Abstract
The transcription factor hypoxia-inducible factor-1α (HIF-1α) is a master regulator of the cellular response to low oxygen. HIF-1α protein accumulates in hypoxia due to inhibition of prolyl hydroxylase enzymes, which under normoxic conditions use molecular oxygen to hydroxylate HIF-1α on two conserved proline residues (Pro402 and Pro564), thus targeting the protein for 26 S proteasome-dependent degradation. A functional mitochondrial electron transport chain is known to be necessary for HIF-1α stabilization in hypoxia. It has been reported that reactive oxygen species (ROS), produced under hypoxia by complex III of the mitochondrial electron transport chain, play a critical role in the stabilization of the HIF-1α protein, possibly by directly inhibiting prolyl hydroxylase enzymes. In contrast, we found that ROS production by complex III is not required for hypoxia-induced HIF-1α stabilization. Thus, reestablishing mitochondrial oxygen consumption in the presence of a complex III inhibitor by using an artificial electron donor to complex IV or by overexpressing Ciona intestinalis alternative oxidase results in HIF-1α protein stabilization in hypoxia. Furthermore, five inhibitors that target different sites of the mitochondrial electron transport chain have similar effects on the HIF-1α protein half-life in hypoxia but vary in their effects on mitochondrial ROS production. Finally, ROS do not regulate prolyl hydroxylase activity directly. We conclude that HIF-1α protein stabilization in hypoxia occurs independently of mitochondrial ROS production. However, mitochondria can modulate the cellular hypoxic response through altered respiratory activity, likely by regulating the cellular oxygen availability.
Keywords: Mitochondria, Reactive Oxygen Species, Hypoxia-inducible Factor, Alternative Oxidase, Prolyl Hydroxylase
Introduction
Hypoxia induces a complex transcriptional program designed to improve cellular oxygen supply and adapt cellular metabolism to the lack of oxygen. This hypoxic response is mediated by hypoxia-inducible factor (HIF),2 a heterodimeric transcription factor consisting of an oxygen-regulated α subunit (HIF-1α and HIF-2α) and a constitutively expressed β subunit. The oxygen-dependent regulation of the α subunit occurs at the level of its protein stability. At normal oxygen concentration, a family of prolyl hydroxylase (PHD) enzymes catalyzes the hydroxylation of two conserved proline residues in HIF-1α and HIF-2α (1, 2). This reaction is dependent on both oxygen, which is a cosubstrate, and non-heme-reduced Fe2+, bound to the catalytic center of PHD enzymes. Hydroxylation of HIF-1α and HIF-2α at either of the two proline residues creates a recognition site for binding to the Von Hippel-Lindau (pVHL) protein. Binding of HIF-1α and HIF-2α to pVHL results in polyubiquitination by the pVHL-associated E3 ubiquitin ligase and degradation by the 26 S proteasome. Under hypoxic conditions, lack of oxygen inhibits PHD activity, thus leading to HIF-1α and HIF-2α protein stabilization and accumulation in the nucleus.
The mitochondrial electron transport chain (ETC), which consumes the vast majority of cellular oxygen and therefore directly controls intracellular oxygen tension, is known to play an important role in cellular oxygen sensing (3, 4). Indeed, inhibition of mitochondrial respiration by pharmacological or genetic means prevents the accumulation of both HIF-1α and HIF-2α upon exposure of cells to hypoxia (5, 6). This effect is due to PHD-dependent HIF-1α degradation (7). Thus, it has been suggested that mitochondrial activity regulates the HIF-1α protein by determining the amount of oxygen available for PHD enzymes (7, 8).
However, given that the mitochondrial ETC is also the major source of cellular reactive oxygen species (ROS) in most cell types, an alternative mechanism has been proposed (9–11). According to this model, increased amounts of ROS produced under hypoxia by complex III of the mitochondrial ETC are required for hypoxia-dependent HIF-1α stabilization. The absence of a functional, ROS-producing ETC therefore prevents hypoxia-dependent HIF-1α stabilization.
Given the universal importance of the cellular hypoxic response for normal cell physiology and the critical involvement of HIF in ischemic diseases and in cancer, we studied in detail the mechanism through which mitochondria regulate the HIF-1α protein. To this end, we used a number of approaches, including manipulations that specifically alter mitochondrial oxygen consumption without affecting ROS production, to show that the primary mechanism through which the mitochondrial ETC controls HIF-1α stability is by regulating the cellular oxygen availability.
EXPERIMENTAL PROCEDURES
Cell Culture
Human osteosarcoma 143B cells were cultured in RPMI (Roswell Park Memorial Institute) 1640 medium, whereas human kidney epithelial HEK293 cells and HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM). Both media were supplemented with 10% fetal bovine serum, 2 mm l-glutamine, and 1% penicillin-streptomycin (Invitrogen).
Oxygen Conditions
Hypoxic condition (1% O2, 94% N2, and 5% CO2) was achieved in a humidified variable aerobic work station (Coy Laboratories Products, Grass Lake, MI) or a Pro-ox 110 oxygen controller and Pro-ox in vitro chamber (BioSpherix, Redfield, NY).
Immunoblotting
Cell lysis and Western blotting were carried out as previously described (39). The following antibodies were used: mouse anti-β-actin (Sigma-Aldrich), mouse anti-α-tubulin (Invitrogen), mouse anti-HIF-1α (BD Pharmingen), rabbit anti-alternative oxidase (AOX) (18), mouse anti-VHL (BD Pharmingen), goat anti-GST (Amersham Biosciences), and anti-catalase (Abcam). Western blots shown are representative of at least two independent experiments.
Detection of Cellular Superoxide
Superoxide production was estimated using dihydroethidium. Cells were plated in 12-well plates and grown to 70% confluence and cotreated with 20 μm dihydroethidium and mitochondrial inhibitors for 30 min. Subsequently, the cells were rinsed with PBS and kept in PBS for analysis of fluorescence using a multiwell plate reader (Molecular Devices, excitation and emission wavelengths were 520 nm and 610 nm, respectively). The validity of this assay has previously been verified by using menadione and 2-methoxyestradiol as positive controls (40).
Measurement of Oxygen Consumption
Oxygen consumption was measured with a Clark-type oxygen electrode (Oxygraph; Hansatech) in Krebs buffer using cells at a density of 1.6 × 108/ml.
Generation of HEK293T Cells Expressing AOX
HEK293T cells were infected with 4 μl of concentrated pWPI-AOX-IRES-GFP lentiviral preparation (18, 41) leading to ∼30% of cell stably expressing the green fluorescence as measured with COULTER®EPICS®-XL_MCLTM flow cytometer and Cyflogic 1.2.1 analysis software. HEK293T-infected cells were subjected to two successive rounds of purification, first plating the infected population in 96-well plates at a density of one cell/well and then subjecting each of the single clones obtained from this seeding to limited dilution using 1/3 serial dilutions. AOX expression in subclones was verified by polarography using a Clark-type oxygen electrode.
Preparation of Mitochondria
Mitochondria were isolated from mouse liver by differential centrifugation in mitochondria isolation buffer containing 280 mm sucrose, 10 mm Tris (pH 7.4), and 1 mm EDTA.
Mitochondrial Complex I Activity
Measurement of complex I activity was carried out as previously described (42) using 0.1 mg of mouse liver mitochondria. Mitochondria were subjected to freeze-thawing prior to the assay.
Overexpression of Cytosolic and Mitochondrially Targeted Catalase
Murine catalase was PCR-amplified from I.M.A.G.E. clone (ID 4241333) and inserted in the retroviral expression vector Puro-MaRX (a gift from Dr. David Beach). It has been previously shown that a robust overexpression of catalase results in accumulation of the protein in the cytoplasm. The N-terminal mitochondrial targeting sequence of SOD2 (comprising the sequence corresponding to residues 1–28) was used to target catalase to mitochondria. This construct also included a C-terminal V5 epitope tag. Retroviral particles were generated using catalase, mitochondrial catalase-V5, or enhanced GFP (control) containing Puro-MaRX retroviral expression vector in 293-gag-pol cells and pseudotyped with vesicular stomatitis virus-G. Protein expression was assessed by immunofluorescence (enhanced GFP) or immunoblotting with catalase antibody.
Detection of H2O2 Scavenging Capacity
H2O2 scavenging capacity was estimated using DCFDA. 143B cells resuspended in Krebs buffer containing 10 mm glucose were loaded with 20 μm DCFDA for 15 min at 37 °C and analyzed by flow cytometry using the fluorescein isothiocyanate (FITC) channel (log mode) both before and after treating with 200 μm H2O2 for 5 min. In each analysis, 10,000 events were recorded.
pVHL Binding Assays
GST-HIF-1α fusion protein plasmid was generated by inserting the HIF-1α coding sequence corresponding to 521–652 amino acids into the pGEX-KG vector. The GST fusion protein was expressed in BL21 cells and purified using GSH-agarose beads. The immobilized GST-HIF was subjected to prolyl hydroxylation assays in the presence of cell lysate from HEK293 cells transfected with PHD2 and VHL and 2 mm 2-oxoglutarate and 2 mm ascorbate for 1 h at room temperature. After the assay, the cell lysate was removed, and the beads were washed 3 times using Nonidet P-40 wash buffer (containing 0.5% Nonidet P-40, 5% glycerol, 0.5 mm EDTA, and 50 mm NaCl) and washed once using PBS. pVHL bound to GST-HIF fusion protein was analyzed by Western blotting.
Statistical Analysis
Data are expressed as the mean ± S.E. Differences in measured variables were assessed with Student's t test. p < 0.05 was considered to be statistically significant.
RESULTS
All Inhibitors of Mitochondrial Electron Transport Have Similar Effects on HIF-1α Protein Half-life in Hypoxia
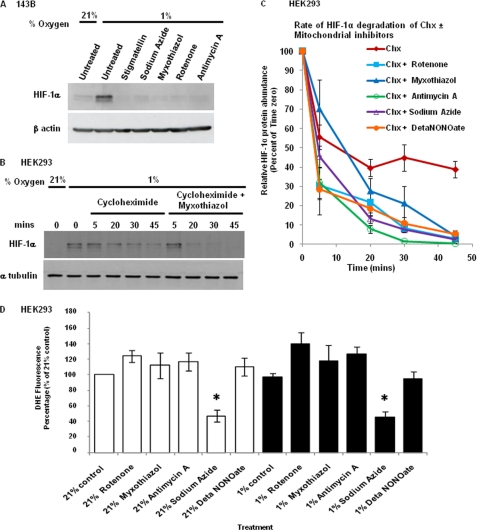
ROS are produced to different degrees at the various complexes of the mitochondrial ETC. We initially determined whether any specific site within the mitochondrial ETC is required for hypoxia-dependent HIF-1α stabilization. To this end, we tested the effects of various inhibitors of the ETC on 143B human osteosarcoma cells grown under hypoxic conditions (1% oxygen) for 4 h. HIF-1α protein accumulation was prevented upon treatment with the different inhibitors (rotenone, myxothiazol, antimycin A, stigmatellin, sodium azide) at concentrations that completely inhibit ETC activity (Fig. 1A). Thus, the requirement of a functional ETC is not restricted to complex III.
FIGURE 1.
Inhibition of mitochondrial electron transport at different sites prevents hypoxia-dependent HIF-1α stabilization and has similar effects on HIF-1α half-life in hypoxia. A, 143B cells were incubated at 1% O2 for 4 h in the presence of the mitochondrial inhibitors stigmatellin (1 μm), sodium azide (2 mm), myxothiazol (1 μm), rotenone (0.5 μm), and antimycin A (1 μm), as indicated. HIF-1α was analyzed by Western blotting. B, after incubation at 1% O2 for 3 h in a hypoxia chamber, HEK293 cells were treated with cycloheximide (Chx, 40 μm) and myxothiazol (1 μm) as indicated, within the chamber without introducing atmospheric oxygen, and cell lysates were obtained after 5, 20, 30, and 45 min followed by immunoblotting using HIF-1α antiserum. C, summary of immunoblot analysis of HIF-1α in cell lysates of HEK293 cells incubated under the same conditions as those described in B with mitochondrial inhibitors rotenone (0.5 μm), myxothiazol (1 μm), antimycin A (1 μm), sodium azide (2 mm) and DETA-NONOate (100 μm) present. The band intensity, as determined by densitometry, was normalized to the α-tubulin signal and expressed as percent of the HIF-1α abundance of time zero. The results shown represent the means ± S.E. (error bars) of three independent experiments. The decrease in HIF-1α protein expression in mitochondrial inhibitor treated versus control cells was statistically significant (p < 0.05) for all inhibitors at 45 min, for all inhibitors except myxothiazol at 30 min, for sodium azide and DETA-NONOate at 20 min. D, HEK293 cells were incubated in either 21% O2 or 1% O2 for 3 h followed by treatment with mitochondrial inhibitors used at the same concentration as in B and C and 20 μm dihydroethidium dye (DHE) for 30 min to measure superoxide concentration. The fluorescence results shown represent the average of three independent experiments (mean ± S.E.). Only sodium azide caused a statistically significant (p < 0.05) decrease in the superoxide concentration in normoxia and hypoxia (as indicated by an asterisk).
To examine this further, we obtained a more quantitative measurement of the effect of the mitochondrial inhibitors by measuring the rate at which the HIF-1α protein is degraded. HEK293 cells were subjected to hypoxic conditions for 3 h followed by incubation with the protein synthesis inhibitor cycloheximide in the presence or the absence of inhibitors for different durations. In the presence of cycloheximide, there was a slow gradual decrease in the HIF-1α protein concentration. However, in the presence of both cycloheximide and the complex III inhibitor myxothiazol, HIF-1α protein was degraded at a much faster rate (Fig. 1B). We repeated the same experiment using different mitochondrial inhibitors (rotenone, antimycin A, sodium azide, DETA-NONOate), as summarized in Fig. 1C. The HIF-1α half-life in hypoxia in control cells was 32.99 min. All inhibitors of the mitochondrial ETC decreased the HIF-1α half-life to similar degrees (rotenone t½ = 8.94 min; myxothiazol t½ = 9.95 min; antimycin A t½ = 5.74 min; sodium azide t½ = 8.59 min; DETA-NONOate t½ = 10.72 min). These results therefore indicate that inhibition of any site in the mitochondrial ETC inhibits hypoxia-dependent HIF-1α protein induction by decreasing its half-life.
Next, we also tested the effects of the inhibitors on ROS production in both normoxia and hypoxia by measuring the superoxide-dependent oxidation of the fluorescent dye dihydroethidium. In the untreated cells, there was no significant increase in the superoxide concentration in hypoxia compared with normoxia. Of the mitochondrial ETC inhibitors, only sodium azide reduced superoxide production in both hypoxia and normoxia significantly. There was no significant change with nitric oxide donor and a slight increase was observed with the other inhibitors (Fig. 1D). Thus, there is no correlation between the effects of the various inhibitors on superoxide production and hypoxia-dependent HIF-1α protein stabilization, suggesting that superoxide may not play an important role in HIF-1α protein stabilization.
Reestablishing Oxygen Consumption in the Presence of Complex III Inhibitor Restores Hypoxia-dependent HIF-1α Stabilization
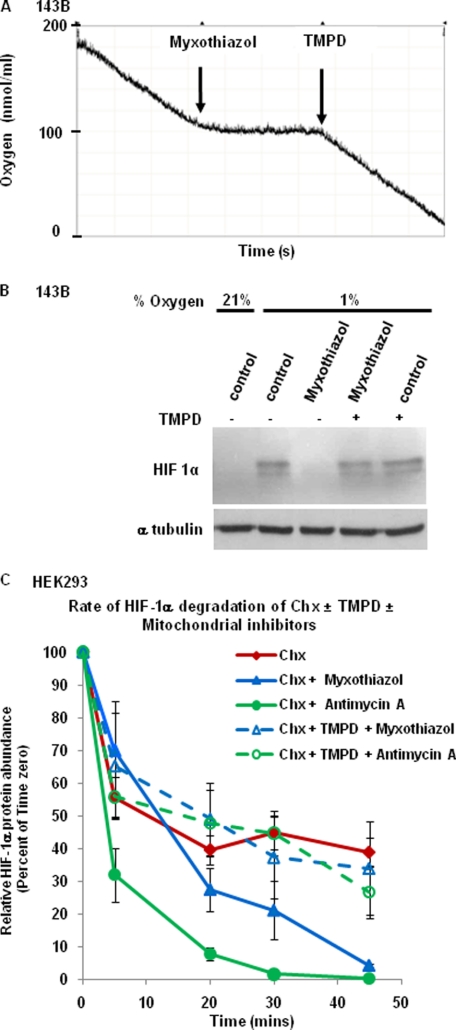
To establish further whether the effects of mitochondrial inhibitors on HIF-1α are due to inhibition of mitochondrial oxygen consumption or inhibition of ROS production from complex III, we investigated the effect of myxothiazol on HIF-1α protein accumulation in the presence of 2,2,4-trimethyl-1,3-pentanediol (TMPD). The complex III inhibitor myxothiazol has been reported to inhibit complex III-dependent superoxide production, whereas TMPD is an artificial electron donor that donates electrons to cytochrome c and complex IV downstream of complex III. As a result, oxygen consumption is reactivated even in the presence of myxothiazol as illustrated in Fig. 2A. We then determined the effect of TMPD on hypoxia-dependent HIF-1α stabilization. TMPD by itself had no effect on the HIF-1α protein concentration in hypoxia (Fig. 2B). However, TMPD completely rescued myxothiazol-induced inhibition of HIF-1α stabilization. This indicates that in the presence of an inhibitor of complex III, reestablishing oxygen consumption via complex IV is sufficient for HIF-1α stabilization. The result suggests that electron transport via complex III and superoxide derived from complex III are not required for HIF-1α induction. Therefore, the effect of myxothiazol is likely due to inhibition of mitochondrial oxygen consumption.
FIGURE 2.
Reestablishing oxygen consumption under condition when complex III is inactive is sufficient for hypoxia-dependent HIF-1α stabilization. A, oxygen consumption of 143B cells was measured using a Clark-type oxygen electrode at 37 °C. Myxothiazol (1 μm) and TMPD (0.2 mm) addition is indicated. B, 143B cells were incubated at 1% O2 for 3 h with myxothiazol in the absence or presence of TMPD. HIF-1α protein abundance was determined by Western blotting. C, HEK293 cells were incubated at 1% O2 for 3 h, followed by treatment with cycloheximide (Chx, 40 μm) and the complex III inhibitors myxothiazol (1 μm) and antimycin A (1 μm), as indicated, in the absence or presence of TMPD. Cell lysates were obtained after 5, 20, 30, and 45 min, followed by immunoblotting using HIF-1α antiserum. The results were calculated and are presented as in Fig. 1C (n = 3, means ± S.E. (error bars)). With antimycin, TMPD caused a statistically significant (p < 0.05) increase in HIF-1α protein expression at 20, 30, and 45 min. With myxothiazol, the TMPD-induced increase in HIF-1α protein expression was not statistically significant.
Next, we repeated the HIF-1α half-life measurements in the presence of TMPD. As shown in Fig. 2C, addition of TMPD increased the HIF-1α half-life in the presence myxothiazol from 9.95 min to 28.91 min, which is similar to the untreated control (t½ = 32.99 min). A very similar trend was observed with a different complex III inhibitor, antimycin A (increase in the HIF-1α half-life from 5.74 min to 23.70 min in the absence versus presence of TMPD). These results further suggested that HIF-1α protein stabilization is independent of ROS produced by complex III.
Alternative Oxidase-dependent Oxygen Consumption Is Sufficient to Reactivate the Hypoxia Response When Complex III Is Inhibited
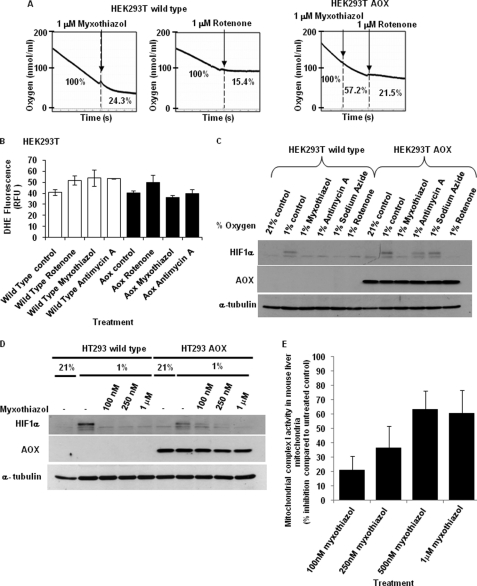
AOX is a mitochondrial enzyme that is expressed in plants, some fungi, protists, and lower metazoa (12). It has two reported physiological functions: to carry out thermogenesis and to lower the ROS production during mitochondrial electron transport (13–16). The AOX protein is localized in the inner mitochondrial membrane. It accepts electrons from coenzyme Q and uses the electrons to reduce oxygen to water directly, therefore bypassing complex III and IV. As a consequence, electron transport via complex III is decreased, resulting in lower superoxide production from complex III via the Q cycle while maintaining mitochondrial oxygen consumption. We have previously shown that AOX derived from Ciona intestinalis is well tolerated in mammalian cells, functional, and can confer complex III and complex IV inhibitor-resistant respiration (17–19). To genetically determine whether HIF-1α stabilization is dependent on mitochondrial ROS from complex III, we utilized HEK293T cells with stable AOX expression. We first compared the respiratory properties of both wild type and AOX-expressing HEK293T cells by measuring their oxygen consumption polarographically using a Clark-type oxygen electrode. When subjected to myxothiazol and rotenone, respiration of the wild type cells was inhibited by 75.7% and 84.8%, respectively (Fig. 3A). However, in the AOX-expressing cells, respiration was inhibited by only 42.8% when myxothiazol was present (Fig. 3A). Subsequent addition of rotenone resulted in similar inhibition (78.5%) as observed in control cells. Thus, the AOX-expressing cells still consume oxygen even in the presence of myxothiazol, due to their ability to bypass both complex III and IV and utilize AOX to consume oxygen.
FIGURE 3.
AOX expression reactivates the hypoxic response when complex III is inhibited. A, both HEK293T wild type and AOX-overexpressing cells were treated overnight in the presence of 2 mm pyruvate to ensure maximum AOX activity, followed by measurement of oxygen consumption in the presence of the mitochondrial inhibitors myxothiazol (1 μm) or rotenone (0.5 μm), as indicated. B, HEK293T wild type (open bars) and AOX-overexpressing (filled bars) cells were treated with the mitochondrial inhibitors rotenone (0.5 μm), myxothiazol (1 μm), and antimycin A (1 μm) and 20 μm dihydroethidium dye (DHE) for 1 h to measure cellular superoxide concentrations. The fluorescence results shown are expressed in relative fluorescence units (RFU) and represent the average of two independent experiments (mean ± S.E. (error bars)). C, HEK293T wild type and AOX-overexpressing cells were exposed to 1% O2 for 3 h in the presence of the mitochondrial inhibitors myxothiazol (1 μm), antimycin A (1 μm), sodium azide (5 mm), and rotenone (0.5 μm). HIF-1α was detected by Western blotting. D, both HEK293T wild type and AOX-overexpressing cells were exposed to 1% O2 for 3 h in the presence myxothiazol (100 nm, 250 nm, and 1 μm). HIF-1α protein abundance was determined by Western blotting. E, activity of mitochondrial complex I was determined in mouse liver mitochondria treated with myxothiazol (100 nm, 250 nm, 500 nm, and 1 μm) using NADH as substrate as described under “Experimental Procedures.”
Next, we determined the effect of AOX protein expression on the cellular superoxide concentration in the presence of different mitochondrial ETC inhibitors. AOX reduced superoxide production in the presence of the complex III inhibitors myxothiazol and antimycin A but had no effect on superoxide when the complex I inhibitor rotenone was present (Fig. 3B). Thus, as expected, AOX only inhibits complex III-dependent superoxide production. To investigate whether reestablishing oxygen consumption under conditions where complex III is inactive is sufficient for hypoxia-dependent HIF-1α stabilization, we treated wild type and AOX-expressing HEK293T cells with mitochondrial inhibitors under hypoxic conditions. In wild type cells, HIF-1α protein accumulation was prevented in the presence of inhibitors of complex I, III, and IV (Fig. 3C). In AOX-expressing cells, HIF-1α induction was also inhibited with complex I inhibitor, rotenone. This result is expected because rotenone inhibits electron transport upstream of AOX. In contrast, antimycin A and sodium azide had no inhibitory effect on HIF-1α expression in AOX-expressing cells. These cells still consume oxygen via the AOX protein, but do not produce increased superoxide because electron transport via complex III is prevented. These results suggest that mitochondrial ETC regulates HIF-1α stability by controlling intracellular oxygen but not ROS concentration.
Unexpectedly, HIF-1α expression was still significantly inhibited when AOX-expressing cells were treated with myxothiazol. Myxothiazol is also a complex III inhibitor. Therefore, its inhibitory effect on hypoxia-dependent HIF-1α accumulation would be expected to be reversed when AOX is expressed and oxygen can be consumed independently from complex III. A possible explanation for this finding is that the concentration of 1 μm myxothiazol that was used is high enough to inhibit complex I also. Indeed, it has been previously reported that at higher concentrations myxothiazol also functions as complex I inhibitor (see Ref. 20). To confirm this hypothesis, both wild type and AOX-expressing cells were treated with different concentrations of myxothiazol under hypoxic conditions (Fig. 3D). In the wild type cells, hypoxia-dependent HIF-1α stabilization was prevented at any concentration of myxothiazol. In contrast, in AOX-expressing cells, HIF-1α could be detected at the lower myxothiazol concentrations (100 nm and 250 nm) but not at the higher concentration of 1 μm. When measuring the effects of the different myxothiazol concentrations on mitochondrial complex I activity in mouse liver mitochondria, significant inhibition was observed with 500 nm and 1 μm, but not with lower concentrations of myxothiazol (Fig. 3E). The results indicate that the inhibition of HIF-1α accumulation in AOX-expressing cells by 1 μm myxothiazol (as observed in Fig. 3C) is due to an inhibitory effect on complex I. In contrast, AOX expression prevents the inhibition of hypoxia-dependent HIF-1α stabilization by concentrations of myxothiazol which specifically inhibit complex III.
Regulation of HIF-1α Protein Stabilization Is Likely Independent of Mitochondrial ROS Produced during Hypoxia
Our results indicate that increasing mitochondrial oxygen consumption in the absence of electron transport via complex III is sufficient for hypoxia-dependent HIF-1α accumulation. To assess the role of ROS in mediating HIF-1α stabilization directly, we used glucose oxidase/glucose to produce constant amounts of H2O2. At glucose oxidase concentrations of 250 milliunits/ml and higher, toxic effects on 143B cells were observed. We therefore used a range of lower glucose oxidase concentration of 25, 100, and 200 milliunits/ml. Based on dichlorofluorescein fluorescence calibration with known amounts of H2O2, addition of 100 milliunits/ml glucose oxidase to 143B cells over a 15-min period resulted in DCF fluorescence that was similar to exposure of cells to 200 μm H2O2. We found that glucose oxidase/glucose-derived H2O2 at any concentration was without effect on HIF-1α protein accumulation during hypoxia (supplemental Fig. S1A). To test further whether H2O2 is involved in HIF-1α stabilization, we retrovirally overexpressed both mitochondria-targeted and cytosolic catalase in 143B cells. The catalase-transduced cells had markedly increased catalase protein expression and H2O2 scavenging capacity (supplemental Fig. S1B). However, HIF-1α protein in cells that overexpressed either form of catalase was still induced in hypoxia to the same degree despite lower amounts of mitochondrial ROS release (supplemental Fig. S1B). These results suggest that mitochondrial or exogenous ROS are not involved in the stabilization of the HIF-1α protein.
Hydrogen Peroxide Does Not Directly Inhibit Prolyl Hydroxylase Activity
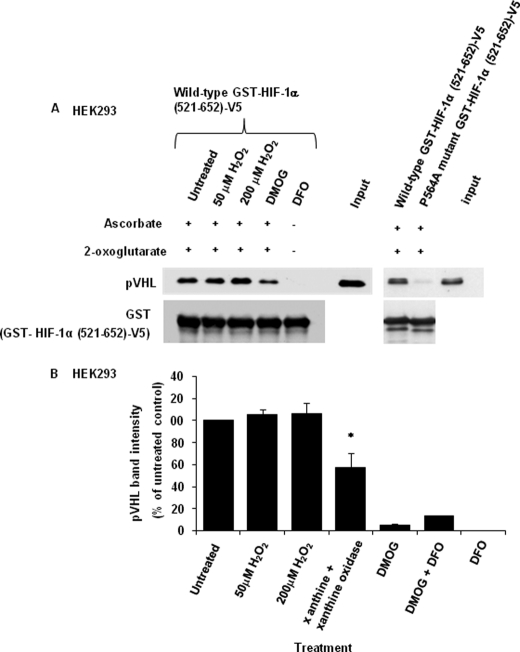
According to the ROS hypothesis, increased mitochondrial superoxide released from complex III will be converted to H2O2 which inhibits the PHD enzyme in the cytoplasm, possibly by oxidizing the non-heme-bound Fe2+ in the PHD active site (3). PHD enzymes in the presence of oxygen hydroxylate Pro402 and Pro564 residues in HIF-1α, leading to binding of the pVHL/Cul2 E3 ligase and consequently ubiquitination and degradation of HIF-1α. To determine directly the effect of H2O2 on the enzymatic activity of PHD, an in vitro PHD assay was carried out using cell lysate to mimic physiological conditions. In this assay, the C-terminal oxygen-dependent degradation domain of HIF-1α containing proline 564 was fused to GST [GST-HIF-1α(521–652)-V5] and immobilized on GSH agarose beads. Cell lysate containing PHD2 and pVHL was used to hydroxylate the recombinant GST-HIF-1α protein. Binding of pVHL to hydroxylated HIF-1α was used as an indicator of Pro564 hydroxylation. pVHL bound to HIF-1α in the presence of the PHD cosubstrate 2-oxoglutarate and the cofactor ascorbate but showed reduced binding in the presence of the PHD inhibitor dimethyloxaloylglycine. As expected, there was no binding of pVHL to HIF-1α upon treatment with the Fe2+ chelator deferoxamine mesylate in the absence of 2-oxoglutarate and ascorbate. There was also no significant pVHL binding to GST-HIF-1α(521–652)-V5 after mutation of Pro564 to Ala in the recombinant HIF-1α fusion protein (Fig. 4A, right panel). In the presence of 50 μm and 200 μm H2O2, pVHL binding was not affected, suggesting that H2O2 does not directly inhibit PHD (Fig. 4A). We also tested whether superoxide affects PHD activity by using xanthine/xanthine oxidase as a superoxide-generating system. In contrast to H2O2, superoxide caused an ∼50% reduction in binding of pVHL to HIF-1α (Fig. 4B), suggesting that superoxide inhibits PHD activity partially. Although H2O2, after conversion from complex III-derived superoxide, has been proposed as the mediator that causes PHD inhibition under hypoxia (10, 11), we wanted to test for any effect of cellular superoxide on PHD activity in cells. To this end, cells were treated with SOD1 inhibitor diethyldithiocarbamate (DCC), the SOD mimetic MnTBAP and the antioxidants N-acetylcysteine and ascorbate which are known to scavenge superoxide (21). Reducing the superoxide concentration via increased conversion of superoxide to H2O2 by MnTBAP or treatment of cells with the N-acetylcysteine and ascorbate did not significantly affect the hypoxia-dependent HIF-1α stabilization (supplemental Fig. S2), suggesting that superoxide is not required. When using DCC to inhibit SOD1, we also observed no significant effect on HIF-1α stabilization in hypoxia. However, DCC markedly increased HIF-1α protein expression in normoxia (supplemental Fig. S2). This effect of DCC was not reversed when adding SOD mimetic MnTBAP to dismutate the accumulating superoxide, suggesting that DCC increases HIF-1α protein expression in normoxia via a superoxide-independent mechanism. Therefore, taken together, our results suggest that neither H2O2 nor superoxide is involved in the regulation PHD activity in vivo.
FIGURE 4.
Hydrogen peroxide does not directly inhibit PHD activity. Cell lysate of HEK293 cells transfected with PHD2 and pVHL was incubated with wild type or P564A mutant GST-HIF-1α(521–652)-V5 immobilized on GSH agarose beads in the presence of 2 mm 2-oxoglutarate and 2 mm ascorbate to detect pVHL binding by Western blotting. The reaction was performed in the presence of H2O2 (50 μm and 200 μm), dimethyloxaloylglycine (DMOG; 2 mm), and deferoxamine mesylate (DFO; 200 μm) as described under “Experimental Procedures.” After the incubation, the lysate was removed, the GSH-agarose beads were washed, and bound pVHL was analyzed by Western blotting. Densitometric analysis of three independent pVHL binding assays in the presence of 50 μm or 200 μm H2O2, superoxide-generating system xanthine/xanthine oxidase, 2 mm dimethyloxaloylglycine is presented in B (percent of untreated control; mean ± S.E. (error bars)). The decrease in the PHD activity in the presence of xanthine/xanthine oxidase compared with the untreated control reached statistical significance (p < 0.05), as indicated by an asterisk.
DISCUSSION
At a given atmospheric oxygen concentration, the intracellular oxygen concentration varies widely in different human tissues as a function of both oxygen supply (i.e. tissue oxygenation) and oxygen consumption. Given that mitochondria consume more than 95% of cellular oxygen, it can be assumed that the mitochondrial metabolic activity in a cell is a major factor in regulating the cellular hypoxic response. Thus, fluctuations in mitochondrial respiration, for instance due to altered metabolic activity (e.g. resting versus contracting muscle cells) or due to intracellular signaling events mediated via NO and CO (which both can inhibit the terminal oxygen acceptor in the mitochondrial ETC, cytochrome c oxidase), may play an important role in regulating HIF-1α stability. Furthermore, mitochondrial ETC activity is reduced under pathological conditions, such as in genetic mitochondrial ETC complex deficiencies or under conditions of septic shock, and the inability of cells to elicit an appropriate hypoxic response may contribute to the pathology in these conditions. Studies by several groups have indeed provided strong evidence that by regulating intracellular oxygen availability for HIF-1α PHD enzymes, mitochondria are important regulators of the cellular hypoxic response (7, 8, 22, 23). Alternatively, a number of studies have proposed that the role of mitochondria is to release ROS, derived from complex III of the ETC, under hypoxic conditions (9–11). The complex III-derived ROS were reported to be required for hypoxia-dependent HIF-1α stabilization, and this effect has been suggested to be due to ROS-mediated inhibition of PHD enzymes. In this study, we evaluated the validity of the two proposed mechanisms by using a number of approaches that differentially affect mitochondrial oxygen consumption and ROS production.
In this study, we provide evidence that mitochondria regulate HIF-1α protein accumulation by controlling the intracellular oxygen availability and not by releasing ROS from complex III. Thus, we show that all ETC inhibitors tested, regardless of which complex they inhibit, decrease the HIF-1α protein half-life under hypoxia to a very similar degree. This suggests that inhibition at any site in the ETC has the same effect on HIF-1α degradation under hypoxia. On the other hand, the inhibitors are known to have varying effects on superoxide production at complex III and had different effects on the cellular superoxide concentrations. This suggests that effects on mitochondrial superoxide production are unlikely to account for the stimulation of HIF-1α degradation by the mitochondrial inhibitors.
Conflicting reports have been published on whether hypoxia increases or decreases cellular ROS. Although a number of reports demonstrated increased ROS under hypoxia from mitochondrial (24, 25) or nonmitochondrial sources (26), numerous studies reported that decreased oxygen availability in hypoxia leads to lower ROS concentrations (27–33). Results in which antioxidant compounds or enzymes were used to prevent ROS-dependent effects on HIF-1α are equally contradictory. Therefore, in this study we used approaches that would specifically change mitochondrial oxygen consumption rates without affecting ROS production. Previous studies supporting the ROS hypothesis relied partially on the inhibitory effects of the ETC inhibitors rotenone and myxothiazol on both hypoxia-dependent HIF-1α stabilization and hypoxia-induced ROS production (5, 10, 34, 35). However, these ETC inhibitors are not well suited because they also inhibit mitochondrial oxygen consumption. Here, we demonstrate that in the presence of inhibitors of complex III, reestablishing oxygen consumption by using an electron donor that provides electrons to complex IV or by bypassing complex III with AOX is sufficient for hypoxia-dependent HIF-1α stabilization. Thus, electron transport via complex III- and complex III-mediated superoxide production is not required for HIF-1α stabilization. We also did not detect a direct inhibitory effect of H2O2 on PHD activity. Although superoxide caused partial inhibition of PHD activity, superoxide released from complex III is quickly converted to H2O2 via SOD1 in the mitochondrial intermembrane space and SOD2 in the mitochondrial matrix. Thus, most signaling functions of superoxide, including the proposed inhibition of prolyl hydroxylases, are believed to be due to its conversion into H2O2 (10, 11). Finally, we also found that manipulating cellular superoxide production using a SOD1 inhibitor or SOD mimetic was without effect on hypoxia-dependent HIF-1α stabilization.
Various studies have shown that ROS, either added exogenously or produced by manipulating the activity of intracellular ROS-producing or -inactivating enzymes, can under certain conditions stabilize HIF-1α. However, this by itself does not provide proof that mitochondria-derived ROS are required for hypoxia-dependent HIF-1α stabilization. In support of a complex III- and ROS-mediated regulation of HIF-1α in hypoxia, several studies have used different pharmacological and genetic approaches. For instance, a number of studies have relied on the mitochondria-targeted antioxidant MitoQ to inactivate superoxide produced by complex III in hypoxia (35, 36). However, more recent studies suggest that in addition to its antioxidant function, this compound also inhibits mitochondrial oxygen consumption (35) and can under certain conditions give rise to ROS via redox cycling (37, 38). Similarly, a number of genetic approaches using ETC-deficient Rho0 cells, siRNA-mediated knockdown of the Rieske iron-sulfur protein of complex III (10, 11), loss of cytochrome c (11), and complex I-deficient Chinese hamster fibroblast CCL16-B2 cells (9) are expected to inhibit both mitochondrial oxygen consumption and superoxide production, thus could not distinguish between the two potential mechanisms. In a different genetic model used by Bell et al. (36), cells deficient in cytochrome b were shown to produce increased amounts of ROS in hypoxia and have markedly reduced mitochondrial oxygen consumption, yet are still able to stabilize HIF-1α in hypoxia. Although we are currently unable to explain this discrepancy with our data, none of the data presented in the current study support a role of mitochondrial ROS in hypoxia-induced HIF-1α stabilization. Thus, we did not observe an increase in cellular superoxide in hypoxia and found no effect on HIF-1α when ROS were reduced or increased using different approaches. We also found no evidence for H2O2-dependent inhibition of PHD activity. We observed that even though different mitochondrial inhibitors varied in their effects on superoxide production, they all had very similar effects on HIF-1α protein half-life in hypoxia. Finally, reestablishing mitochondrial oxygen consumption under conditions where complex III remains inhibited is sufficient to induce HIF-1α stabilization in hypoxia. Therefore, our results do not support a role for complex III-derived ROS in hypoxia-dependent HIF-1α stabilization. In contrast, the data presented in this study indicate that the mitochondrial oxygen consumption rate is the main determinant of HIF-1α stability in hypoxia. Mitochondrial regulation of the intracellular oxygen concentration may thus be a major mechanism that regulates the cellular hypoxic response.
In conclusion, our study characterizes the mechanism through which mitochondria regulate hypoxia-dependent HIF-1α stabilization. We provide further evidence that inhibiting cellular oxygen consumption prevents HIF-1α accumulation in hypoxia and that this is the result of increased cellular oxygen availability. The increased oxygen concentration leads to activation of oxygen-dependent prolyl hydroxylase enzymes that target the HIF-1α protein for ubiquitination and proteasome-dependent degradation.
Supplementary Material
Acknowledgments
We thank David Beach (Institute of Cell and Molecular Science, London, United Kingdom) for providing the retroviral expression vector Puro-MaRX and James Whelan (University of Western Australia) for helpful discussions.
This work was supported by grants from the Singapore National Medical Research Council and National University of Singapore, the School of Medicine, Academy of Finland, INSERM, the Tampere University Hospital Medical Research Fund, and the Sigrid Juselius Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- HIF
- hypoxia-inducible factor
- AOX
- alternative oxidase
- DCC
- diethyldithiocarbamate
- DCFDA
- 2′-7′-dichlorodihydrofluorescein diacetate
- ETC
- electron transport chain
- PHD
- prolyl hydroxylase
- pVHL
- Von Hippel-Lindau protein
- ROS
- reactive oxygen species
- SOD
- superoxide dismutase
- TMPD
- 2,2,4-trimethyl-1,3-pentanediol.
REFERENCES
- 1.Epstein A. C., Gleadle J. M., McNeill L. A., Hewitson K. S., O'Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., Tian Y. M., Masson N., Hamilton D. L., Jaakkola P., Barstead R., Hodgkin J., Maxwell P. H., Pugh C. W., Schofield C. J., Ratcliffe P. J. (2001) Cell 107, 43–54 [DOI] [PubMed] [Google Scholar]
- 2.Bruick R. K., McKnight S. L. (2001) Science 294, 1337–1340 [DOI] [PubMed] [Google Scholar]
- 3.Tormos K. V., Chandel N. S. (2010) J. Cell. Mol. Med. 14, 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor C. T. (2008) Biochem. J. 409, 19–26 [DOI] [PubMed] [Google Scholar]
- 5.Chandel N. S., McClintock D. S., Feliciano C. E., Wood T. M., Melendez J. A., Rodriguez A. M., Schumacker P. T. (2000) J. Biol. Chem. 275, 25130–25138 [DOI] [PubMed] [Google Scholar]
- 6.Agani F. H., Pichiule P., Chavez J. C., LaManna J. C. (2000) J. Biol. Chem. 275, 35863–35867 [DOI] [PubMed] [Google Scholar]
- 7.Hagen T., Taylor C. T., Lam F., Moncada S. (2003) Science 302, 1975–1978 [DOI] [PubMed] [Google Scholar]
- 8.Doege K., Heine S., Jensen I., Jelkmann W., Metzen E. (2005) Blood 106, 2311–2317 [DOI] [PubMed] [Google Scholar]
- 9.Brunelle J. K., Bell E. L., Quesada N. M., Vercauteren K., Tiranti V., Zeviani M., Scarpulla R. C., Chandel N. S. (2005) Cell Metab. 1, 409–414 [DOI] [PubMed] [Google Scholar]
- 10.Guzy R. D., Hoyos B., Robin E., Chen H., Liu L., Mansfield K. D., Simon M. C., Hammerling U., Schumacker P. T. (2005) Cell Metab. 1, 401–408 [DOI] [PubMed] [Google Scholar]
- 11.Mansfield K. D., Guzy R. D., Pan Y., Young R. M., Cash T. P., Schumacker P. T., Simon M. C. (2005) Cell Metab. 1, 393–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDonald A. E. (2008) Funct. Plant Biol. 35, 535–552 [DOI] [PubMed] [Google Scholar]
- 13.Van Aken O., Giraud E., Clifton R., Whelan J. (2009) Physiol. Plant. 137, 354–361 [DOI] [PubMed] [Google Scholar]
- 14.Maxwell D. P., Wang Y., McIntosh L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 8271–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dufour E., Boulay J., Rincheval V., Sainsard-Chanet A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4138–4143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rustin P., Jacobs H. T. (2009) Physiol. Plant. 137, 362–370 [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Ayala D. J., Sanz A., Vartiainen S., Kemppainen K. K., Babusiak M., Mustalahti E., Costa R., Tuomela T., Zeviani M., Chung J., O'Dell K. M., Rustin P., Jacobs H. T. (2009) Cell Metab. 9, 449–460 [DOI] [PubMed] [Google Scholar]
- 18.Dassa E. P., Dufour E., Gonçalves S., Paupe V., Hakkaart G. A., Jacobs H. T., Rustin P. (2009) EMBO Mol. Med. 1, 30–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hakkaart G. A., Dassa E. P., Jacobs H. T., Rustin P. (2006) EMBO Rep. 7, 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Degli Esposti M., Ghelli A., Crimi M., Estornell E., Fato R., Lenaz G. (1993) Biochem. Biophys. Res. Commun. 190, 1090–1096 [DOI] [PubMed] [Google Scholar]
- 21.Yang S., Ma H. W., Yu L., Yu C. A. (2008) J. Biol. Chem. 283, 28767–28776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong Y., Agani F. H. (2005) Am. J. Physiol. Cell Physiol. 288, C1023–C1029 [DOI] [PubMed] [Google Scholar]
- 23.Brown S. T., Nurse C. A. (2008) Am. J. Physiol. Cell Physiol. 294, C1305–C1312 [DOI] [PubMed] [Google Scholar]
- 24.Chandel N. S., Budinger G. R. S. (2007) Free Radic. Biol. Med. 42, 165–174 [DOI] [PubMed] [Google Scholar]
- 25.Dirmeier R., O'Brien K. M., Engle M., Dodd A., Spears E., Poyton R. O. (2002) J. Biol. Chem. 277, 34773–34784 [DOI] [PubMed] [Google Scholar]
- 26.Grishko V., Solomon M., Breit J. F., Killilea D. W., Ledoux S. P., Wilson G. L., Gillespie M. N. (2001) FASEB J. 15, 1267–1269 [DOI] [PubMed] [Google Scholar]
- 27.Hoffman D. L., Brookes P. S. (2009) J. Biol. Chem. 284, 16236–16245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang W., Fang H., Groom L., Cheng A., Zhang W., Liu J., Wang X., Li K., Han P., Zheng M., Yin J., Wang W., Mattson M. P., Kao J. P., Lakatta E. G., Sheu S. S., Ouyang K., Chen J., Dirksen R. T., Cheng H. (2008) Cell 134, 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung S. N., Yang W. K., Kim J., Kim H. S., Kim E. J., Yun H., Park H., Kim S. S., Choe W., Kang I., Ha J. (2008) Carcinogenesis 29, 713–721 [DOI] [PubMed] [Google Scholar]
- 30.Mehta J. P., Campian J. L., Guardiola J., Cabrera J. A., Weir E. K., Eaton J. W. (2008) Chest 133, 1410–1414 [DOI] [PubMed] [Google Scholar]
- 31.Hoffman D. L., Salter J. D., Brookes P. S. (2007) Am. J. Physiol. Heart Circ. Physiol. 292, H101–H108 [DOI] [PubMed] [Google Scholar]
- 32.Tuttle S. W., Maity A., Oprysko P. R., Kachur A. V., Ayene I. S., Biaglow J. E., Koch C. J. (2007) J. Biol. Chem. 282, 36790–36796 [DOI] [PubMed] [Google Scholar]
- 33.Fandrey J., Frede S., Jelkmann W. (1994) Biochem. J. 303, 507–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schroedl C., McClintock D. S., Budinger G. R., Chandel N. S. (2002) Am. J. Physiol. Lung Cell Mol. Physiol. 283, L922–L931 [DOI] [PubMed] [Google Scholar]
- 35.Pan Y., Mansfield K. D., Bertozzi C. C., Rudenko V., Chan D. A., Giaccia A. J., Simon M. C. (2007) Mol. Cell. Biol. 27, 912–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bell E. L., Klimova T. A., Eisenbart J., Moraes C. T., Murphy M. P., Budinger G. R. S., Chandel N. S. (2007) J. Cell Biol. 177, 1029–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doughan A. K., Dikalov S. I. (2007) Antioxid. Redox Signal 9, 1825–1836 [DOI] [PubMed] [Google Scholar]
- 38.James A. M., Cochemé H. M., Smith R. A., Murphy M. P. (2005) J. Biol. Chem. 280, 21295–21312 [DOI] [PubMed] [Google Scholar]
- 39.Chew E. H., Matthews C. S., Zhang J., McCarroll A. J., Hagen T., Stevens M. F., Westwell A. D., Bradshaw T. D. (2006) Biochem. Biophys. Res. Commun. 346, 242–251 [DOI] [PubMed] [Google Scholar]
- 40.Chua Y. S., Chua Y. L., Hagen T. (2010) Mol. Cancer Ther. 9, 224–235 [DOI] [PubMed] [Google Scholar]
- 41.Dassa E. P., Dufour E., Goncalves S., Jacobs H. T., Rustin P. (2009) Physiol. Plant. 137, 427–434 [DOI] [PubMed] [Google Scholar]
- 42.Birch-Machin M. A., Briggs H. L., Saborido A. A., Bindoff L. A., Turnbull D. M. (1994) Biochem. Med. Metab. Biol. 51, 35–42 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.