Abstract
Human induced pluripotent stem (iPS) cells have the potential to establish a new field of promising regenerative medicine. Therefore, the safety and the efficiency of iPS-derived cells must be tested rigorously using appropriate animal models before human trials can commence. Here, we report the establishment of rabbit iPS cells as the first human-type iPS cells generated from a small laboratory animal species. Using lentiviral vectors, four human reprogramming genes (c-MYC, KLF4, SOX2, and OCT3/4) were introduced successfully into adult rabbit liver and stomach cells. The resulting rabbit iPS cells closely resembled human iPS cells; they formed flattened colonies with sharp edges and proliferated indefinitely in the presence of basic FGF. They expressed the endogenous pluripotency markers c-MYC, KLF4, SOX2, OCT3/4, and NANOG, whereas the introduced human genes were completely silenced. Using in vitro differentiating conditions, rabbit iPS cells readily differentiated into ectoderm, mesoderm, and endoderm. They also formed teratomas containing a variety of tissues of all three germ layers in immunodeficient mice. Thus, the rabbit iPS cells fulfilled all of the requirements for the acquisition of the fully reprogrammed state, showing high similarity to their embryonic stem cell counterparts we generated recently. However, their global gene expression analysis revealed a slight but rigid difference between these two types of rabbit pluripotent stem cells. The rabbit model should enable us to compare iPS cells and embryonic stem cells under the same standardized conditions in evaluating their ultimate feasibility for pluripotent cell-based regenerative medicine in humans.
Keywords: Cell Differentiation, Embryonic Stem Cell, Gene Transfer, Mammal, Microarray, Stem Cell, Transcription Factors, iPS Cell, Pluripotency, Rabbit
Introduction
Mammalian pluripotent stem cells are those cells capable of differentiating into all germ layers (ectoderm, mesoderm, and endoderm) but not into the extraembryonic tissues (e.g. placenta). They can be generated from blastocyst inner cell mass cells, epiblast cells, primordial germ cells, or male germ line stem cells (for review, see Ref. 1). These are now designated as embryonic stem (ES) cells, epiblast pluripotent stem cells, embryonic germ cells, and multipotent germ line stem cells, respectively. They are invaluable research resources for the study of cell and embryonic development as well as practical tools for the production of genetically engineered animals and for future therapeutic purposes. All of these pluripotent stem cell types were founded by exploiting the epigenetic status of the original undifferentiated cells, yet endowing them with the capacity for self-proliferation. Therefore, the landmark discovery that fully differentiated cells can be reprogrammed directly to a pluripotent state by exogenous transcription factors inevitably revised our views on the nature of pluripotency. Such induced pluripotent stem (iPS)3 cells were first established in mice followed by humans (2–4). Human iPS cells are expected to open a new frontier in human regenerative medicine because they can overcome two major issues associated with human ES cells. In theory, they can avoid the potential issues of allogeneic immune rejection if they are derived from the patient's own cells when used for gene therapy, and they can also bypass the ethical problems of using human embryos for establishing ES cell lines.
Thus, human iPS cells might represent an ideal source for patient-specific cell-based regenerative medicine. However, they are uniquely distinct in their origin compared with other pluripotent stem cells, and they might express unidentified or uncontrollable characters that do not exist in “normal” ES cells (5–7). These problems include the accidental reactivation of introduced genes and persistent donor cell gene expression, as reported (6). Furthermore, as in the case of ES cell-based therapies, the possibility of tumor formation in transplanted tissues caused by dedifferentiation of the donor cells or arising from residual undifferentiated cells should be examined vigorously. In this context, animal models are expected to play important roles before any clinical trials of iPS-based therapies can be approved ethically (8). Indeed, mice expressing human disease phenotypes were treated successfully and safely with genetically corrected autologous iPS cells (9). However, any direct extrapolation of such results to humans is problematic because mice differ considerably from humans in their physiology and life span, and the pluripotency regulatory systems also differ between mouse and human iPS cells (10). In addition to mouse iPS cells, monkey and pig iPS cells were established successfully in several laboratories (11–15). As expected, the iPS cells from these two species resemble human iPS cells more than do the mouse cells and thus should provide superior experimental models to assess therapeutic applications of iPS cells. However, if human-type iPS cells could be made available from smaller laboratory species, they could become attractive alternatives with easy accessibility in most biomedical laboratories.
The laboratory rabbit (Oryctolagus cuniculus) has long been used for developing new reproductive and stem cell-related technologies since the first embryo transfer experiments in 1897 (16). Basic reproductive engineering techniques have long been established, including in vitro fertilization and embryo cryopreservation (17–19). Now, more advanced techniques, including transgenesis, nuclear transfer, and intracytoplasmic sperm injection, are available (20–25). Rabbits are easy to maintain and handle, are larger than mice or rats, enabling us to perform surgical operations on any tissues and organs. Several ES cell lines were developed and characterized as showing major important characters in common with human ES cells (26–30). Rabbit genomic information is accumulating rapidly and is now available to the public at http://www.ncbi.nlm.nih.gov/projects/genome/guide/rabbit/. In this study, we sought to generate rabbit iPS cells, supposing that they would show close resemblance with their ES cell counterparts and therefore with human iPS/ES cells. We expect that rabbit iPS cells will provide unique, easily accessible animal models for assessing the efficacy and safety of new iPS-based treatment. Moreover, this rabbit model should enable us to compare iPS cells and ES cells under the same standardized conditions in evaluating their ultimate feasibility for cell-based regenerative medicine.
EXPERIMENTAL PROCEDURES
Culture of Adult Rabbit Somatic Cells
Sexually mature Dutch Belted rabbits were purchased from Kitayama Labes (Nagano, Japan). After euthanizing the animals, the stomach and liver were extirpated, incised, washed with Hanks' balanced salt solution, and bluntly dissected using 1 mg/ml collagenase, 1.4 mg/ml DNase, and 0.2% trypsin. After termination of enzyme treatment by adding fetal bovine serum (FBS), loosened tissues were dissected into single cells and small clumps by gentle pipetting. These cells were plated and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS, 10 mg/liter insulin, 5.5 mg/liter transferrin, 6.7 μg/liter selenium, 40 ng/ml hepatocyte growth factor (Peprotech, Rocky Hill, NJ), 20 ng/ml epidermal growth factor (Peprotech), penicillin, and streptomycin. Rabbit ES cells (rES8-2), described previously (28), were used as a control for iPS cell evaluation. For DNA microarray analysis, rabbit ES cell lines (rdES4 and rdES2-1) were derived from Dutch Belted rabbits according to established methods (28).
iPS Cell Induction and Culture
Rabbit somatic cells were cultured overnight in DMEM/Ham's F-12 (Invitrogen) supplemented with 10% FBS-containing lentiviruses at a multiplicity of infection of 30 in flat-bottomed 24-well plates at 37 °C under 6% CO2 in air. Two days after transduction, the cells were harvested by trypsinization; 1 × 103 cells were replated into 100-mm culture dishes and cultured on mitomycin C-treated mouse embryonic fibroblasts at a concentration of 6 × 103/cm2 at 38 °C under 6% CO2 and 5% O2 in air. Hypoxic conditions were sustained for 14 days. The culture medium (iPSM) consisted of 78% DMEM/Ham's F-12 supplemented with 20% knock-out serum replacement (KSR), 2 mm GlutaMax (Invitrogen), 1% nonessential amino acids, 0.1 mmol/liter β-mercaptoethanol, 103 units/ml ESGRO (murine leukemia inhibitory factor; Invitrogen), and 4 ng/ml human recombinant basic fibroblast growth factor (bFGF) (Wako, Osaka, Japan). On days 12–18, rabbit ES-like cell colonies were isolated mechanically and replated onto mouse embryonic fibroblasts. Passage of iPS cells was performed by incubating the cells with 0.1% trypsin for 1 min at room temperature and disaggregating the resulting small clumps mechanically into single cells. Cells were then counted in a hemocytometer, resuspended, and plated in iPSM supplemented with 8 ng/ml bFGF. Fresh medium was added daily and cells were passaged every 3–4 days.
Lentiviral Vectors
Human OCT3/4, SOX2, KLF4, and c-MYC (human “four-factors”) cDNAs were amplified by reverse transcription-PCR (RT-PCR) using mRNA prepared from human ES cells as a template with the primer sets as described (3). The cDNAs were inserted into the pENTR/D-TOPO entry vector plasmid (Invitrogen) and verified by sequencing. The cDNAs in pENTR/D-TOPO were then transferred to the pCSII-EF-MCS-IRES2-Venus lentiviral vector plasmid using Gateway LR clonase (Invitrogen). Lentiviral vectors pseudotyped with the vesicular stomatitis virus G glycoprotein (VSV-G) were produced by transient transfection of three plasmids into 293T cells: the packaging plasmid (pCAG-HIVgp), the vesicular stomatitis virus G glycoprotein- and Rev-expressing plasmid (pCMV-VSV-G-RSV-Rev), and the lentiviral vector plasmid. The culture supernatant was concentrated by ultracentrifugation, and the viral pellet was resuspended in Hanks' balanced salt solution. The titers of vectors were determined by infection of HeLa CD4+ cells with serial dilutions of the vector stocks followed by fluorescence-activated cell sorting analysis for Venus+ cells.
RT-PCR Analysis
Total RNA was isolated using ISOGEN (Nippon Gene, Toyama, Japan) from cells cultured under different conditions. After DNase treatment to prevent genomic DNA contamination, first-strand cDNA was synthesized using an RNA PCR kit (TaKaRa, Shiga, Japan) using an oligo(dT)-3 site adaptor primer. Synthesized cDNA was subjected to PCR using the specific primers listed in supplemental Table S1 with a program of 94 °C for 3 min, 35 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s.
DNA Microarray Analysis
The rabbit 44,000 oligonucleotide array, G2519F (Agilent Technologies, Santa Clara, CA), was used throughout this study. DNase-treated total RNA was labeled with Cy3 dye with Quick Amp labeling kit (Agilent Technologies). Hybridized slides were scanned using a microarray scanner (Agilent Technologies), and the signals were processed with the Feature Extraction software version GX (Agilent Technologies). Spots flagged as bad or not found were excluded. The ratios of signal intensities between the eight cell types studied were calculated by Lowess normalization using GeneSpring GX (Agilent Technologies), resulting in 32,474 reporters in the final analysis. Unsupervised hierarchical clustering, principal component analysis, and Venn diagrams were used for assessment.
Detection of Undifferentiated Markers
Marker expression was analyzed by fixing iPS cells attached to the bottom of the culture plates in 4% paraformaldehyde for 30 min at room temperature and then washing three times (5 min each) with 1% bovine serum albumin (BSA) containing Tris-buffered saline (wash buffer). For permeabilization, cells were treated with 0.1% Triton X-100 in wash buffer for 10 min. Cells were incubated in blocking solution (10% normal donkey serum and 1% BSA in wash buffer) for 30 min. The following primary antibodies were used: anti-SSEA1, anti-SSEA3, and anti-SSEA4 from the Developmental Studies Hybridoma Bank (Iowa City, IA), anti-OCT3/4 from Santa Cruz Biotechnology (Santa Cruz, CA), and anti-NANOG from COSMOBIO (Tokyo, Japan). All antibodies were diluted in blocking solution and incubated with samples overnight at 4 °C. The next day, cells were washed three times with wash buffer and incubated with secondary antibodies at room temperature for 1 h. Cells were washed three times with wash buffer and covered with 50% glycerol containing 4′,6-diamidino-2-phenylindole (DAPI). The fluorescent signals were analyzed using a BZ-9000 fluorescence microscope (Keyence, Osaka, Japan).
To detect alkaline phosphatase activity, rabbit iPS cells were stained using an alkaline phosphatase kit (Sigma-Aldrich) according to the manufacturer's protocol. Quantitative analysis of relative activity of alkaline phosphatase was assessed using BZ-9000 fluorescence microscope and BZII-analyzer (Keyence).
Detection of Telomerase Activity
Telomerase activity was detected using a telomeric repeat amplification (TRAP)eze telomerase detection kit (Chemicon, Temecula, CA) according to the manufacturer's protocol. The samples were separated using Tris-buffered EDTA-based 8% acrylamide nondenaturing gel electrophoresis. The gels were stained with SYBR Green I (1:10,000; TaKaRa).
Karyotype Analysis
Karyotyping of rabbit iPS cells in the log growth phase was assessed by previously described methods (28). At least 20 metaphase spreads were counted for each iPS cell line.
Bisulfite Sequencing
Bisulfite treatment was performed using a CpGenome modification kit (Chemicon) according to the manufacturer's protocol. The PCR primers are listed in supplemental Table S1 with a program of 94 °C for 3 min, 35 cycles of 94 °C for 30 s, 50 °C for 30 s, and 72 °C for 30 s. The amplified products were cloned into T vectors (TaKaRa) and then sequenced.
Differentiation in Vitro and in Vivo
To test for embryoid body formation, iPS cells were digested with 0.1% trypsin, resuspended in a solution containing 78% DMEM/Ham's F-12, 20% KSR, 2 mm GlutaMax, 1% nonessential amino acids, and 0.1 mm β-mercaptoethanol, and cultured in Petri dishes. Embryoid bodies were collected after 4–7 days in suspension culture and transferred to plastic dishes coated with gelatin to promote adherence. Culture continued for an additional 14–21 days to promote further differentiation. For endodermal differentiation, rabbit iPS cells were cultured using 78% DMEM/Ham's F-12, 20% KSR, 2 mm GlutaMax, 1% nonessential amino acids, 0.1 mm β-mercaptoethanol, 4 ng/ml bFGF, and 50 ng/ml activin A (R & D Systems, Minneapolis, MN) on feeder free gelatin-coated dishes for 5 days. The outgrowths were fixed with 4% paraformaldehyde for 30 min at room temperature and then washed three times (5 min each) with 1% BSA in wash buffer. For permeabilization, cells were treated with 0.1% Triton X-100 in wash buffer for 10 min. Cells were incubated with blocking solution (10% normal donkey serum and 1% BSA in wash buffer) for 30 min. The following primary antibodies were used: anti-βIII-tubulin, anti-glial fibrillary acidic protein (R & D Systems), anti-α-smooth muscle actin (Abcam, Cambridge, MA), and anti-GATA4 (Santa Cruz Biotechnology). All antibodies were diluted in blocking solution and incubated with samples overnight at 4 °C. The next day, cells were washed three times with wash buffer and incubated with secondary antibodies at room temperature for 60 min. Cells were washed three times with wash buffer and covered with 50% glycerol containing DAPI. The fluorescent signals were analyzed using a BZ-9000 fluorescence microscope (Keyence, Osaka, Japan).
For teratoma formation, 1–2 × 106 iPS cells were injected under the kidney capsule of 5–8-week-old severe combined immunodeficient (SCID) mice. After 4–8, teratomas were dissected and fixed in paraformaldehyde as above. Paraffin wax sections were stained with hematoxylin and eosin.
RESULTS
Generation of iPS Cell Lines from Rabbit Somatic Cells
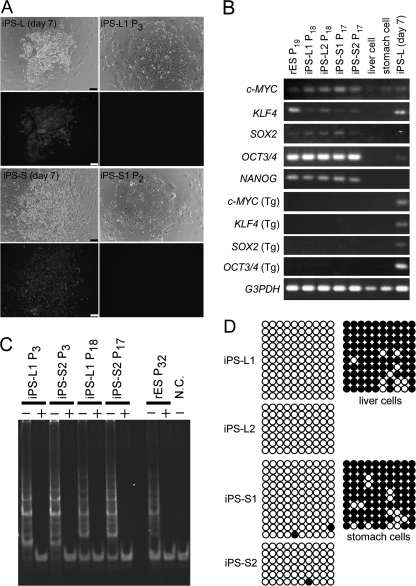
To generate iPS cell lines from rabbit somatic cells, we cultured cells freshly collected from the liver and stomach of an adult female rabbit (Fig. 1A). Four genes for human transcription factors (c-MYC, KLF4, SOX2, and OCT3/4) were introduced by lentiviral vectors at a multiplicity of infection of 30. To monitor the efficiency of infection and the silencing of exogenous genes, we used lentivirus vectors carrying a green fluorescent protein (GFP) cassette, expressed ubiquitously under the control of its CAG promoter. Two days after transduction, the transgenes had integrated successfully into both the somatic cell types, as indicated by GFP fluorescence (Fig. 1B). After being replated onto mitomycin C-treated mouse embryonic fibroblast feeder cell layers at 1 × 103 cells/100-mm dish, the growing cells were cultured in KSR medium. In this early phase of iPS generation, we employed a hypoxic (5% O2) culture condition because it was reported that hypoxia enhances the generation of human iPS cells and prevents the differentiation of human ES cells (31, 32). Eight to 15 days after transduction, colonies resembling rabbit ES cells were observed. Eleven ES-like colonies from liver cells and five colonies from stomach cells were obtained and expanded further in KSR medium. These corresponded to 0.55% and 0.25% of the initial donor cell populations, respectively, as measures of the efficiency of establishment. The expanded ES-like colonies showed a flat and tightly packed morphology, large nuclei, and scant cytoplasm, similar to rabbit and human ES cells (Fig. 1C). The cells were designated iPS-L (liver) cells and iPS-S (stomach) cells, respectively. These colonies each gave rise to iPS lines, 7 from liver cells and 5 from stomach cells, except for 4 of the 11 liver cell-derived colonies, which were lost because of accidental microbiological contamination (supplemental Table S2). Doubling times in the early phase of generation were 16.07 ± 2.35 h (iPS-L) and 14.30 ± 1.51 h (iPS-S). However, after several passages (∼10), the cell cycles accelerated to 13.93 ± 1.21 h and 12.34 ± 1.50 h, respectively, thus reaching a level similar to that of rabbit ES cells (12.82 ± 0.48 h). Most (>80%) of the cells from either of the iPS cell lines showed a normal karyotype (2n = 44) at passages 17 (iPS-L) and 16 (iPS-S) despite complete dissociation by conventional trypsin treatment at their passages (Fig. 1D). As with the rabbit ES cells we had established already, the rabbit iPS cells could form colonies most efficiently when they were plated onto feeder layers at a cell density of 6 × 103/cm2, one-sixth of the density at confluence (28). Culture of rabbit iPS cells without feeder cells or with feeder cells at a higher density (36 × 103/cm2) resulted in their immediate differentiation or arrest of division (supplemental Fig. S1). Our group has shown that bFGF and activin but not leukemia inhibitory factor signaling are the primary mechanisms for maintaining rabbit ES cell pluripotency, in common with human ES cells (30). To examine whether this was also the case for rabbit iPS cells, we cultured iPS-L cells and iPS-S cells without bFGF or leukemia inhibitory factor, or with SB431542, a potential inhibitor of the activin signaling pathway. Their undifferentiated status was assessed by staining for alkaline phosphatase activity. As expected, rabbit iPS cells lost most alkaline phosphatase activity when the bFGF or activin pathways were inhibited, whereas no such loss of status was observed in a leukemia inhibitory factor-free condition (supplemental Fig. S1).
FIGURE 1.
Generation of iPS cells from adult rabbit liver and stomach cells. A, morphology of the primary cultures of somatic cells prepared from the liver (left panel) and stomach (right panel). B, expression of GFP at 2 days after transduction in human 4 factor-infected liver cells (left panel) and stomach cells (right panel). C, appearance of iPS cell colonies at passages 18 (liver, iPS-L1 cells) and 17 (stomach, iPS-S1 cells). D, normal number (2n = 44) of metaphase chromosomes confirmed in rabbit iPS cells arising from iPS-L1 cells at passage 18 (left panel) and iPS-S1 at passage 17 (right panel). Scale bar, 100 μm.
Expression of Pluripotent Markers in Rabbit iPS Cells
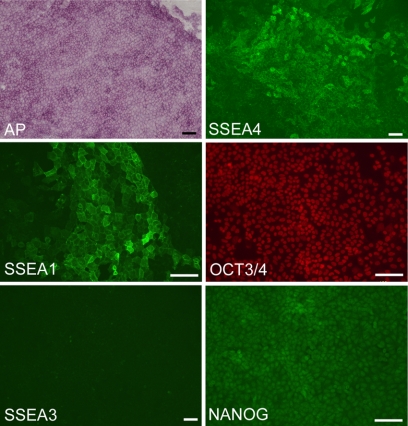
Although the GFP fluorescence signal was observed in the liver and stomach cells at 7 days after infection, all iPS cell lines established became negative for GFP fluorescence as early as the second or third passage, suggesting that the exogenous lentiviral transcripts were silenced, at least at the level of expression of this fluorescent marker (Fig. 2A). To examine more precisely exogenous gene silencing, we applied RT-PCR, using primers specific for the human transgenes, together with primers for the corresponding endogenous rabbit genes. Although low expression levels of the four transgenes were found at passages 6 (iPS-S) or 7 (iPS-L) (supplemental Fig. S2), they disappeared completely at passages 17 or 18, respectively, indicating that exogenous reprogramming genes could be successfully silenced with repeated passages (Fig. 2B). By contrast, all endogenous pluripotency-related genes, namely rabbit c-MYC, KLF4, SOX2, OCT3/4, and NANOG, were detected simultaneously by RT-PCR (Fig. 2B). To investigate the epigenetic control status of OCT3/4 expression in rabbit iPS cells, DNA methylation of the promoter region of the OCT3/4 gene was analyzed by bisulfite genomic sequencing. In mice, the Oct3/4 gene has a GC-rich and TATA-poor minimal promoter (33), and most CpG islands in this region were hypomethylated in mouse ES cells but hypermethylated in liver cells (34). Our results demonstrated that the promoter region of OCT3/4 in rabbit iPS cells was hypomethylated, whereas it was hypermethylated in the parental somatic cells (Fig. 2D). Rabbit iPS cells also showed a high alkaline phosphatase activity (Fig. 3 and supplemental Fig. S1) and expressions of some ES cell-related markers, SSEA1, SSEA4, OCT3/4, and NANOG (Fig. 3). Taken together, rabbit iPS cells were indistinguishable from their ES cell counterparts in their patterns of pluripotent marker expression as well as their colony morphology and proliferation characteristics.
FIGURE 2.
Characterization of rabbit iPS cells by their gene expression, telomerase activity, and DNA methylation of the OCT3/4 promoter. A, GFP fluorescence signal observed in the cells at 7 days after infection (iPS-L day 7 and iPS-S day 7). However, it became undetectable in iPS-L1 cells and iPS-S1 cells as early as passages 3 and 2, respectively. B, RT-PCR analysis of the expression of selected pluripotency-related genes and transgenes (Tgs) in rabbit ES cells, rabbit iPS cells, the original rabbit liver and stomach cells, and liver cells at 7 days after infection (iPS-L day 7). Five selected endogenous genes were expressed in all rabbit iPS cell lines as well as their ES cell counterparts. The four human transgenes were silenced in the iPS cells by passages 17 or 18, whereas they were slightly expressed at earlier passages (passages 6 and 7; see supplemental Fig. S2). C, detection of the telomerase activity of rabbit iPS cells using the telomeric repeat amplification protocol. The iPS cells showed high telomerase activity, similar to that of rabbit ES cells. Heat-inactivated samples (+) were used as negative controls. N.C., negative control with lysis buffer only. D, bisulfite genomic sequencing of CpG-enriched regions of the rabbit OCT3/4 promoter in iPS cells and their original (parental) somatic cells. Both liver-derived (iPS-L1 and 2, passage 7) and stomach cell-derived (iPS-S1 and 2, passage 6) lines showed a highly unmethylated pattern whereas their parental cells showed a highly methylated pattern. Open circles and filled circles represent unmethylated and methylated CpG sites, respectively. Scale bar, 100 μm.
FIGURE 3.
Expression of pluripotency markers in rabbit iPS cells detected by staining for alkaline phosphatase (AP) activity and by immunostaining. The iPS cells were positive for alkaline phosphatase, SSEA1, SSEA4, OCT3/4, and NANOG, but not for SSEA3. Representative images were prepared from iPS-L1 cells at passage 12. Scale bars, 100 μm.
Telomerase Activity of Rabbit iPS Cells
The rabbit iPS cells we generated had apparently indefinite proliferation potential because they could be passaged without losing their proliferation ability for at least 7 months (50 passages, at the time of preparing this paper). Consistent with this feature, all rabbit iPS cells examined showed high telomerase activity (Fig. 2C and supplemental Table S2), a hallmark of ES cells in several species (28, 35–37).
Differentiation Ability of Rabbit iPS Cells in Vitro and in Vivo
We then tested the differentiation ability of rabbit iPS cells in vitro and in vivo to confirm their potential as pluripotent stem cells. All of the iPS lines we established formed embryoid bodies readily under a feeder-free differentiation culture condition. After replating of embryoid bodies onto gelatin-coated culture dishes or after directly inducing differentiation from iPS cells, at least four of the lines tested differentiated into ectodermal, mesodermal, and endodermal derivatives, as evidenced by positive staining for βIII-tubulin (ectoderm), glial fibrillary acidic protein (ectoderm), α-smooth muscle actin (mesoderm), and GATA4 (endoderm) (Fig. 4A and supplemental Table S2). The in vivo differentiation ability was assessed by transplantation under the kidney capsule of SCID mice. At 4–8 weeks after transplantation, all iPS cell lines generated successfully formed typical teratomas in recipient mice. They contained tissues derived from all three germ layers, including epidermis (ectoderm), neural tissues (ectoderm), bone (mesoderm), muscle fibers (mesoderm), exocrine glands (endoderm), and epithelium with goblet cells (endoderm) (Fig. 4B and supplemental Table S2).
FIGURE 4.
Differentiation of rabbit iPS cells in vitro (A) and in vivo (B). Aa, embryoid bodies formed from iPS cells under differentiation conditions at day 5. After replating on gelatin-coated dishes, they further differentiated into a variety of cell types from the three basic germ layers expressing specific markers, as detected by immunostaining. Ab, neural cells (ectoderm) expressing βIII-tubulin (green) and glial fibrillary acidic protein (red). Ac, smooth muscle cells (mesoderm) expressing α-smooth muscle myosin (green). Ad, endodermal cells expressing GATA4 (red). The cells were counterstained with DAPI. Representative images were prepared from iPS-L1 cells at passage 13. B, teratoma formation by rabbit iPS cells (iPS-L1 at passage 8). Various tissues of the three germ layer origins are identified: epidermis (a, ectoderm), bone (b, mesoderm), glands (c, endoderm), neural tissue (d, ectoderm), muscle fibers (e, mesoderm), and epithelium with goblet cells (f, endoderm). Scale bar, 100 μm.
DNA Microarray Analysis
To investigate whether the transcriptional properties of rabbit iPS cells generated from somatic cells might differ from those of rabbit ES cells, we analyzed their global gene expression profiles using a microarray system. Unsupervised hierarchical clustering analysis clearly distinguished a donor cell group (liver and stomach cells) and a stem cell group (Fig. 5A). The stem cell group could be divided roughly into three categories: early passage iPS cells (P6 and P7), late passage iPS cells (P22 and P23), and ES cells. This indicated that the rabbit iPS cells were more similar to each other according to their passage number rather than according to their original cell type. Interestingly, these clustering patterns suggested that the late passage iPS cells were more similar to ES cells than were early passage cells. This tendency was confirmed by principal component analysis, which was used to present the relative distances between samples graphically. The iPS cells and ES cells were linearly mapped on the three-dimensional transcript profiles, the late passage iPS cells being closer to ES cells (Fig. 5B). These findings indicate that the iPS cells continued to be reprogrammed even after a number of passages, while becoming more similar to ES cells in their gene expression profiles. We then identified differentially expressed genes in iPS cells at early or late passages compared with those of ES cells. The statistical analysis was performed for three category groups (early passage iPS cells, late passage iPS cells, and ES cells) according to the result from the hierarchical clustering. In all, 220 genes were identified as differentially expressed genes between early passage iPS cells and ES cells. Of these, 175 were non-differentially expressed genes in late passage iPS cells compared with ES cells (supplemental Fig. S3 and supplemental Table S3). Thus, late passage iPS cells were very similar to ES cells in their gene expression profiles, although there still remained a minor difference even after 22 or 23 passages.
FIGURE 5.
DNA microarray analysis demonstrating the global gene expression profiles of iPS cells at different passage numbers, of ES cells, and of parental somatic cells. A, unsupervised hierarchical clustering. Rabbit iPS cells were more similar to each other according to their passage number rather than by origin. B, three-dimensional principal component analysis mapping. Consistent with the result of hierarchical clustering in A, iPS cells at similar passage numbers are close to each other. This map clearly demonstrates that these rabbit iPS cells started to resemble ES cells as passage proceeded. Scale bar, 100 μm.
DISCUSSION
Since the first discovery that four transcriptional factors could reprogram somatic cells into iPS cells in mice, essentially similar strategies have been applied successfully to human, rat, primate, and porcine cells (3, 4, 12–15, 38). The present study for the first time has demonstrated generation of iPS cells in the rabbit, with a long history as a non-rodent laboratory animal species. The rabbit iPS cells established in this study fulfilled all of the requirements for the acquisition of the fully reprogrammed state and showed high similarity to their ES cell counterparts.
Assessment of the reprogramming state of the somatic cell genome is essential for experiments in the generation of iPS cells. The fully reprogrammed state is of practical importance because iPS cells are expected to differentiate properly into certain cell types provided their genomes are set initially at the epigenetic “ground state.” Although the most convincing evidence for this state in ES or iPS cells is the generation of germ line-competent chimeric offspring, this experimental system has been validated only in mice and rats (39–41). Instead, in other species, including rabbits, the generation of teratomas with confirmation of the formation of all three germ layers (endoderm, mesoderm, and ectoderm) from ES or iPS cells has to be used as a validation system (supplemental Table S4). Recent molecular analyses suggest that the latter type of pluripotent cells correspond to mouse epiblast stem cells, which are established from epiblast cells of postimplantation embryos (42, 43). These two types of the pluripotent state with distinct natures were termed “naïve” and “primed,” respectively (44) (supplemental Table S4). Therefore, although we could not expect the production of chimeric embryos from our rabbit iPS cells, they formed teratomas containing these three germ layers and thus can be considered to have been fully reprogrammed in this sense.
Furthermore, we could also generate several cell types of three germ layer origins in vitro following differentiation stimulus. Chan et al. have now reported several markers for “fully reprogrammed” human iPS cells, which can distinguish them from “partially reprogrammed” iPS-like cells (45). Although some of these markers are unavailable for rabbit iPS cells, other markers such as demethylation of the OCT3/4 promoter and silencing of viral transgenes were common to the features of our rabbit iPS cells and therefore imply that they exhibited full reprogramming status. Additionally, we have examined other ES cell-related markers including alkaline phosphatase activity, telomerase activity, and OCT3/4, SSEA1, SSEA4, and NANOG expression levels. The endogenous expressions of rabbit OCT3/4, SOX2, KLF4, NANOG, and c-MYC were also confirmed. Taken together, we conclude that the rabbit somatic cell genome can be reprogrammed fully to the pluripotent state by these human transcription factors.
It is generally assumed that there must be some technical keys to successful iPS cell generation for each animal species. For rabbits, the choice of somatic donor cells seemed to us to be crucial. Initially, we attempted to reprogram fetal and adult fibroblast cells because they are the most commonly used cell types in other species. It was also reported that human iPS cells derived from fetal fibroblast cells were more similar to ES cells in their global gene expression profiles than those from other cell types (6). However, contrary to our initial ideas, we obtained no success with rabbit fibroblasts. That was probably caused by an exceptionally high proliferation rate of rabbit fibroblasts in vitro: they immediately reached confluence and discontinued dedifferentiation, just like rabbit ES cells when cultured on a high density of feeder cells (28). We next examined liver and stomach cells, which have been proven to be reprogrammed efficiently in the mouse (46). During primary culture, they proliferated moderately, but only when they responded to the ectopic expression of the transgenes did they start to grow rapidly and form ES cell-like colonies. It is therefore probable that the initial proliferation rate of somatic cells is a defining factor for iPS cell establishment in rabbits. However, once colonies were formed by successful gene transduction, it was relatively easy to maintain their undifferentiated status. Indeed, all the initial ES cell-like colonies continued to proliferate and gave rise to iPS cell lines. This is in sharp contrast to iPS generation in other species, which shows diminution of colonies with prolonged culture. For example, in human iPS cells colonies of partially reprogrammed cells appeared among fully reprogrammed iPS cell colonies and failed to differentiate into endodermal tissues in teratomas (45).
Rabbits have several advantages over other laboratory species as models for human cell-based regenerative medicine. First, they are evolutionarily closer to primates than are rodents (47) and have long been used in biomedical research as experimental models for human diseases (48, 49). Rabbit models are important because the etiologies of some human diseases are more similar to those in rabbits than those in mice (e.g. myocardial infarction). Second, rabbits are larger than mice, making surgery easier and enabling large samples to be obtained. They have a longer life span than mice or rats (7–8 years) and thus can be used for long term testing of cell-based therapies. Despite many promising strategies presented so far for treating degenerative diseases using ES cells or iPS cells, further studies will be needed to address the long term efficacy and safety of using these cells. As advised from a historical overview of gene therapy clinical trials, long term examinations using animal models should be conducted before bringing the new cell/tissue-based strategies to human patients (50). Rabbits are cheaper, more accessible, and more easily maintained than primates or pigs. Therefore, we expect that rabbits will be very well suited for the long term assessment of iPS-based therapies for future clinical applications in humans.
Besides their advantages in terms of physiology, longevity, and size, as described above, the rabbit has a further important merit as an animal model for human iPS research. Stable rabbit ES cell lines can be established easily, and several lines are now available from laboratories including ours (27–29). Importantly, they are very similar to human ES cells in their shape, biochemical characters, and molecular mechanisms for self-renewal and pluripotency (30). As has been suggested, epigenetic and phenotypic comparisons between iPS cells and ES cells should be done carefully for assessing the extent of reprogramming of iPS cells and for cell selection and applications in humans. We anticipate that the same side-by-side analyses might be performed more rapidly and more extensively when using rabbits as models. In this study, we have shown that rabbit iPS cells are indistinguishable from their embryonic counterparts in their biochemical characteristics, but they still had a minor difference in their global gene expression patterns even after complete silencing of the introduced genes. However, the numbers of genes differentially expressed between early passage iPS cells and ES cells decreased by about 80% after further passages (from 220 genes to 45 genes) (supplemental Fig. S3). We are still continuing passage of these rabbit iPS cell lines, and those with higher passage numbers will be examined appropriately, as reported for human and mouse iPS cells (5). These small transcriptional differences might diminish with time, allowing the rabbit iPS cells to become more ES cell-like. It would be interesting to compare iPS cells from different passage numbers and ES cells in their ability to serve as cell resources for regenerative medicine. Such assessment using rabbits would provide invaluable information for human medicine as it offers a substantial statistical basis. Furthermore, we can add rabbit ES cells derived from somatic cell nuclear transfer (SCNT) for comparison because it is relatively easy to obtain blastocysts by SCNT in rabbits (21, 27). In primates including humans, the generation of SCNT blastocysts is very difficult for unknown reasons, and only two SCNT-ES cell lines have been established (51). However, rabbit SCNT-ES cells are free of transgenes, and once reprogrammed to the totipotent state, they should be considered as cell resource candidates for future regenerative medicine.
In conclusion, stable rabbit iPS cells were generated from somatic cells by the introduction of four human transcription factors. The fully pluripotent state of the iPS cells established was confirmed by complete silencing of the introduced genes, formation of teratomas with the three basic germ layers, and expression of other pluripotency-related markers. Phenotypically, they were very similar to rabbit ES cells and to human iPS/ES cells. Rabbits are very common, easily accessible, and suitable for long term safety assessments. Therefore, we expect that rabbit iPS cells, together with their ES cell counterparts, will provide invaluable experimental models for assessing the efficacy and safety of new cell-based treatments for degenerative diseases in humans.
Supplementary Material
Acknowledgments
We thank Drs. Naomi Yoshida and Kazuhiro Sudo for valuable discussions of this study, Sumie Togayachi for assistance in lentivirus preparation, and Dr. Kuniya Abe and Rieko Ikeda for assistance in DNA microarray analysis.
This work was supported by PRESTO of the Japan Science and Technology Agency and a grant-in-aid for scientific research (B).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3, Tables S1–S4, and additional references.
- iPS
- induced pluripotent stem
- bFGF
- basic FGF
- iPSM
- iPS medium
- KSR
- knock-out serum replacement
- SCID
- severe combined immunodeficient
- SCNT
- somatic cell nuclear transfer.
REFERENCES
- 1.Martins-Taylor K., Xu R. H. (2010) J. Cell. Biochem. 109, 16–25 [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- 4.Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin II, Thomson J. A. (2007) Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- 5.Chin M. H., Mason M. J., Xie W., Volinia S., Singer M., Peterson C., Ambartsumyan G., Aimiuwu O., Richter L., Zhang J., Khvorostov I., Ott V., Grunstein M., Lavon N., Benvenisty N., Croce C. M., Clark A. T., Baxter T., Pyle A. D., Teitell M. A., Pelegrini M., Plath K., Lowry W. E. (2009) Cell. Stem Cell 5, 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghosh Z., Wilson K. D., Wu Y., Hu S., Quertermous T., Wu J. C. (2010) PLoS One 5, e8975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchetto M. C., Yeo G. W., Kainohana O., Marsala M., Gage F. H., Muotri A. R. (2009) PLoS One 4, e7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Familari M., Selwood L. (2006) Mol. Reprod. Dev. 73, 123–131 [DOI] [PubMed] [Google Scholar]
- 9.Kazuki Y., Hiratsuka M., Takiguchi M., Osaki M., Kajitani N., Hoshiya H., Hiramatsu K., Yoshino T., Kazuki K., Ishihara C., Takehara S., Higaki K., Nakagawa M., Takahashi K., Yamanaka S., Oshimura M. (2010) Mol. Ther. 18, 386–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koestenbauer S., Zech N. H., Juch H., Vanderzwalmen P., Schoonjans L., Dohr G. (2006) Am. J. Reprod. Immunol. 55, 169–180 [DOI] [PubMed] [Google Scholar]
- 11.Liu H., Zhu F., Yong J., Zhang P., Hou P., Li H., Jiang W., Cai J., Liu M., Cui K., Qu X., Xiang T., Lu D., Chi X., Gao G., Ji W., Ding M., Deng H. (2008) Cell. Stem Cell 3, 587–590 [DOI] [PubMed] [Google Scholar]
- 12.Esteban M. A., Xu J., Yang J., Peng M., Qin D., Li W., Jiang Z., Chen J., Deng K., Zhong M., Cai J., Lai L., Pei D. (2009) J. Biol. Chem. 284, 17634–17640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Z., Chen J., Ren J., Bao L., Liao J., Cui C., Rao L., Li H., Gu Y., Dai H., Zhu H., Teng X., Cheng L., Xiao L. (2009) J. Mol. Cell. Biol. 1, 46–54 [DOI] [PubMed] [Google Scholar]
- 14.Ezashi T., Telugu B. P., Alexenko A. P., Sachdev S., Sinha S., Roberts R. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 10993–10998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Y., Zhang Y., Mishra A., Tardif S. D., Hornsby P. J. (2010) Stem Cell Res. 4, 180–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biggers J. D. (1991) J. Reprod. Fertil. 93, 173–186 [DOI] [PubMed] [Google Scholar]
- 17.Chang M. C. (1959) Nature 184, 466–467 [DOI] [PubMed] [Google Scholar]
- 18.Kasai M., Hamaguchi Y., Zhu S. E., Miyake T., Sakurai T., Machida T. (1992) Biol. Reprod. 46, 1042–1046 [DOI] [PubMed] [Google Scholar]
- 19.Liu J. L., Kusakabe H., Chang C. C., Suzuki H., Schmidt D. W., Julian M., Pfeffer R., Bormann C. L., Tian X. C., Yanagimachi R., Yang X. (2004) Biol. Reprod. 70, 1776–1781 [DOI] [PubMed] [Google Scholar]
- 20.Chesné P., Adenot P. G., Viglietta C., Baratte M., Boulanger L., Renard J. P. (2002) Nat. Biotechnol. 20, 366–369 [DOI] [PubMed] [Google Scholar]
- 21.Inoue K., Ogonuki N., Yamamoto Y., Noguchi Y., Takeiri S., Nakata K., Miki H., Kurome M., Nagashima H., Ogura A. (2002) Cloning Stem Cells 4, 311–317 [DOI] [PubMed] [Google Scholar]
- 22.Yang F., Hao R., Kessler B., Brem G., Wolf E., Zakhartchenko V. (2007) Reproduction 133, 219–230 [DOI] [PubMed] [Google Scholar]
- 23.Ogonuki N., Inoue K., Miki H., Mochida K., Hatori M., Okada H., Takeiri S., Shimozawa N., Nagashima H., Sankai T., Ogura A. (2005) Mol. Reprod. Dev. 72, 411–417 [DOI] [PubMed] [Google Scholar]
- 24.Bosze Z., Hiripi L., Carnwath J. W., Niemann H. (2003) Transgenic Res. 12, 541–553 [DOI] [PubMed] [Google Scholar]
- 25.Fan J., Watanabe T. (2003) Pharmacol. Ther. 99, 261–282 [DOI] [PubMed] [Google Scholar]
- 26.Graves K. H., Moreadith R. W. (1993) Mol. Reprod. Dev. 36, 424–433 [DOI] [PubMed] [Google Scholar]
- 27.Fang Z. F., Gai H., Huang Y. Z., Li S. G., Chen X. J., Shi J. J., Wu L., Liu A., Xu P., Sheng H. Z. (2006) Exp. Cell Res. 312, 3669–3682 [DOI] [PubMed] [Google Scholar]
- 28.Honda A., Hirose M., Inoue K., Ogonuki N., Miki H., Shimozawa N., Hatori M., Shimizu N., Murata T., Hirose M., Katayama K., Wakisaka N., Miyoshi H., Yokoyama K. K., Sankai T., Ogura A. (2008) Reprod. Biomed. Online 17, 706–715 [DOI] [PubMed] [Google Scholar]
- 29.Intawicha P., Ou Y. W., Lo N. W., Zhang S. C., Chen Y. Z., Lin T. A., Su H. L., Guu H. F., Chen M. J., Lee K. H., Chiu Y. T., Ju J. C. (2009) Cloning Stem Cells 11, 27–38 [DOI] [PubMed] [Google Scholar]
- 30.Honda A., Hirose M., Ogura A. (2009) Exp. Cell Res. 315, 2033–2042 [DOI] [PubMed] [Google Scholar]
- 31.Ezashi T., Das P., Roberts R. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4783–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshida Y., Takahashi K., Okita K., Ichisaka T., Yamanaka S. (2009) Cell. Stem Cell 5, 237–241 [DOI] [PubMed] [Google Scholar]
- 33.Okazawa H., Okamoto K., Ishino F., Ishino-Kaneko T., Takeda S., Toyoda Y., Muramatsu M., Hamada H. (1991) EMBO J. 10, 2997–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hattori N., Nishino K., Ko Y. G., Hattori N., Ohgane J., Tanaka S., Shiota K. (2004) J. Biol. Chem. 279, 17063–17069 [DOI] [PubMed] [Google Scholar]
- 35.Armstrong L., Lako M., Lincoln J., Cairns P. M., Hole N. (2000) Mech. Dev. 97, 109–116 [DOI] [PubMed] [Google Scholar]
- 36.Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., Jones J. M. (1998) Science 282, 1145–1147 [DOI] [PubMed] [Google Scholar]
- 37.Sasaki E., Hanazawa K., Kurita R., Akatsuka A., Yoshizaki T., Ishii H., Tanioka Y., Ohnishi Y., Suemizu H., Sugawara A., Tamaoki N., Izawa K., Nakazaki Y., Hamada H., Suemori H., Asano S., Nakatsuji N., Okano H., Tani K. (2005) Stem Cells 23, 1304–1313 [DOI] [PubMed] [Google Scholar]
- 38.Li W., Wei W., Zhu S., Zhu J., Shi Y., Lin T., Hao E., Hayek A., Deng H., Ding S. (2009) Cell. Stem Cell 4, 16–19 [DOI] [PubMed] [Google Scholar]
- 39.Okita K., Ichisaka T., Yamanaka S. (2007) Nature 448, 313–317 [DOI] [PubMed] [Google Scholar]
- 40.Buehr M., Meek S., Blair K., Yang J., Ure J., Silva J., McLay R., Hall J., Ying Q. L., Smith A. (2008) Cell 135, 1287–1298 [DOI] [PubMed] [Google Scholar]
- 41.Li P., Tong C., Mehrian-Shai R., Jia L., Wu N., Yan Y., Maxson R. E., Schulze E. N., Song H., Hsieh C. L., Pera M. F., Ying Q. L. (2008) Cell 135, 1299–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brons I. G., Smithers L. E., Trotter M. W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S. M., Howlett S. K., Clarkson A., Ahrlund-Richter L., Pedersen R. A., Vallier L. (2007) Nature 448, 191–195 [DOI] [PubMed] [Google Scholar]
- 43.Tesar P. J., Chenoweth J. G., Brook F. A., Davies T. J., Evans E. P., Mack D. L., Gardner R. L., McKay R. D. (2007) Nature 448, 196–199 [DOI] [PubMed] [Google Scholar]
- 44.Nichols J., Smith A. (2009) Cell. Stem Cell 4, 487–492 [DOI] [PubMed] [Google Scholar]
- 45.Chan E. M., Ratanasirintrawoot S., Park I. H., Manos P. D., Loh Y. H., Huo H., Miller J. D., Hartung O., Rho J., Ince T. A., Daley G. Q., Schlaeger T. M. (2009) Nat. Biotechnol. 27, 1033–1037 [DOI] [PubMed] [Google Scholar]
- 46.Aoi T., Yae K., Nakagawa M., Ichisaka T., Okita K., Takahashi K., Chiba T., Yamanaka S. (2008) Science 321, 699–702 [DOI] [PubMed] [Google Scholar]
- 47.Graur D., Duret L., Gouy M. (1996) Nature 379, 333–335 [DOI] [PubMed] [Google Scholar]
- 48.Shiomi M., Ito T., Yamada S., Kawashima S., Fan J. (2004) J. Atheroscler. Thromb. 11, 184–189 [DOI] [PubMed] [Google Scholar]
- 49.Weekers F., Van Herck E., Coopmans W., Michalaki M., Bowers C. Y., Veldhuis J. D., Van den Berghe G. (2002) Endocrinology 143, 764–774 [DOI] [PubMed] [Google Scholar]
- 50.Wilson J. M. (2009) Science 324, 727–728 [DOI] [PubMed] [Google Scholar]
- 51.Sparman M., Dighe V., Sritanaudomchai H., Ma H., Ramsey C., Pedersen D., Clepper L., Nighot P., Wolf D., Hennebold J., Mitalipov S. (2009) Stem Cells 27, 1255–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.