Chem. 264, 5352–5357 [PubMed] [Google Scholar]
- 40.Kato J., Zhu J., Liu C., Moss J. (2007) Mol. Cell. Biol. 27, 5534–5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wernick N. L., De Luca H., Kam W. R., Lencer W. I. (2010) J. Biol. Chem. 285, 6145–6152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castro M. G., McNamara U., Carbonetti N. H. (2001) Cell Microbiol 3, 45–54 [DOI] [PubMed] [Google Scholar]
- 43.LaPointe P., Wei X., Gariépy J. (2005) J. Biol. Chem. 280, 23310–23318 [DOI] [PubMed] [Google Scholar]
- 44.Simpson J. C., Roberts L. M., Römisch K., Davey J., Wolf D. H., Lord J. M. (1999) FEBS Lett. 459, 80–84 [DOI] [PubMed] [Google Scholar]
- 45.Yu M., Haslam D. B. (2005) Infect Immun. 73, 2524–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giodini A., Cresswell P. (2008) EMBO J. 27, 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaiser E., Pust S., Kroll C., Barth H. (2009) Cell Microbiol 11, 780–795 [DOI] [PubMed] [Google Scholar]
- 48.Marshall R. S., Jolliffe N. A., Ceriotti A., Snowden C. J., Lord J. M., Frigerio L., Roberts L. M. (2008) J. Biol. Chem. 283, 15869–15877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wójcik C., Rowicka M., Kudlicki A., Nowis D., McConnell E., Kujawa M., DeMartino G. N. (2006) Mol. Biol. Cell 17, 4606–4618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carlson E. J., Pitonzo D., Skach W. R. (2006) EMBO J. 25, 4557–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.