Abstract
TULA-1 (UBASH3A/STS-2) and TULA-2 (p70/STS-1) represent a novel class of protein-tyrosine phosphatases. Previous studies suggest that TULA-2 is sequence-selective toward phosphotyrosyl (Tyr(P)) peptides. In this work the substrate specificity of TULA-1 and -2 was systematically evaluated by screening a combinatorial Tyr(P) peptide library. Although TULA-1 showed no detectable activity toward any of the Tyr(P) peptides in the library, TULA-2 recognizes two distinct classes of Tyr(P) substrates. On the N-terminal side of Tyr(P), the class I substrates contain a proline at the Tyr(P)−1 position, a hydrophilic residue at the Tyr(P)−2 position, and aromatic hydrophobic residues at positions Tyr(P)−3 and beyond. The class II substrates typically contain two or more acidic residues, especially at Tyr(P)−1 to Tyr(P)−3 positions, and aromatic hydrophobic residues at other positions. At the C-terminal side of Tyr(P), TULA-2 generally prefers acidic and aromatic residues. The library screening results were confirmed by kinetic analysis of representative peptides selected from the library as well as Tyr(P) peptides derived from various Tyr(P) proteins. TULA-2 is highly active toward peptides corresponding to the Tyr(P)-323 and Tyr(P)-352 sites of Syk, and the Tyr(P)-397 site of focal adhesion kinase and has lower activity toward other Tyr(P) sites in these proteins. In glycoprotein VI-stimulated platelets, knock-out of the TULA-2 gene significantly increased the phosphorylation level of Syk at Tyr-323 and Tyr-352 sites and to a lesser degree at the Tyr-525/526 sites. These results suggest that Syk is a bona fide TULA-2 substrate in platelets.
Keywords: Chemical Modification, Enzyme Catalysis, Enzyme Kinetics, Peptide Chemical Synthesis, Phosphoprotein Phosphatase, Cell Signaling, Combinatorial Peptide Library, Substrate Specificity
Introduction
The TULA/STS protein family consists of two recently discovered members, UBASH3A/STS-2/TULA (termed TULA-1 thereafter) and p70/STS-1/TULA-2 (1–5). The two proteins share ∼40% sequence identity and a similar multidomain structure, with an N-terminal ubiquitin-association domain, a Src-homology 3 domain, and a C-terminal phosphatase domain. The phosphatase domain of TULA-2 appears to be tyrosine-specific, exhibiting high catalytic activity toward para-nitrophenyl phosphate, phosphotyrosyl (Tyr(P)) peptides, and Tyr(P) proteins such as protein-tyrosine kinases ZAP70, Syk, and certain members of the Src family but has no detectable activity against phosphoseryl or phosphothreonyl substrates (6, 7). TULA-1 has also been reported to have protein-tyrosine phosphatase (PTP)3 activity, although its activity is orders of magnitude lower than that of TULA-2 (6, 7). The TULA family of phosphatases is distinct from the classical PTPs, which utilize a conserved cysteine residue to catalyze the dephosphorylation reaction (8). The PTP domain of TULA-1/2 shares considerable sequence similarity to members of the histidine phosphatase superfamily, which consists of numerous proteins including phosphoglycerate mutase (2, 6, 9). Histidine phosphatases critically depend on a conserved histidine residue in their active site, which acts as the catalytic nucleophile (10). Structural studies revealed that the PTP domain of TULA-2 contains all of the catalytic residues in histidine phosphatases including His-380, the putative catalytic nucleophile (6). Mutation of His-380 to an alanine abolishes the phosphatase activity of TULA-2 (6).
TULA-1 and TULA-2 have been shown to play a role in regulating receptor-mediated signaling in T cells (3, 6) as well as other signaling pathways (4, 5, 11, 12). Interestingly, despite their similarity in protein sequence and structure, TULA-1 and TULA-2 have very different cellular functions. For example, TULA-2 decreases the kinase activity of Syk, which is critical for lymphoid cell signaling, whereas TULA-1 increases it (7). The ability of TULA-2 to suppress T-cell receptor signaling depends on its phosphatase activity (6). Previous studies have indicated that TULA-2 dephosphorylates various Tyr(P) proteins, in particular protein-tyrosine kinases of the Syk and Src families, and demonstrates a certain degree of discrimination between them in in vitro assays (6, 7, 13). These observations raise several important questions about the TULA enzymes. Do they recognize specific Tyr(P) substrates or simply act as nonspecific antagonists of protein-tyrosine kinases? And if they are specific enzymes, what dictates their substrate specificity? These issues are especially important considering that TULA-1 exhibits much lower phosphatase activity than TULA-2, despite its high homology to TULA-2 and other members of the histidine phosphatase superfamily (6, 7, 9). Is the poorer activity of TULA-1 an intrinsic property of its PTP domain or simply because the substrates tested are better substrates of TULA-2?
The classical, cysteinyl PTPs were once thought to be nonspecific housekeeping enzymes. It is now clear that they have exquisite substrate specificity in vivo, and their specificity is determined by a combination of the sequence specificity of the PTP active/substrate binding site and the utilization of specific targeting domains (14). It is conceivable that the TULA family of PTPs employs the same two mechanisms to control their substrate specificity. Indeed, both the ubiquitin association and Src homology 3 domains are protein-protein interaction domains, with the former binding to mono- and polyubiquitinated proteins and the latter recognizing proline-rich sequences (15, 16). In this work we set out to determine the sequence specificity of TULA-1 and TULA-2 PTP domains. Historically, this has been a challenging problem because a phosphatase (e.g. a classical PTP) may interact with 3–5 amino acid residues on either side of the Tyr(P) during substrate binding/catalysis (17, 18) and a prohibitively large number of Tyr(P) peptides (206-2010) needs to be tested against each PTP in question to gain a comprehensive profile of its substrate specificity. This necessitates the use of combinatorial peptide libraries of some sort. However, screening peptide libraries against a PTP is itself technically challenging, as one needs to differentiate a small amount of reaction product (i.e. tyrosine) from a complex mixture of unreacted Tyr(P) substrates. Very recently one of our laboratories developed a novel peptide library method to profile the sequence specificity of classical PTPs (19, 20). In this study we applied this method to determine the sequence specificity of TULA-1/2. Our results show that TULA-1 has intrinsically low phosphatase activity, whereas TULA-2 is a much more active enzyme of considerable sequence specificity. Subsequently, the specificity data were used to identify several potential TULA-2 substrates including Syk for further biochemical and genetic experiments. These studies confirmed Syk as a bona fide TULA-2 substrate in vivo. To our knowledge this is the first time that the complete specificity profile of a PTP has been reported.
EXPERIMENTAL PROCEDURES
Materials
Reagents for peptide synthesis were from Advanced ChemTech (Louisville, KY), Peptides International (Louisville, KY), or Novabiochem. N-(9-Fluorenylmethoxycarbonyloxy)succinimide (Fmoc-OSu) was from Advanced ChemTech. Mushroom tyrosinase, α-cyano-4-hydroxycinnamic acid, phenyl isothiocyanate, and 3-methyl-2-benzothiazolinonehydrazone were obtained from Sigma). Nα-Fmoc-O-t-butyl-3,5-difluorotyrosine (Fmoc-F2Y(tBu)-OH) was chemoenzymatically synthesized as previously described (20). Antibodies to Syk and its phosphosites as well as that to the ERK and PKCδ phosphosites were from Cell Signaling Technology (Boston, MA). Antibody to Bruton's tyrosine kinase Tyr-551 was from Abcam (Cambridge, MA), and to focal adhesion kinase Tyr-397 was from Millipore, Billerica, MA).
Synthesis of Tyr(P) Peptide Libraries
Each peptide library was synthesized on 5 g of amino polyethylene glycol polyacrylamide resin (0.4 mmol/g, 150–300 μm in diameter when swollen in water). All of the manipulations were performed at RT unless otherwise noted. The invariant linker sequence was synthesized using 4 eq. of Fmoc-amino acids and O-benzotriazole-N,N,N′,N′-tetramethyluronium hexafluorophosphate/1-hydroxybenzotriazole hydrate/N-methylmorpholine as the coupling reagents. The coupling reaction was typically allowed to proceed for 1.5 h, and the beads were thoroughly washed with N,N-dimethylformamide and dichloromethane. For the synthesis of random residues, the resin was split into 19 equal portions (by weight), and each portion was coupled twice, each with 5 eq. of a different Fmoc-amino acid for 1 h (except for Fmoc-F2Y, which was coupled once at 2.5 eq). To facilitate sequence determination by mass spectrometry, 5% (mol/mol) deuterated acetic acid was added to the coupling reactions of leucine and lysine, whereas 5% alpha-deuterated propionic acid was added to the coupling reaction of norleucine (21). The resin-bound library was washed with dichloromethane and side chain-deprotected using a modified reagent K (7.5% phenol, 5% water, 5% thioanisole, 2.5% ethanedithiol, and 1% anisole in trifluoroacetic acid) at RT for 2 h. The library was washed with trifluoroacetic acid, dichloromethane, and N,N-dimethylformamide and stored in N,N-dimethylformamide at −20 °C until use.
Library Screening
In a typical screening experiment, 14 mg of the proper peptide library (∼40,000 beads) was placed in a plastic micro-BioSpin column (0.8 ml, Bio-Rad) and washed exhaustively with N,N-dimethylformamide and double-distilled H2O. The resin was blocked for 1 h with a blocking buffer (30 mm Hepes, pH 7.4, 150 mm NaCl, 0.01% Tween 20, and 0.1% gelatin). The library was then incubated with TULA-1 (final concentration 1.0 μm) or TULA-2 (0.5 μm) in the blocking buffer plus 5 mm tris(carboxyethyl)phosphine (total volume 0.8 ml) at RT for 1 h with gentle mixing. The resin was drained, washed with 0.1 m KH2PO4 (pH 6.8, 3 × 1 ml), and resuspended in 1.6 ml of 0.1 m KH2PO4 (pH 6.8) containing 1.2 μm tyrosinase and 6 mm 3-methyl-2-benzothiazolinonehydrazone. The resulting mixture was incubated at RT with gentle mixing and exposure to air. An intense pink/red color typically developed on positive beads within 30 min. The positive beads were removed from the library manually using a micropipette under a dissecting microscope and individually sequenced by partial Edman degradation-MS (21). Control experiments without phosphatase produced no colored beads under otherwise identical conditions.
Synthesis of Individual Peptides
Individual peptides were each synthesized on 100 mg of CLEAR-amide resin using standard Fmoc-based peptide chemistry. For the coupling reaction of Tyr(P), 2.0 eq of Fmoc-amino acid were employed, whereas 4.0 eq were used for all other amino acids. The resin-bound peptides were cleaved from the resin and side-chain deprotected using the modified reagent K at RT for 2 h. The solvents were removed by evaporation under a N2 stream and trituration with cold diethyl ether. The precipitate was collected and dried under a vacuum. The crude peptides were purified by reversed-phase HPLC on a semipreparative C18 column. The identity of each peptide was confirmed by MALDI-TOF mass spectrometric analysis.
Enzyme Assay with Peptide Substrates
TULA-2 activity assay was performed with synthetic Tyr(P) peptides as substrates in a quartz microcuvette. The reaction mixture contained 100 mm Hepes, pH 7.4, 50 mm NaCl, 2 mm EDTA, 1 mm tris(carboxyethyl)phosphine, 0.1 mg/ml bovine serum albumin (BSA), and 0.05–1.3 mm Tyr(P) peptide. The reaction was initiated by the addition of TULA-2 (final concentration 47 nm) and monitored continuously at 282 nm (for Tyr, Δϵ = 1102 m−1cm−1) on a UV-visible spectrophotometer. The initial velocities were determined from the initial linear region of the reaction progress curves and fitted to the Michaelis-Menten equation to determine the kinetic parameters kcat and kcat/Km.
In Vitro Phosphatase Assay with Syk as Substrate
The assay was carried out essentially as described (7) with minor modifications. Syk produced in insect cells as a GST fusion protein was purchased from Invitrogen. The TULA-2 phosphatase domain was produced in Escherichia coli (6). Syk was diluted to a final concentration of 0.5 μg/ml in a reaction buffer containing 25 mm HEPES, pH 7.2, 50 mm NaCl, 2 mm EDTA, 5 mm DTT, 200 μg/ml BSA, and 0.01% Triton X-100. The resultant reaction mix (total volume 100 μl) was prewarmed at 37 °C for 2 min. TULA-2 was added to the above mixture where indicated at a final concentration of 0.12 or 0.03 μg/ml. The reaction was allowed to proceed at 37 °C. At the indicated time points, 30-μl sample aliquots were withdrawn from the reaction mixture, mixed with 4× SDS-PAGE sample buffer, heated to 90 °C for 5 min, and analyzed by SDS-PAGE and immunoblotting on nitrocellulose membranes.
Preparation and Stimulation of Mouse Platelets
Generation of p70 (STS-1/TULA-2)-knock-out (KO) mice on a mixed genetic background (C57BL/6xSv129) was described previously (2). Subsequently, they were back-crossed with wild-type C57B/L6 mice to generate KO mice on the standard C57BL/6 background. Blood was collected from the vena cava of anesthetized mice into syringes containing a one-tenth blood volume of 3.8% sodium citrate as anticoagulant. Red blood cells were removed by centrifugation at 100 × g for 10 min at RT. Platelet-rich plasma was recovered, and platelets were pelleted at 400 × g for 10 min at RT. The platelet pellet was resuspended in Tyrode buffer (138 mm NaCl, 2.7 mm KCl, 2 mm MgCl2, 0.42 mm NaH2PO4, 5 mm glucose, 10 mm HEPES, pH 7.4, and 0.2% bovine serum albumin at pH 7.4) containing 0.05 units/ml apyrase to a density of 2 × 108 cell/ml. Platelets were stimulated with 100 ng/ml convulxin (Centerchem, Norwalk, CT) under stirring conditions at 37 °C for the time indicated, and the reaction was stopped by adding 3× SDS-PAGE sample buffer. Samples were heated and processed for SDS-PAGE separation and immunoblotting on PVDF membranes.
Immunoblotting
Membranes with transferred proteins were probed with the indicated primary antibodies against phosphosites/proteins at appropriate concentrations. Nitrocellulose membranes were blocked with phosphate-buffered saline (PBS; Na2HPO4 10 mm, NaH2PO4 1.76 mm, NaCl 137 mm, KCl 2.7 mm at pH 7.4) containing 5% (w/v) dry milk for 1 h, probed with antibodies in PBS for 1 h, washed with PBS containing 0.2% (v/v) Tween 20 (PBST) 4 times for 5 min, then probed with near-infrared-labeled secondary antibodies (LI-COR Biosciences, Lincoln, NE) in PBST with 5% dry milk for 45 min, washed 4 times for 5 min (all at RT), and finally visualized and quantified using a LI-COR Odyssey system.
PVDF membranes were blocked in Tris-buffered saline-Tween (TBST; 20 mm Tris, 140 mm NaCl, 0.1% (v/v) Tween 20 at pH 7.6) containing 0.5% (w/v) dry milk and 3% (w/v) BSA for 30 min at RT, and membranes were incubated with primary antibody in TBST, 2% BSA with gentle agitation at 4 °C overnight. After 3 washes for 5 min each with TBST, membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibody in TBST, 2% BSA for 1 h at RT and washed again as described above. Immunoreactive bands were visualized with an HRP chemiluminescent substrate and quantified using Fujifilm LAS-3000 Luminescent Image Analyzer (Fuji, Tokyo, Japan).
RESULTS
Determination of Substrate Specificity of TULA-2 by Library Screening
Two one-bead-one-compound Tyr(P) peptide libraries, N-terminal library SAXXXXXpYAABBRM resin, and C-terminal library Alloc-AApYXXXXXNNBBRM resin (where B is β-alanine, and X is F2Y, norleucine, or any of the 17 proteinogenic amino acids excluding Met, Cys, and Tyr) were designed to examine the sequence specificity of PTPs on the N- and C-terminal side of Tyr(P), respectively. A linker sequence BBRM was added at the C terminus to facilitate mass spectrometric analysis (positively charged Arg) and cleavage from the resin (after Met with CNBr) (22). Two invariant residues (SA) or an allyloxycarbonyl group was added to the N terminus of all library peptides to minimize any bias caused by the positive charge associated with a free N terminus. Substitution of norleucine for methionine in the random region prevents internal cleavage during peptide release with CNBr, whereas replacement of tyrosine with F2Y is necessary to avoid false positives during library screening (see below). We have previously shown that substitution of F2Y for Tyr does not significantly alter the catalytic activity of PTPs (20). Each library has a theoretical diversity of 2.48 × 106 and was synthesized on 5 g of polyethylene glycol polyacrylamide resin, which has superior permeability to macromolecules such as enzymes as compared with polystyrene-based resins (23).
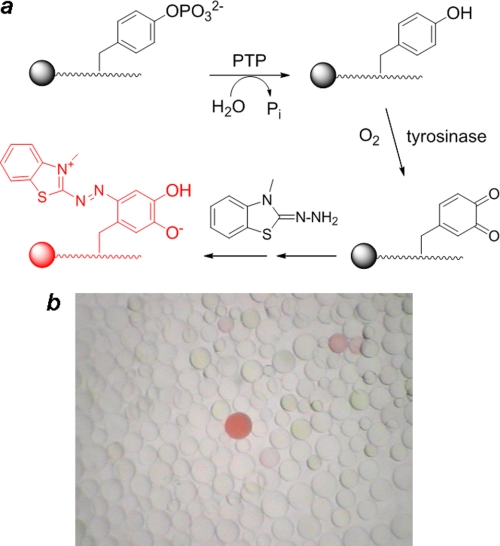
Library screening was based on an on-bead enzyme-linked colorimetric assay previously developed in one of our laboratories (19, 20). Dephosphorylation of a Tyr(P) peptide by a PTP exposes a free tyrosine side chain that is specifically and quantitatively oxidized into an orthoquinone by exhaustive treatment with a tyrosinase in the presence of atmospheric O2 (Fig. 1). The orthoquinone is then trapped in situ with 3-methyl-2-benzothiazolinonehydrazone to form an intensely pink/red pigment. Thus, as a result of the reaction cascade, a bead carrying a preferred PTP substrate turns pink/red, whereas beads carrying poor substrates do not. It should be pointed out that the on-bead reaction does not follow the simple kinetics of a solution-phase reaction, as the substrates are immobilized on the solid phase. Under the conditions employed for this study, the peptide “concentration” on each bead was ∼50 mm (calculated on the basis of 200 pmol of peptides/bead and an average bead diameter of 200 μm), a condition that is expected to render most PTPs operate at the Vmax value (and, thus, incapable of differentiating substrates with the same Vmax but different Km values). In practice, however, library screening against PTPs usually results in beads of a wide range of color intensities (Fig. 1). We have previously shown that the color intensity on a bead is correlated with the catalytic activity of the peptide substrate displayed on the bead (with beads that carry the more reactive substrates being more intensely colored), although the correlation between color intensity and activity (in terms of the kcat/Km value) is not linear (19, 20). In the current study we controlled the screening reaction condition (by adjusting the PTP concentration and reaction time) to allow ∼0.1% of the library beads to turn intensely pink/red. The most colored beads were retrieved from the library with a micropipette and individually sequenced by partial Edman degradation-MS (21).
FIGURE 1.
a, reactions involved in the on-bead screening of a Tyr(P) peptide library against a PTP are shown. b, shown is a view of a small portion of the stained library under a dissecting microscope.
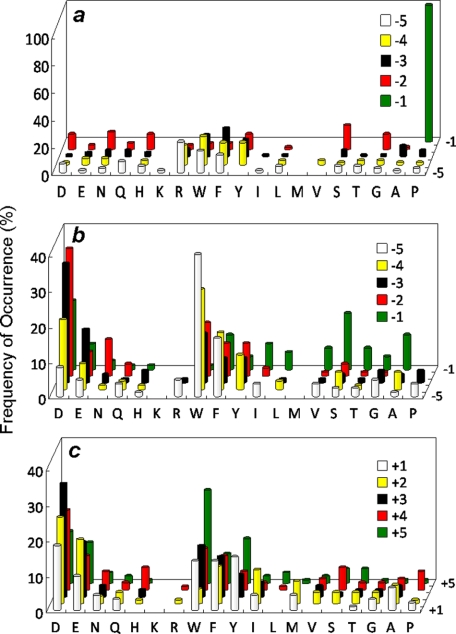
A total of 42 mg of the N-terminal library (∼120,000 beads) was screened against TULA-2 in three separate experiments. Sequencing of the most intensely colored beads produced 152 sequences, which should represent the most efficient substrates of TULA-2. These sequences were separated into two different classes on the basis of sequence similarities (supplemental Table I). The class I substrates (66 total) all contain a Pro at the Tyr(P)−1 position. There is also a strong preference for a small hydrophilic residue (e.g. Ser, Asn, Asp, Gly, and His) at the Tyr(P)−2 position (Fig. 2a). At the Tyr(P)−3 and Tyr(P)−4 positions, TULA-2 shows a strong selectivity for hydrophobic especially aromatic residues (Trp, Phe, and Tyr); of the 66 class I substrates, 57 (87%) has an aromatic residue at one or both of the positions, whereas the remaining peptides usually contain an aliphatic hydrophobic residue (e.g. norleucine and Val). There is some preference for Trp, Arg, Phe, and Tyr at the Tyr(P)−5 position, although most amino acids are tolerated. The class II sequences display an overall abundance of acidic (Asp and Glu) and aromatic hydrophobic residues (Trp, Phe, and F2Y) (Fig. 2b). Of the 86 class II sequences, 52 (60%) contain two or more acidic residues at various positions, most frequently at the Tyr(P)−2 and Tyr(P)−3 positions, whereas every sequence contains at least one but usually two aromatic residues (e.g. 71% of all identified class II sequences contain at least 1 aromatic residue at the Tyr(P)−4 or Tyr(P)−5 position; of sequences having only a single aromatic residue, 89% contain it at these positions). Overall, the hydrophilic and hydrophobic residues generally occur at alternate positions (supplemental Table II). At the Tyr(P)−1 to Tyr(P)−3 positions, most of the selected sequences feature some type of combination of one hydrophobic and two hydrophilic residues (e.g. YDD, DYD, and DDY). Interestingly, whereas the class I sequences contain a fair number of positively charged residues (Arg and Lys), especially at the Tyr(P)−5 position, the class II substrates had only Arg in four of them.
FIGURE 2.
Substrate specificity of TULA-2. a, shown are most preferred class I substrates selected from the N-terminal library (X5pY). b, shown are most preferred class II substrates selected from the N-terminal library (X5pY). (c) Most preferred substrates selected from the C-terminal library (pYX5). The histograms represent the amino acids identified at each position from Tyr(P)−5 to Tyr(P)+5 (z axis). Frequency of occurrence on the y axis represents the percentage of selected sequences that contained a particular amino acid at a certain position. M, norleucine; Y, F2Y.
A total of 97 hits were obtained by screening 42 mg of the C-terminal library (supplemental Table II). TULA-2 has a broad specificity on the C-terminal side of Tyr(P) and is less selective than the N-terminal side, with an overall preference for acidic and aromatic hydrophobic residues (Fig. 2c). The acidic residues are most frequently present at the Tyr(P)+2 to Tyr(P)+4 positions, whereas the hydrophobic residues are most frequently found at the Tyr(P)+1 and Tyr(P)+5 positions. The most striking feature is the near complete absence of positively charged residues among the preferred substrates, suggesting that TULA-2 strongly disfavors positively charged residues at the C-terminal side of Tyr(P).
To examine whether the invariant Ala-Ala motif causes any sequence bias on the other side of Tyr(P), a third library containing five random residues on each side of Tyr(P), AX5pYX5NNBBRM resin (where X is norleucine or any of the 17 proteinogenic amino acids excluding Met, Cys, and Tyr), was synthesized and screened against TULA-2. Screening of a total of 36 mg of the library identified 86 most preferred substrates (supplemental Table III). Note that this library has a theoretical diversity of 1810 or 3.57 × 1012, and the amount of resin screened represented only a small fraction of the total sequence space. Nonetheless, the data from this library confirmed the results derived from the N- and C-terminal libraries. Both classes of substrates were selected, although the class I sequences were observed at a lower frequency as compared with the N-terminal library (24 versus 43%). Again, TULA-2 strongly prefers acidic residues on both sides of Tyr(P), especially for the class II substrates. No obvious covariance between N- and C-terminal sequences was observed, as both class I and II substrates contained similar, acidic residues on the C-terminal side of Tyr(P). Finally, 32 colorless beads, which represented the poor substrates of TULA-2, were randomly selected from the library and sequenced by partial Edman degradation-MS to give 30 sequences (supplemental Table IV). The poor substrates contained a drastically lower abundance of Pro at the Tyr(P)−1 position (1/30 sequences) and acidic residues at all positions. In contrast, each of the 30 poor substrates contained at least one basic residue (Arg, Lys, and/or His).
TULA-1 Has Intrinsically Low Phosphatase Activity
Screening 14 mg of the N-terminal library (∼40,000 beads) against TULA-1 (at 1.0 μm) under otherwise the same conditions as TULA-2 produced no colored bead at all. This result indicates that TULA-1 has intrinsically low phosphatase activity against all Tyr(P) peptides.
Kinetic Properties of TULA-2 toward Tyr(P) Peptide Substrates
Ten representative peptides selected from the N-terminal (Table 1, peptides 1–8) and the N+C-terminal libraries (peptides 9 and 10) were individually synthesized and assayed against TULA-2 in solution to validate the screening results. Because most of the peptides failed to saturate the enzyme at 1 mm (supplemental Fig. 1), only the kcat/Km values are reported. Nine of the peptides were efficient substrates of TULA-2, with kcat/Km values of 1.3 × 104–4.2 × 105 m−1 s−1. As a comparison, phenyl phosphate had a kcat/Km value of 200 m−1 s−1. Peptide 4, which has an Arg as the Tyr(P)−2 residue, was less efficient (kcat/Km = 1.4 × 103 m−1 s−1). This is not surprising, as only two of the 152 peptides selected from the N-terminal library contained an Arg at the Tyr(P)−2 position (supplemental Table I). To assess the significance of the selected amino acids at each position, alanine-scanning studies were carried out for peptides 3 and 10 (Table 1). Substitution of Ala for the Pro at the Tyr(P)−1 position of peptide 3 decreased the TULA-2 activity by 4-fold. Replacement of the Pro by arginine resulted in even greater reduction in the activity (∼20-fold) (Table 1, compare peptides 10 and 16). At the Tyr(P)−2 position, replacement of Asp by Ala reduced the activity by 3.5-fold (Table 1, compare peptides 3 and 12). Removal of the aromatic side chain at the Tyr(P)−3 position decreased the activity by 2.6-fold. On the other hand, conversion of Phe to Ala at the Tyr(P)−4 position had little effect, probably because peptide 3 already had an aromatic residue at the Tyr(P)-3 position (many of the selected peptides had an aromatic residue at either the Tyr(P)-3 or −4 position (supplemental Table I)). Replacement of Ser at the Tyr(P)−5 position by Ala increased the activity by ∼2-fold, in keeping with the observed preference for hydrophobic residues at this position. Finally, the overwhelming preference for acidic residues at the C-terminal side of Tyr(P) was confirmed by replacing the acidic residues at Tyr(P)+2 and Tyr(P)+4 positions of peptide 10 by Ala or Arg. Removal of the acidic residues from peptide 10 (replacement by Ala) resulted in a 7-fold decrease in activity, whereas introduction of basic residues caused an additional 5-fold reduction in catalytic activity (Table 1, compare peptides 10 versus 17 and 18).
TABLE 1.
Catalytic activities of TULA-2 against Tyr(P) peptides
FAK, focal adhesion kinase.
| Peptide | Source | Peptide sequencea | kcat/Km |
|---|---|---|---|
| ×103m−1s−1 | |||
| 1 | Library | RLFSPpYAA | 15.0 ± 0.3 |
| 2 | Library | ITFNPpYAA | 33 ± 1 |
| 3 | Library | SFYDPpYAA | 148 ± 6 |
| 4 | Library | GYARPpYAA | 1.4 ± 0.1 |
| 5 | Library | WDGDFpYAA | 77 ± 38 |
| 6 | Library | WQDDFpYAA | 28 ± 2 |
| 7 | Library | TYYDDpYAA | 10 ± 1 |
| 8 | Library | IWPSApYAA | 20 ± 1 |
| 9 | Library | AAPQNPpYFQPTR | 20 ± 1 |
| 10 | Library | ANDVDPpYLEFDI | 420 ± 100 |
| 11 | SFYDApYAA | 37 ± 5 | |
| 12 | SFYAPpYAA | 42 ± 6 | |
| 13 | SFADPpYAA | 58 ± 18 | |
| 14 | SAYDPpYAA | 140 ± 12 | |
| 15 | AFYDPpYAA | 280 ± 30 | |
| 16 | ANDVDRpYLEFDI | 23 ± 1 | |
| 17 | ANDVDPpYLAFAI | 61 ± 33 | |
| 18 | ANDVDPpYLRFRI | 12 ± 1 | |
| 19 | Syk | 346EVYESPpYADPE356 | 290 ± 100 |
| 20 | 346EVYESApYADPE356 | 71 ± 17 | |
| 21 | Syk | 317TVSFNPpYEPEL327 | 21 ± 1 |
| 22 | 317TVSFNApYEPEL327 | 4.1 ± 0.1 | |
| 23 | Syk | 520RADENYpYKAQT530 | 0.39 ± 0.04 |
| 24 | Syk | 520RADENpYYKAQT530 | 0.57 ± 0.08 |
| 25 | Syk | 520RADENpYpYKAQT530 | 1.1 ± 0.4 |
| 26 | PEAR1 | 919LSSENPpYATIR929 | 4.5 ± 0.7 |
| 27 | 919LSSENApYATIR929 | 0.20 ± 0.01 | |
| 28 | Erk1/2 | 198NGFLTEpYVATR208 | 0.28 ± 0.01 |
| 29 | Src | 413LIEDNEpYTARQ423 | 0.92 ± 0.01 |
| 30 | FAK | 391VSETDDpYAEII401 | 12 ± 2 |
| 31 | BTK | 544YVLDDEpYTSSV554 | 4.6 ± 0.2 |
| 32 | PKCδ | 149KQAKIHpYIKNH159 | 0.29 ± 0.07 |
| 33 | PKCδ | 305TESVGIpYQGFE315 | 0.85 ± 0.08 |
| pNPP | 100 ± 10 | ||
| phenyl phosphate | 0.20 ± 0.06 |
a All peptides were acetylated at the N terminus and contained a C-terminal amide. Peptides 1–8 also contained a mini-PEG followed by a glycine at the C-terminus.
Next, we synthesized a series of Tyr(P) peptides corresponding to the phosphorylation sites of proteins involved in GPVI-induced signaling in platelets. This system was selected because our previous study suggested that the lack of TULA-2 alone was sufficient to facilitate receptor-mediated signaling in platelets (25). A search of the PhosphoSitePlus data base for convulxin-induced phosphorylation sites in platelets retrieved 12 Tyr(P) proteins. Eight of the proteins including Syk, PEAR1, Erk1/2, Src, focal adhesion kinase, BTK, and PKCδ contained one or more Tyr(P) motifs that showed similarity with either the class I or II consensus of TULA-2 (Table 1). A total of 15 Tyr(P) peptides were synthesized and assayed against TULA-2. Three of the peptides (peptides 19, 21, and 26) matched the class I consensus sequence, and others were related to the class II consensus (peptides 23, 24, 25, 29, 30, and 31). The rest of the peptides were variants of the above peptides (peptides 20, 22, and 27), designed to further test the significance of Pro at the Tyr(P)−1 position, or did not match either class consensus (peptides 28, 32, and 33). Overall, the three class I peptides had the highest activities toward TULA-2, with kcat/Km values in the range of 104-105 m−1s−1 and comparable with those of the preferred substrates selected from the libraries. Replacement of the prolyl residue with an alanine decreased the activity by 4–22-fold (Table 1, compare peptides 19 versus 20, 21 versus 22, and 26 versus 27). Among the class II peptides, peptides 30 and 31 had relatively high activity (kcat/Km values in the range of 103-104 m−1s−1). The other class II peptides had lower activities, apparently due to the presence of positively charged residues (Arg and Lys) at the C-terminal side of Tyr(P) (peptides 23–25 and 29). The activity difference among the three class I peptides (peptides 19, 21, and 26) can also be partially explained by the presence of an Arg residue in peptide 26. Peptides 28, 32, and 33, which did not match either consensus motif, had significantly poorer activities. Thus, the kinetic data determined with synthetic peptide substrates in the solution phase were in excellent agreement with the on-bead library screening results.
Finally, to determine whether the selected substrates are specific for TULA-2, we compared the catalytic activities of peptides 19 and 20 against TULA-2, PTP1B, and SHP-1 (Table 2). PTP1B and SHP-1 are prototypical members of the classical PTP family. Peptide 19, which contains a Pro at the Tyr(P)−1 position and is one of the most active substrates of TULA-2, had a 4.5-fold lower activity toward SHP-1, but a ∼7-fold higher activity toward PTP1B. Interestingly, although substitution of Ala for the Tyr(P)−1 Pro (peptide 20) reduced the activity of TULA-2 by 4-fold, it increased the activities of PTP1B and SHP-1 by 1.5- and 7.7-fold, respectively. These results allow us to draw two conclusions. First, the preference for a Pro at the Tyr(P)−1 site may be a unique feature of TULA-2, certainly not a common feature of all PTPs. It also provides further support for the validity of our library screening method. Second, the most preferred TULA-2 peptide substrates are not specific for TULA-2, suggesting that other factors must also be involved in controlling the substrate specificity of TULA-2 in vivo (see below).
TABLE 2.
Activity comparison of TULA-2, PTP1B, and SHP-1
| Peptide sequence | kcat | Km | kcat/Km | |
|---|---|---|---|---|
| s−1 | μm | ×103m−1s−1 | ||
| EVYESPpYADPE | TULA-2 | 26 ± 3 | 90 ± 26 | 290 |
| SHP1 | 14 ± 1 | 220 ± 20 | 65 | |
| PTP1B | 26 ± 1 | 13 ± 2 | 2000 | |
| EVYESApYADPE | TULA-2 | 27 ± 3 | 380 ± 70 | 71 |
| SHP1 | 48 ± 3 | 95 ± 18 | 500 | |
| PTP1B | 26 ± 1 | 8.6 ± 0.9 | 3100 |
Dephosphorylation of Syk Protein by TULA-2
To test whether the specificity profile obtained with peptide substrates is predictive of the reactivity of protein substrates, we examined the ability of TULA-2 to dephosphorylate the various Tyr(P) sites of Syk and monitored the reaction progress by immunoblotting with antibodies specific for each Tyr(P) site. Our previous study has shown that tyrosyl-phosphorylated recombinant GST-Syk acts as a TULA-2 substrate in vitro (7). The human Syk produced in baculovirus-infected insect cells has high levels of phosphorylation on Tyr-525/526 and Tyr-352. The Tyr-323 site was poorly phosphorylated in this Syk preparation and, therefore, was not analyzed. As described above, kinetic studies against phosphopeptides suggest that the Tyr(P)-352 site should be efficiently dephosphorylated by TULA-2, whereas the Tyr(P)-525/526 site would be a poor substrate.
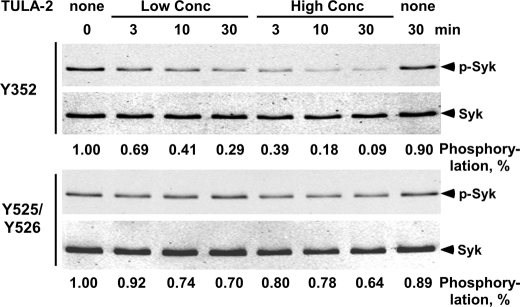
Because the Tyr(P)-525/526 site can be dephosphorylated under forcing conditions (at high concentrations of TULA-2) (7), we utilized a significantly lower concentration of TULA-2 in our current study to reveal any differential reactivity toward the Tyr(P) sites. Under our assay conditions, TULA-2 dephosphorylated Tyr(P)-352 with half-lives of ∼10 and ∼3 min at 0.03 and 0.12 μg/ml TULA-2, respectively, whereas the Tyr(P)-525/526 site was dephosphorylated at a much lower rate (half-life >30 min at 0.12 μg/ml TULA-2) (Fig. 3). Thus, dephosphorylation of Tyr(P) sites on protein substrates follow the same sequence selectivity rules as peptide substrates.
FIGURE 3.
In vitro dephosphorylation of Syk Tyr(P) sites by TULA-2. Recombinant human Syk (0.5 μg/ml) and TULA-2 PTP domain (0, 0.03, or 0.12 μg/ml) were incubated at 37 °C for the indicated time periods, and the amounts of phosphorylation at Tyr-352 and Tyr-525/526 and total Syk in the samples were detected by immunoblotting. The phosphorylation level was quantified and normalized for the amount of total Syk in each sample and is shown at the bottom of the panel. A value of 1.0 was assigned to the sample in the absence of TULA-2.
Increased Tyrosine Phosphorylation of Syk in TULA-2 KO Cells
To determine whether Syk and several other Tyr(P) proteins are bona fide in vivo substrates of TULA-2, we examined their tyrosine phosphorylation levels induced by the ligation of GPVI receptor on platelets. GPVI is a key collagen receptor on platelets, and GPVI-mediated signaling is essential for platelet activation at the site of endothelial damage, which exposes the collagen layer. GPVI-mediated signaling induces platelet responses such as aggregation and secretion, thus triggering thrombus formation (24). For the current study, we selected the candidate proteins on the basis of the following criteria. First, the protein is phosphorylated on tyrosine in response to GPVI ligation. Second, the phosphorylation event is critical for the physiological function of platelets (25–28). Third, the protein should contain at least one Tyr(P) site that matches the substrate specificity profile of TULA-2 as revealed by the library screening and kinetic analysis.
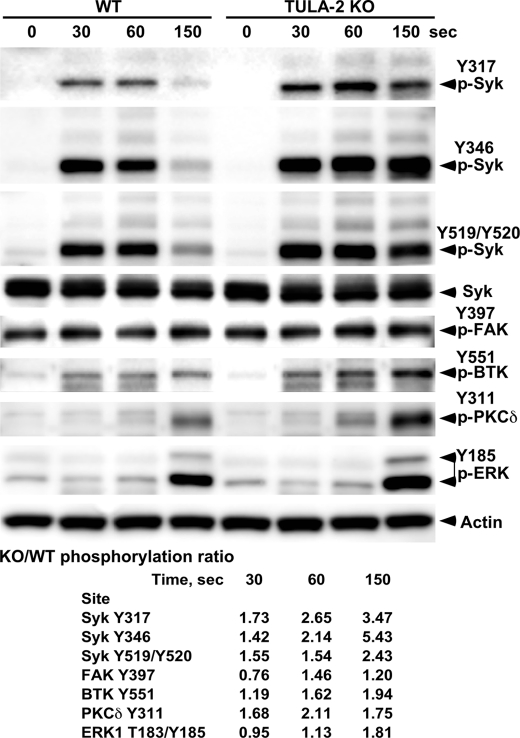
Platelets derived from wild-type and TULA-2 KO mice were stimulated with convulxin, a strong agonist of GPVI routinely used to ligate this receptor in signaling studies (29–31), and tyrosine phosphorylation of the candidate proteins was monitored using antibodies against their individual Tyr(P) sites. A significant increase in the tyrosine phosphorylation of several proteins was observed in TULA-2-deficient platelets (Fig. 4). Thus, in wild-type platelets, GPVI induces a transient phosphorylation of all of the Syk sites examined (Tyr(P)-317, Tyr(P)-346, and Tyr(P)-519/520; note that these are mouse Syk sites, which correspond to Tyr(P)-323, Tyr(P)-352, and Tyr(P)-525/526 in human Syk, respectively); they peak at 30 to 60 s post-stimulation and then decrease to essentially the basal level by 150 s. Phosphorylation of all these sites was increased in TULA-2 KO platelets, and this increase was especially pronounced for the descending part of the time course, rendering their phosphorylation persistent. Of the three sites, Tyr-346 showed the most profound difference between TULA-2 KO and WT platelets. First, the phosphorylation level of Tyr-346 did not decline during the time course of the experiment in TULA-2 KO platelets, whereas those of Tyr-317 and Tyr-519/520 decreased, albeit more slowly than in WT platelets. Second, the ratio between the levels of phosphorylation in TULA-2 KO and WT platelets at 150 s was 5.4, 3.5, and 2.4 for Tyr-346, Tyr-317, and Tyr-519/520, respectively. These results are consistent with the kinetic analysis using synthetic phosphopeptides and recombinant Syk, indicating that the reactivity of Syk Tyr(P) sites toward TULA-2 in platelets follows the order of Tyr(P)-346 > Tyr(P)-317 > Tyr(P)-519/520.
FIGURE 4.
Effect of TULA-2 on protein tyrosine phosphorylation in GPVI-stimulated platelets. Platelets isolated from WT and TULA-2 KO mice were stimulated with convulxin for the indicated periods of time or left untreated. The amounts of phosphorylation at various Y sites and total proteins were detected by immunoblotting with appropriate antibodies. The ratio of tyrosine phosphorylation for each phosphosite in WT and KO platelets was determined at each time point and is shown at the bottom. A ratio of 1.0 indicates equal level of phosphorylation of a site in WT and KO platelets. BTK, Bruton's tyrosine kinase; FAK, focal adhesion kinase.
Protein-tyrosine kinases focal adhesion kinase and BTK were also examined for differential phosphorylation in WT and TULA-2 KO platelets (Fig. 4). Focal adhesion kinase Tyr-397 exhibited substantial phosphorylation in both types of platelets even without stimulation, and the phosphorylation level was essentially unchanged upon knock-out of TULA-2. Similarly, phosphorylation of Src at Tyr-416 did not differ significantly in WT and KO platelets (data not shown).
BTK Tyr-551 showed a ∼2-fold increase in the phosphorylation level, but the increase occurred at a later time (at 150 s) relative to the Syk sites, suggesting that its increased phosphorylation might be the result of a downstream event instead of the direct effect of TULA-2 KO. Similar effects were also observed for PKCδ Tyr-311 and ERK1 Tyr-185; these sites were phosphorylated late in the course of activation, and their phosphorylation levels were increased by ∼2-fold in TULA-2 KO platelets.
DISCUSSION
Our current study has established that the PTP domain of TULA-2, like classical cysteinyl PTPs, exhibits strong sequence specificity, as reflected by the 2100-fold activity difference between peptide 10 and phenyl phosphate (Table 1). It recognizes two distinct classes of Tyr(P) substrates; the class I substrates all feature a proline at the Tyr(P)−1 position, whereas the class II substrates contain acidic residues near the Tyr(P) (Tyr(P)−1 to Tyr(P)-3). Both classes of substrates have a high percentage of aromatic hydrophobic residues at more distal positions (Tyr(P)−3 to Tyr(P)−5). The x-ray crystal structure of TULA-2 revealed that the Tyr(P) binding pocket consists of three Arg residues, two His residues and a Glu residue (6). The positive charges in the active site facilitate the binding of the negatively charged Tyr(P) phosphate group and are likely also responsible for interacting with the acidic residues surrounding the Tyr(P). Outside the positively charged area, the substrate binding surface is hydrophobic, providing an explanation for TULA-2 preference for hydrophobic amino acids at positions beyond Tyr(P)−3 and Tyr(P)+2 (Fig. 2). The observation that in most of the selected substrates, hydrophobic and hydrophilic residues occur at alternate positions suggests that the peptides bind to TULA-2 in an extended conformation, with its hydrophobic face interacting with the enzyme surface and the hydrophilic residues pointing to the solvent. The long range nature of charge-charge interactions permits the acidic residues, even when facing away from the protein surface, to contribute to the overall binding affinity and specificity. The structural basis for the overwhelming selection of a proline as the Tyr(P)−1 residue among the class I substrates is not immediately obvious from the reported structures of the free protein or the TULA-2-phosphate complex (6).
Importantly, the phosphatase activity of TULA-2 toward the intact Syk protein largely correlates with the kinetic data obtained with synthetic Tyr(P) peptides. Thus, the three peptides corresponding to the Tyr(P)-352, Tyr(P)-323, and Tyr(P)-525/526 sites of Syk had kcat/Km values of 2.9 × 105, 2.1 × 104, and 1 × 103 m−1 s−1, respectively (Table 2). Correspondingly, in vitro phosphatase assay with recombinant GST-Syk showed that the Tyr(P)-352 site was much more rapidly dephosphorylated by TULA-2 than the Tyr(P)-525/526 site (Fig. 3). Likewise, a comparison of GPVI-stimulated WT and TULA-2 KO platelets shows that the site most profoundly affected by the lack of TULA-2 activity is Syk Tyr(P)-352 (>5-fold increase at 150 s) followed by the Tyr(P)-323 site (>3-fold increase) and the Tyr(P)-525/526 site (<3-fold). These results together with the earlier in vitro observations (7) provide a compelling argument that Syk is a bona fide substrate protein of TULA-2 during GPVI-mediate receptor signaling in platelets. Similarly, TULA-2 has previously been shown to negatively regulate TCR signaling and able to dephosphorylate ZAP70 in vitro (6). ZAP70 is structurally similar to Syk, and the peptide sequence surrounding Tyr(P)-319 is very similar to the Tyr(P)-352 site of Syk. We hypothesize that ZAP70 is a direct substrate of TULA-2 during TCR signaling and the Tyr(P)-319 site is the primary site of action by TULA-2.
Interestingly, the phosphorylation levels of PKCδ Tyr-311 and ERK1 Tyr-185 also increased in the absence of TULA-2, although the synthetic Tyr(P) peptides corresponding to these sites are rather poor substrates. We suggest that these are likely indirect effects caused by the up-regulation of Syk activity in TULA-2 KO platelets, as Syk, as a GPVI-proximal kinase, initiates the signaling pathway that results in phosphorylation of PKCδ and ERK1 as well as many other proteins (25, 32). We also noted that although the synthetic peptide corresponding to focal adhesion kinase Tyr(P)-397 is a fairly active TULA-2 substrate (kcat/Km = 1.2 × 104 m−1 s−1), its phosphorylation level is increased by only 1.5-fold upon loss of TULA-2 activity. However, phosphorylation of this site is largely constitutive under conditions of our experiments and, thus, may be regulated quite differently from the other sites studied. Another possible reason for the observed discrepancies might be the contribution of the N-terminal ubiquitin-association and Src homology 3 domains, which mediate protein-protein interactions (4, 5, 7, 11, 12, 33–36). These protein binding domains may target the catalytic domain of TULA-2 to specific protein substrates (e.g. Syk and ZAP70) in the cellular context.
Another apparent discrepancy between in vitro and in vivo results is the relative activity of TULA-2 and PTP1B. In vitro, the Syk Tyr(P)-352 motif (peptide 19), which is one of the most active substrates of TULA-2 known to date, is 7-fold more reactive toward PTP1B than TULA-2 (Table 2). This suggests that the PTP domain of TULA-2 is intrinsically less active than PTP1B. And yet, TULA-2 KO resulted in dramatically increased phosphorylation level of Syk Tyr(P)-352 in mouse platelets, which express both TULA-2 and PTP1B (37). Again, the simplest explanation is that the ubiquitin-association and/or Src homology 3 domains of TULA-2 may have acted to target the PTP domain to Syk, although other factors such as differential expression levels may also play a role. It also explains why TULA-2 KO increased the phosphorylation levels of all Syk Tyr(P) sites examined in this study, and the difference in the magnitudes of increase at the different Tyr(P) sites was much smaller than would be predicted from the catalytic activities of the Tyr(P) peptides toward the TULA-2 catalytic domain (Table 1).
In summary, our study has demonstrated that the catalytic domain of TULA-2 exhibits strong sequence specificity. There is a good correlation between the catalytic activity of TULA-2 against Tyr(P) peptides and their rate of dephosphorylation in intact protein substrates. At the meantime, there is also circumstantial evidence that the PTP domain of TULA-2 is targeted to physiological substrates by its other structural elements outside the PTP active site. We hypothesize that like the classical PTPs, the in vivo substrate specificity of TULA-2 is controlled by both its PTP active site and the targeting domain(s). The sequence specificity profile of TULA-2 should facilitate the identification of its additional protein substrates.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant CA132855. This work was also supported by the Pennsylvania Department of Health (to A. Y. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables I–IV and Fig. 1.
- PTP
- protein-tyrosine phosphatase
- F2Y
- 3,5-difluorotyrosine
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- Fmoc-OSu
- N-(9- Fmoc-oxy)succinimide
- GPVI
- glycoprotein VI.
REFERENCES
- 1.Wattenhofer M., Shibuya K., Kudoh J., Lyle R., Michaud J., Rossier C., Kawasaki K., Asakawa S., Minoshima S., Berry A., Bonne-Tamir B., Shimizu N., Antonarakis S. E., Scott H. S. (2001) Hum. Genet 108, 140–147 [DOI] [PubMed] [Google Scholar]
- 2.Carpino N., Kobayashi R., Zang H., Takahashi Y., Jou S. T., Feng J., Nakajima H., Ihle J. N. (2002) Mol. Cell. Biol. 22, 7491–7500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carpino N., Turner S., Mekala D., Takahashi Y., Zang H., Geiger T. L., Doherty P., Ihle J. N. (2004) Immunity 20, 37–46 [DOI] [PubMed] [Google Scholar]
- 4.Kowanetz K., Crosetto N., Haglund K., Schmidt M. H., Heldin C. H., Dikic I. (2004) J. Biol. Chem. 279, 32786–32795 [DOI] [PubMed] [Google Scholar]
- 5.Feshchenko E. A., Smirnova E. V., Swaminathan G., Teckchandani A. M., Agrawal R., Band H., Zhang X., Annan R. S., Carr S. A., Tsygankov A. Y. (2004) Oncogene 23, 4690–4706 [DOI] [PubMed] [Google Scholar]
- 6.Mikhailik A., Ford B., Keller J., Chen Y., Nassar N., Carpino N. (2007) Mol. Cell 27, 486–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agrawal R., Carpino N., Tsygankov A. (2008) J. Cell. Biochem. 104, 953–964 [DOI] [PubMed] [Google Scholar]
- 8.Burke T. R., Jr., Zhang Z. Y. (1998) Biopolymers 47, 225–241 [DOI] [PubMed] [Google Scholar]
- 9.Chen Y., Jakoncic J., Parker K. A., Carpino N., Nassar N. (2009) Biochemistry 48, 8129–8135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rigden D. J. (2008) Biochem. J. 409, 333–348 [DOI] [PubMed] [Google Scholar]
- 11.Collingwood T. S., Smirnova E. V., Bogush M., Carpino N., Annan R. S., Tsygankov A. Y. (2007) J. Biol. Chem. 282, 30920–30928 [DOI] [PubMed] [Google Scholar]
- 12.Smirnova E. V., Collingwood T. S., Bisbal C., Tsygankova O. M., Bogush M., Meinkoth J. L., Henderson E. E., Annan R. S., Tsygankov A. Y. (2008) Virology 372, 10–23 [DOI] [PubMed] [Google Scholar]
- 13.Raguz J., Wagner S., Dikic I., Hoeller D. (2007) FEBS Lett. 581, 4767–4772 [DOI] [PubMed] [Google Scholar]
- 14.Tonks N. K., Neel B. G. (2001) Curr. Opin. Cell Biol. 13, 182–195 [DOI] [PubMed] [Google Scholar]
- 15.Hicke L., Schubert H. L., Hill C. P. (2005) Nat. Rev. Mol. Cell Biol. 6, 610–621 [DOI] [PubMed] [Google Scholar]
- 16.Musacchio A. (2002) Adv. Protein Chem. 61, 211–268 [DOI] [PubMed] [Google Scholar]
- 17.Jia Z., Barford D., Flint A. J., Tonks N. K. (1995) Science 268, 1754–1758 [DOI] [PubMed] [Google Scholar]
- 18.Yang J., Cheng Z., Niu T., Liang X., Zhao Z. J., Zhou G. W. (2000) J. Biol. Chem. 275, 4066–4071 [DOI] [PubMed] [Google Scholar]
- 19.Garaud M., Pei D. (2007) J. Am. Chem. Soc. 129, 5366–5367 [DOI] [PubMed] [Google Scholar]
- 20.Gopishetty B., Ren L., Waller T. M., Wavreille A. S., Lopez M., Thakkar A., Zhu J., Pei D. (2008) Org. Lett. 10, 4605–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thakkar A., Wavreille A. S., Pei D. (2006) Anal. Chem. 78, 5935–5939 [DOI] [PubMed] [Google Scholar]
- 22.Sweeney M. C., Wavreille A. S., Park J., Butchar J. P., Tridandapani S., Pei D. (2005) Biochemistry 44, 14932–14947 [DOI] [PubMed] [Google Scholar]
- 23.Leon S., Quarrell R., Lowe G. (1998) Bioorg. Med. Chem. Lett. 8, 2997–3002 [DOI] [PubMed] [Google Scholar]
- 24.Watson S. P., Auger J. M., McCarty O. J., Pearce A. C. (2005) J. Thromb. Haemost. 3, 1752–1762 [DOI] [PubMed] [Google Scholar]
- 25.Thomas D. H., Getz T. M., Newman T. N., Dangelmaier C. A., Carpino N., Kunapuli S. P., Tsygankov A. Y., Daniel J. L. (2010) Blood, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zahedi R. P., Begonja A. J., Gambaryan S., Sickmann A. (2006) Biochim. Biophys. Acta 1764, 1963–1976 [DOI] [PubMed] [Google Scholar]
- 27.Shankar H., Kahner B., Kunapuli S. P. (2006) Curr. Drug Targets 7, 1253–1263 [DOI] [PubMed] [Google Scholar]
- 28.Wee J. L., Jackson D. E. (2006) Curr. Drug Targets 7, 1265–1273 [DOI] [PubMed] [Google Scholar]
- 29.Polgár J., Clemetson J. M., Kehrel B. E., Wiedemann M., Magnenat E. M., Wells T. N., Clemetson K. J. (1997) J. Biol. Chem. 272, 13576–13583 [DOI] [PubMed] [Google Scholar]
- 30.Quinton T. M., Ozdener F., Dangelmaier C., Daniel J. L., Kunapuli S. P. (2002) Blood 99, 3228–3234 [DOI] [PubMed] [Google Scholar]
- 31.Dangelmaier C. A., Quinter P. G., Jin J., Tsygankov A. Y., Kunapuli S. P., Daniel J. L. (2005) Blood 105, 3918–3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhavaraju K., Kim S., Daniel J. L., Kunapuli S. P. (2008) Eur. J. Pharmacol. 580, 285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoeller D., Crosetto N., Blagoev B., Raiborg C., Tikkanen R., Wagner S., Kowanetz K., Breitling R., Mann M., Stenmark H., Dikic I. (2006) Nat. Cell Biol. 8, 163–169 [DOI] [PubMed] [Google Scholar]
- 34.Bertelsen V., Breen K., Sandvig K., Stang E., Madshus I. H. (2007) Exp. Cell Res. 313, 1696–1709 [DOI] [PubMed] [Google Scholar]
- 35.Tsygankov A. Y. (2008) IUBMB Life 60, 224–231 [DOI] [PubMed] [Google Scholar]
- 36.Tsygankov A. Y. (2009) Cell. Mol. Life Sci. 66, 2949–2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frangioni J. V., Oda A., Smith M., Salzman E. W., Neel B. G. (1993) EMBO J. 12, 4843–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.