Abstract
Chromatin undergoes a variety of changes in response to UV-induced DNA damage, including histone acetylation. In human and Drosophila cells, this response is affected by mutations in the tumor suppressor p53. In this work, we report that there is a global decrease in trimethylated Lys-9 in histone H3 (H3K9me3) in salivary gland cells in wild type flies in response to UV irradiation. In contrast, flies with mutations in the Dmp53 gene have reduced basal levels of H3K9me3, which are then increased after UV irradiation. The reduction of H3K9me3 in response to DNA damage occurs preferentially in heterochromatin. Our experiments demonstrate that UV irradiation enhances the levels of Lys-9 demethylase (dKDM4B) transcript and protein in wild type flies, but not in Dmp53 mutant flies. Dmp53 binds to a DNA element in the dKdm4B gene as a response to UV irradiation. Furthermore, heterozygous mutants for the dKdm4B gene are more sensitive to UV irradiation; they are deficient in the removal of cyclobutane-pyrimidine dimers, and the decrease of H3K9me3 levels following DNA damage is not observed in dKdm4B mutant flies. We propose that in response to UV irradiation, Dmp53 enhances the expression of the dKDM4B histone demethylase, which demethylates H3K9me3 preferentially in heterochromatin regions. This mechanism appears to be essential for the proper function of the nucleotide excision repair system.
Keywords: Chromatin Immunoprecipitation (ChIP), Chromatin Histone Modification, DNA Damage, DNA Repair, Drosophila Genetics, Histone Methylation, p53
Introduction
In eukaryotic cells, the dynamics of chromatin structure play a central role in all processes involving nuclear DNA, including transcription, replication, recombination, and repair. It has been shown that ATP-dependent complexes that alter the structure and array of nucleosomes, together with complexes containing enzymes that modify different histone residues, are the primary modulators of chromatin structure.
The most extensively studied histone modifications include acetylation, methylation, phosphorylation, ubiquitination, sumoylation, and ADP-ribosylation (1, 2). It is possible to correlate the status of the chromatin with the residue that is modified and the type of modification, in a manner that is dependent on the specific histone involved. Some histone modifications are prevalent in heterochromatic regions, and others are preferentially linked to euchromatin. For instance, in the case of DNA transcription and replication, it is generally accepted that relaxed chromatin facilitates the incorporation of factors that recognize elements in the DNA, allowing multisubunit complexes to assemble and achieve these functions. A similar situation may occur during DNA repair, because the DNA repair machinery has to repair DNA in the context of chromatin (3).
DNA damage by UV irradiation in eukaryotic cells is repaired via the nucleotide excision repair (NER)3 mechanism. In vitro reconstituted assays have demonstrated that removal of a lesion requires recognition by XPC-HR23b and subsequent unwinding of the DNA duplex by TFIIH (3, 4). The resulting single strands of DNA are then stabilized by xeroderma pigmentosum A and replication protein A, and the margins of the resulting DNA bubble are recognized by XPG and ERCC1-XPF, which generate 3′ and 5′ incisions on the damaged strand, respectively (3, 4). However, in vivo, the NER machinery must recognize DNA damage within the context of chromatin. In the case of transcribed regions, the chromatin is accessible to the recruitment of the NER machinery without the need for XPC-HR23b, because a stalled RNA polymerase II is also capable of recruiting the NER factors (3, 4). In silenced chromatin, the DNA-binding protein complex (DDB) acts in conjunction with XPC-HR23b to identify sites of UV damage (5, 6). DDB consists of a heterodimeric complex of the DDB1 and DDB2 proteins, which interact with the ubiquitin ligases Cul4A or Cul4B and RBX1, respectively (7). The DDB1-DDB2-CUL4-RBX1 complex recognizes UV damage in the chromatin and monoubiquitinates histone H2A at the sites of UV lesions (7). In addition to H2A ubiquitination, a complex composed of Cul4-DDB-ROC1 ubiquitinates histones H3 and H4 at the UV-damaged site in the chromatin (8). Together, these modifications of H3 and H4 weaken the interaction between the histones and the DNA, facilitating the action of the NER machinery (8).
Another histone modification that has been linked to the response to UV damage is histone acetylation. Acetylation of histones generates more relaxed and accessible chromatin, facilitating access by the NER machinery. For example, H3K9 acetylation is increased after UV irradiation. However, in human cancer cell lines carrying mutations that affect the p53 gene, the basal levels and the increase in H3K9 acetylation are affected, together with a general increase in histone acetylation (9, 10). A similar situation has been identified in Drosophila: in Dmp53 mutant flies, the increase in H3K9 acetylation after UV irradiation is dramatically affected (11). Thus, in both human and Drosophila cells, p53 has a direct influence on histone acetylation in response to UV irradiation.
Another important and highly studied covalent modification of histones is methylation. Lysine methylation has been implicated in gene regulation and, depending on the residue and state of methylation (mono-, di-, and trimethylation), has been recognized as a marker for gene repression or activation (12–14). Several histone demethylases have recently been discovered. For instance, LSD1 can demethylate mono- and dimethylated histones H3K4 and H3K9 in a FAD oxidation reaction (15, 16). In addition, a family of histone demethylases containing a JmjC domain has been identified. This family of proteins can remove the methyl group from all three stages of methylated lysines using Fe(II) and α-ketoglutarate as cofactors (17–19).
The effect and role of histone methylation in NER has only recently been investigated in yeast (20, 21). One study observed that H3K79me is important for correct action of the NER mechanism at the HML locus (22). However, the roles of other methylated histone residues in NER remain completely unknown.
In this work, we analyze the effect of UV-induced DNA damage on histone H3K9me3 in wild type and Dmp53 mutant Drosophila. Remarkably, Dmp53 is required to enhance the expression of the H3K9me3 demethylase dKDM4B, which in turn reduces the levels of this mark preferentially in heterochromatin. This process seems to be essential for the proper action of the NER mechanism in the fly.
EXPERIMENTAL PROCEDURES
Drosophila Stocks
The wild type strain was OreR. The Dmp53−ns strain was generated by direct targeting (23). The P element insertion mutant in EP-KDM4B, EP-Kdm4B03531/CyO, was obtained from Bloomington Stock Center at Indiana University (stock number 11336).
UV Irradiation
For UV irradiation, the third instar larvae wild type, Dmp53 null, and EP-Kdm4B03531 were collected and irradiated at UV light 200 J/m2 using a UVB Stratalinker 2400 (Stratagene). After irradiation, the larvae were incubated at 25 °C for 30 min, and the salivary glands were dissected in 0.7% NaCl for polytene chromosomes preparations. Only for smuch preparations were the salivary glands of WT larvae directly irradiated at UV light 100 J/m2, and they were incubated at 25 °C for 60 min before fixation.
Inmunofluorescence of Polytene Chromosomes and H3K9me3 Quantification
Salivary glands were fixed in solution 1 (PBS, 3.7% paraformaldehyde, and 1% Triton X-100) and then in solution 2 (3.7% paraformaldehyde, 50% acetic acid). The chromosomes were spread on poly-l-lysine-coated microscope slides. Anti-H3K9me3 antibody (Upstate) was used at 1:200, and anti-Cy3-conjugated secondary antibody (Rockland) was used at 1:300. The images were taken on a confocal laser scanning microscope (Zeiss LSM 510 META). The pixel intensity distribution was measured along the chromosomes, using the photon counting and length profile programs from the confocal microscope operating system, where the maximum number of pixels is 256, which represents the maximum accumulation of target bound to secondary antibody, and the minimum value is 0, which represents the absence of signal.
Statistical Analysis
The statistical analysis was performed as described in Ref. 11. For chromosome scanning we selected randomly approximately two-thirds of each genome independently of the chromosome arm, obtained in an average of 800 pixels/cell. At last 15 genomes were analyzed of each condition. The values were emptied out in a histogram representing the pixel arbitrary intensity (0–256 pixels) against the frequency. After we calculated the average frequency of each different condition, we performed the X2 test, using the Excel program. The X2 test was used to determine whether there was a significant difference between the frequency distributions. If the p value of each X2 test was <0.05, it was considered that a significant difference exists between the analyzed samples.
Smuch Nuclei Preparations and Nuclei Immunostainings
For salivary gland nuclei smuch preparations, we dissected three pairs of late third instar larval salivary glands in 1× PBS, and after UV irradiation the tissue was placed on a slide in a drop of 1× PBS, covered with a coverslip, and gently squashed (44). The nucleus were then fixed in 4% paraformaldehyde in 2% PBST (PBS with 0.2% of Triton X-100) at 4 °C for 20 min. For washing, the preparations were immersed in 0.4% PBST for 40 min. After they were blocked with 1% normal goat serum in 0.4% PBT for 2 h at 4 °C. Staining was performed as in immunofluorescence of polytene chromosomes, where anti-DmKDM4B was used at 1:50, and anti-CPD (Kamiya labs) was used at 1:25.
Chromatin Immunoprecipitations
The third instar larvae salivary glands were dissected in ice-cold Roberts buffer (87 mm NaCl, 3.2 mm KCl, 1.3 mm CaCl2, 1 mm MgCl2, 10 mm HEPES, pH 7.6), and the salivary glands (∼120 pairs) were fixed for 15 min at room temperature in 1 ml of cross-linking solution (1% formaldehyde/Roberts buffer) with rotation. The reaction was stopped by washing for 5 min in 1 ml of glycine buffer (125 mm glycine/Roberts buffer) after the salivary glands were washed for 5 min in 1 ml of Tris-buffered saline (150 mm NaCl, 20 mm Tris-HCl, pH 7.6) with rotation. The glands were then resuspended in 600 μl of sonication buffer (8 mm KCl, 0.5% Igepal CA-630, 1% SDS, 10 mm EDTA, pH 8) and sonicated 11 min (pulsed 11 times for 30 s, paused 30 s) in an Ultrasonic Processor. The chromatin was precleared in 1 ml of immunoprecipitation buffer (0.01% SDS, 1% Triton X-100, 1 mm EDTA, 15 mm Tris-HCl, 167 mm NaCl) with 40 μl of protein A/G beads (Invitrogen) for 3 h at room temperature. 10% of the chromatin was taken for input reaction. The anti-H3K9me3 (Upstate), anti-H3.1 (Upstate), and anti-Dmp53 (Santa Cruz) antibodies were incubated overnight with rotation at 4 °C. Antibody-chromatin complexes were recovered by incubation with 50 μl of protein A/G beads overnight at 4 °C. The washes were performed four times for 10 min at room temperature with 1 ml of radioimmune precipitation assay buffer, 1 ml of high salt buffer, 1 ml of LiCl buffer, and 1 ml of TE buffer. Then the samples were incubated with RNase A (Sigma) at 2 h, incubated with proteinase K (Roche Applied Science) at 2 h, and incubated at 65 °C overnight to reversal of cross-linking. Finally the DNA was extracted with phenolchloroform and diluted in 40 μl of H2O. To calculate the amount of target sequence (rover, AL1, NippedA, Plc21, Sgs8, Hsp70Aa, rp49, and repaer) in immunoprecipitated chromatin, we performed real time qPCR.
RNA Extraction
Total RNA was isolated from third instar larvae using a TRIzol (Invitrogen), and 10 μg of total RNA was converted to cDNA using reverse transcriptase enzyme (Invitrogen). RNA quantification of the levels of KDM4B transcripts was performed by real time qPCR.
Real Time qPCR Analysis
The primers pairs were designed to amplify 150–200-bp fragments. The qPCR analysis was performed with FastStart DNA MasterPLUS SYBR Green I (Roche Applied Science) in the Light Cycler 1.5 instrument (Roche Applied Science). For the ChIP, the PCR was performed in triplicate, and serial dilutions of pure, input DNA were measured together with the immunoprecipitated DNA samples. This allowed calculation of the amount of target sequence in immunoprecipitated chromatin relative to the amount of target sequence in input chromatin called the percentage of input according to Champion-ChIPTM qPCR primers (quantitative real time PCR analysis of chromatin; SA Biosciences). We used three different ChIP experiments. For the analysis of the RNA levels, we used a constitutive gene as a calibrator (rp49), the PCR for each gene was made in triplicate, and serial dilutions of cDNA of wild type fly were measured together with the cDNA of treated flies. The sequences for the primers used in qPCR were: rover forward, 5′-CAACCAAGACCAACCTACCC-3′; rover reverse, 5′-GCTCATTTTAGTCTGTCCGC-3′; AL1 forward, 5′-CCCACACATTGTCCAGTGCA-3′; AL1 reverse, 5′-CGTAAGATAATCCAGAGCGG-3′; NippedA forward, 5′-CAGCGGTCAAGTTGTTGAAC-3′; NippedA reverse, 5′-GACGGATATGCCGGACTTTG-3′; Plc21 forward, 5′-GTTCATCAGTGGGTCAGTAG-3′; Plc21 reverse, 5′-CAATATCCCCGGACAAAATC-3′; Sgs8 forward, 5′-CTTTACCAGATGGTAACCGT-3′; Sgs8 reverse 5′-GAACGATGACACAGAAAATG-3′; Hsp70Aa promoter forward 5′-CTGCAACTACTGAAATCAAC-3′; Hsp70Aa promoter reverse 5′-GGTCGTTGGCGATAATCTCC-3′; Hsp70Aa 350 bp foward 5′-CCAAGATCGGGGTGGAGTAT-3′; Hsp70Aa 350 bp reverse 5′-TGGGAGTCGTTGAAGTAGGC-3′; rp49 forward 5′-TCAAGATGACCATCCGCCCA-3′; and rp49 reverse 5′-GTTCTCTTGAGAACGCAGGC-3′. For RT-PCR, the sequences were: dKdm4B forward, 5′-CCACTCAGACCGCTCAGACA-3′; and dKdm4B reverse, 5′-CTGCAACATCTGATGGCGTC-3′. For Dmp53 ChIP, the sequences were: Kdm4B forward, 5′-TGTCTGCGAGTGTGTTGTTG-3′; dKm4B reverse, 5′-GCGTCTGATAAGCAGCCATC-3′; reaper forward, 5′-CCTTCTTTGGCACCGTCTAC-3′; and reaper reverse, 5′-CTATGGAAAAAGGGCGAAAA-3′.
UV Irradiation Sensitivity Assays
Third instar wild type and EP-kdm4b03531/+ larvae were irradiated at different UV light dosages (J/m2), using a UVB Stratalinker 2400 (Stratagene). The larvae were then allowed to develop into adults, and the emerged population was counted.
Immunoassay for CPD Repair
The third instar larvae were irradiated at 200 J/m2, and then DNA was isolated at different times after irradiation (5 min, 30 min, 4 h, 6 h, 12 h, and 24 h). One microgram of genomic DNA was denatured by heat and dotted onto a nylon membrane using a vacuum blotter (Bio-Dot microfiltration apparatus; Bio-Rad). To evaluate the CPD, a normal immunodetection blot was performed using an anti-CPD antibody (Kamiya Biomedical) diluted 1:1000.
Western Blots
Salivary glands from the third instar larvae were collected in lysis buffer (250 mm sucrose, 50 mm Tris, pH 7.5, 25 mm KCl, 5 mm MgCl2, 5 mm EDTA, plus protease inhibitors) and loaded in 10% SDS-PAGE gels. After the protein resolved, the gels were transferred to nitrocellulose membranes (Bio-Rad). The membranes were blocked with 10% nonfat milk for 2 h and then incubated overnight with the dCPDKDM4B antibody at 1:1000 dilution and β-tubulin antibody at 1:2000 dilution. After washing, the membranes were incubated with the appropriate HRP-conjugated secondary antibodies (Zymed Laboratories Inc.) for 1 h. Finally the membranes were washed, and the signals were detected with a chemiluminescence Western blot detection kit (Thermo Scientific). The Western blot quantification was performed using special software.4
RESULTS
Global H3K9me3 Levels Are Reduced after UV Irradiation, but This Response Is Affected in Dmp53 Mutant Flies
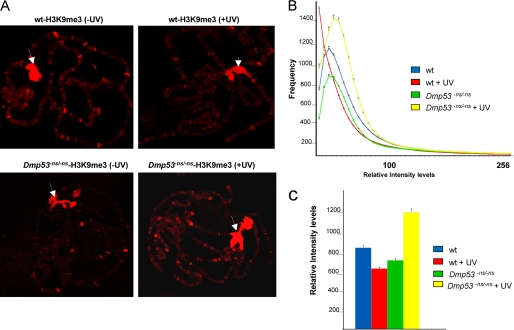
We have recently shown that global H3K9Ac levels are increased after UV irradiation in the Drosophila salivary gland polytene chromosomes and that this response is diminished in Dmp53-deficient organisms (11). Therefore, we decided to investigate the effect of UV irradiation on global H3K9me3 levels using a similar approach (11). An important advantage of these experiments is the use of a whole living organism with only a mutation in the Dmp53 gene, a feature that is uncommon in cell lines derived from cancer patients expressing mutated p53 that have accumulated many mutations. Wild type and Dmp53−ns allele homozygous mutant (23) third instar larvae were irradiated with 200 J/m2 (24, 25). After an average of 60–90 min of recovery, polytene chromosomes were prepared from irradiated and nonirradiated larvae for analysis by confocal microscopy. The signal intensities of ∼20 genomes (60 linear chromosomes) were quantified using photon counting for each condition, as reported previously (11, 26). The chromocenter was not quantified; only the immunofluorescent regions on the chromosome arms were analyzed. In previous works, we demonstrated that the signal obtained by immunostaining of polytene chromosomes could be quantitatively measured (11, 26). This approach is more quantitative than Western blot analysis for two reasons: modified histones that do not form part of the chromatin are present in the nuclear lumen, and the high levels of H3K9me3 at the chromocenter do not interfere with quantification using this method. In addition, this assay allows direct analysis of many chromosomes to obtain hundreds of data points for each condition, thus guaranteeing statistically reproducible results (11, 26).
Examples of nonirradiated and irradiated chromosomes are shown in Fig. 1A. Notably, the irradiated samples show a reduction in the intensity and number of bands stained with the anti-H3K9me3 antibody. Fig. 1B shows the distribution of the relative fluorescent intensities for each condition. The signal distribution of the levels of H3K9me3 is different in the wild type chromosomes before and after UV irradiation (Fig. 1B). These wild type samples show a clear reduction in levels of H3K9me3 after UV-induced DNA damage, as indicated by the quantification of the signal (represented as a bar graph in Fig. 1C). In the case of the chromosomes from the Dmp53−ns mutant, we observed a reduction in the basal levels of H3K9me3, suggesting that chromatin structure and histone modifications are altered in the Dmp53−ns mutant chromosomes (Fig. 1). Intriguingly, after UV irradiation, there is an overall increase in total H3K9me3 levels in the Dmp53 mutant chromosomes, as compared with the basal levels (Fig. 1, B and C), suggesting that, in the absence of Dmp53, there is an increase in heterochromatization after DNA damage. These results, together with our previous work and reports of basal levels of H3K9Ac in human cells, indicate that chromatin in p53 mutants displays several differences from wild type chromatin at the level of histone modification (10, 11). Consequently, the response to UV irradiation is also affected at the level of covalent modifications of the nucleosome. On the other hand, in wild type chromosomes, there is a correlation between the previously reported increase in levels of H3K9Ac (11) and the global decrease in H3K9me3 levels after UV irradiation. A recent report that used human cells to study the DNA damage response on histone modifications using hydroxyurea and phelomycin did not find alterations in the H3K9me3 levels, although a clear reduction in H3K9me2 and H3K9me1 levels was observed in the hydroxyurea-treated cells (27). Taking this difference into account with our findings suggests that the type of DNA damage is important for the type of response, because hydroxyurea generates base oxidation, depurination, and stalled replication forks, phelomycin generates double-strand breaks, and as far as we know, NER is not involved in repairing this kind of damage in the DNA.
FIGURE 1.
Effect of UV irradiation on H3K9me3 global levels in WT and Dmp53 null organisms. A, example of H3K9me3 distribution in polytene chromosomes before and after UV irradiation. Note that in addition to the chromocenter (white arrows), where the signal is saturated, specific regions are stained with the antibody. In wild type organisms, the intensity of the chromocenter signal is practically not affected; however, in other chromosomal regions, a reduction of the signal is observed. In the case of Dmp53 mutant flies, there is an increase in the number of sites stained with the antibody after UV irradiation. B, H3K9me3 quantitative analysis in WT and Dmp53ns/ns mutant polytene chromosomes before and after DNA damage. Note that the distribution of the histogram in the WT flies moves to the left after UV irradiation (red line) when compared with the nonirradiated organisms (blue line; p value = 1.9 × 10−3). This indicates that a decrease in the frequency of pixels with higher intensity occurs after DNA damage. Also note that the distribution of the histogram that represents the H3K9me3 basal levels in the Dmp53 null polytene chromosomes also moves to the left, and its area is also reduced when compared with the WT chromosomes (p value = 3.5 × 10−3). On the contrary, after UV irradiation, the histogram that represents the HK9me3 levels in the Dmp53 mutant chromosomes moves to the right, indicating an increase of the H3K9me3 levels (p value = 3.1 × 10−4). C, column representation of the quantification of the relative intensity of the data presented in the histogram. The phenotypes and the experimental conditions are indicated in the figure.
The Reduction of H3K9me3 Levels after UV Irradiation Occurs Preferentially in Heterochromatin
The confocal microscopy quantification method shows a reduction in the global levels of H3K9me3 after UV irradiation in polytene chromosomes. However, the response to DNA damage could differ, depending on chromatin conformation. Therefore, we decided to investigate the levels of H3K9me3 in constitutive and facultative heterochromatin, as well as in euchromatic regions. To perform this analysis, we selected two known constitutive heterochromatic sequences described in third instar larvae polytene chromosomes: the retrotransposon rover and the AL1 element (supplemental Fig. S1). The promoter regions of the Nipped A and Plc21 genes, which are not expressed in salivary glands, were analyzed as facultative heterochromatin. The promoter region of the sgs8 gene, a region that is highly transcribed in salivary glands, and the Hsp70 promoter represented euchromatin. The Hsp70 promoter is partially inactive without heat shock, but it is known that the nucleosome density is low at this site, because RNA polymerase II is poised on the promoter (28) (supplemental Fig. S1). To measure the levels of H3K9me3 at these regions, we used chromatin immunoprecipitation and real time PCR in both wild type and Dmp53−ns mutant organisms before and after UV irradiation.
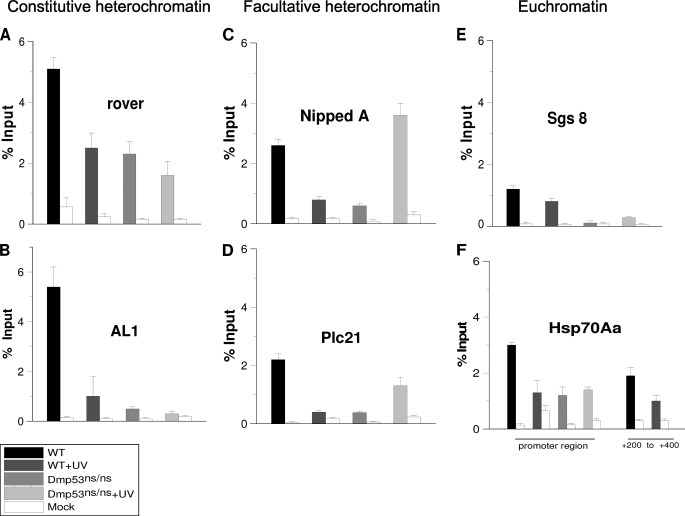
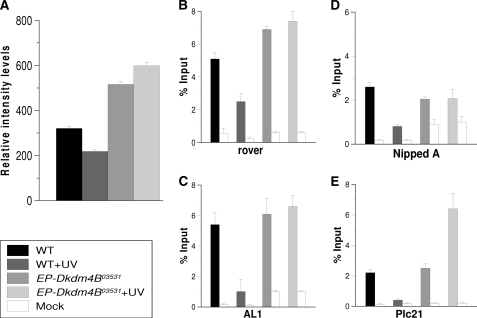
In the case of constitutive heterochromatin (rover and AL1), the basal levels before UV irradiation of H3K9me3 are high in relation to the input (Fig. 2, A and B). However, these levels decrease following induction of DNA damage by UV irradiation (Fig. 2, A and B). These changes occur without the removal of nucleosomes, because ChIP assays using an antibody against canonical H3 do not show any difference between nonirradiated and irradiated organisms (supplemental Fig. S2). In addition, an increase in the levels of H3K9ac is detected at these sites (supplemental Fig. S2). In the facultative heterochromatin (Nipped and Plc21 promoters), the basal levels of H3K9me3 were not as high as in rover and AL1 but were significantly high (Fig. 2, C and D). However, the levels of H3K9me3 also decrease in facultative heterochromatin following UV irradiation (Fig. 2, C and D). When we analyzed the promoters of the sgs8 and Hsp70 genes, we found that the levels of H3K9me3 were low under basal conditions (Fig. 2, E and F). This result agrees with previous reports showing that the density of nucleosomes is low in the Hsp70 promoter region (28). Interestingly, we did observe a small decrease in H3K9me3 levels at the Hsp70Aa promoter after UV irradiation. When we analyzed the levels of H3K9me3 near the site where the first nucleosome is poised in the Hsp70 promoter, +200 to +350 (28), we also detected very low levels of H3K9me3. These levels remained unchanged after UV irradiation in wild type organisms (Fig. 2F). Similarly, we observed a very low density (or a complete absence) of nucleosomes on the sgs8 promoter. In fact, our observations suggest that the entire gene is devoid of nucleosomes, because a similar configuration is present at the 3′end of the gene (Fig. 2E and supplemental Fig. S2). This is an expected result, because sgs8 is highly transcribed in third instar salivary glands. Therefore, no significant changes in the levels of H3K9me3 or histone H3 were observed in the euchromatic regions of the sgs8 and Hsp70 genes in third instar salivary glands after UV irradiation. This finding indicates that these chromosomal regions already show an open chromatin structure; following UV damage, this configuration is maintained, which probably allows for viable recognition by the NER machinery.
FIGURE 2.
H3K9me3 levels in different chromatin regions in WT and Dmp53 null flies before and after UV irradiation. Chromatin was prepared from salivary glands before and after irradiation of third instar larvae at 200 J/m2 and immunoprecipitated using an anti-H3K9me3 antibody. The regions amplified are indicated in supplemental Fig. S1. The graphs show results from independent immunoprecipitation assays (n = 3). ChIP signals quantified by the means of quantitative polymerase chain reaction and are presented as the mean percentages of input chromatin precipitated at each region; the error bars indicate ± S.D. Mock is a ChIP assay from the same salivary glands chromatin using an irrelevant IgG. A, H3K9me3 levels in the rover element. B, H3K9me3 levels in AL1 elements. Note that the basal levels of H3K9me3 are high in these constitutive heterochromatic regions and dramatically decrease after UV irradiation. In the case of the Dmp53ns/ns mutant, the H3K9me3 levels are significantly reduced when compared with the wild type in basal conditions (p value = 0.001) in both heterochromatin regions, and no significant change occurs after UV irradiation. C and D, H3K9me3 levels in two facultative heterochromatin regions in third instar salivary glands. Note that in both regions the H3K9me3 levels decrease after UV induced DNA damage (p value = 0.002). In the case of the Dmp53ns/ns mutant, an increase is observed in both regions after UV irradiation when compared with the nonirradiated Dmp53 mutant flies (NippedA, p value = 0.001; Plc21 = 0.02). E, H3K9me3 levels in the Sgs-8 gene promoter. Note that the levels are very low in all of the tested conditions. The nucleosome density of this region is shown in supplemental Fig. S2. F, H3K9me3 levels in the Hsp70Aa promoter region (around the transcription initiation point) and the +200 to +400 nucleotide positions. A significant reduction in the H3K9me3 levels is observed in the HS70a promoter only after UV irradiation in the wild type flies (p value = 0.01). All of the phenotypes and conditions are indicated in the figure.
In Dmp53 mutant flies, the basal levels of H3K9me3 are clearly reduced in both types of heterochromatin (Fig. 2, A–D). These results correlate with the global status of H3K9me3 in the homozygous Dmp53−ns flies (Fig. 1). Intriguingly, in the facultative heterochromatic regions analyzed, the levels of H3K9me3 rose above basal levels after UV irradiation (Fig. 2, C and D). This observation correlates with the global analysis of H3K9me3 levels after UV irradiation in the Dmp5−ns mutant (Fig. 1). However, levels of H3K9me3 remained low at the rover and AL1 sites (Fig. 2, A and B). Taken together, the results of the analysis using signal intensity from confocal microscopy and the ChIP experiments suggest that the increase in the levels of H3K9me3 after UV irradiation in Dmp53 mutant flies occurs preferentially in facultative heterochromatin but not in constitutive heterochromatin. On the other hand, the levels of H3K9me3 are reduced in the Dmp53−ns mutant flies under basal conditions. This result, together with previous reports on acetylated levels of histones affected by a lack of p53 in human and Drosophila cells, indicates broad effects on nucleosome modifications under basal conditions that may contribute to genome instability.
Dmp53 Is Required for the Increase in dKDM4B Levels after UV Irradiation and dKdm4B Mutant Flies Are More Sensitive to UV Irradiation
Recently, histone demethylases of the JmjC family have been identified in different eukaryotes. In particular, the Drosophila ortholog of the hKDM4 demethylase (dKDM4A) was linked to H3K36me3 and H3K36me2 demethylation (29, 30). In the same studies, another hKDM4 ortholog (dKDM4B) was identified as an H3K9me3 and H3K36me3 demethylase in vitro (29, 30). Because we observed a reduction in the levels of H3K9me3 after UV-induced DNA damage, we hypothesized that a demethylase might be activated or overexpressed to remove the methyl group from H3K9. On the other hand, the global levels of H3K9me3 are lower in Dmp53 mutant flies, and a response to UV irradiation was not observed in these flies, suggesting that the mechanisms induced to demethylate H3K9me3 are not fully operational if Dmp53 is not present. We analyzed the mRNA levels of dKDM4B by quantitative RT-PCR before and after UV irradiation at 200 J/m2 in both wild type and homozygous Dmp53ns third instar larvae. Interestingly, in wild type flies, the transcript levels of the dKDM4B demethylase are increased ∼3-fold after UV irradiation (Fig. 3A). The increase in dKDM4B mRNA levels after UV irradiation was not observed in the homozygous Dmp53 mutant organisms. In contrast, a reduction of dKDM4B mRNA was observed before and after UV irradiation in the mutant flies. Next, we determined the protein levels of dKDM4B in similar samples using a polyclonal antibody against dKDM4B generously donated by Prof. J. Workman. We observed an increase of ∼2.5-fold in the levels of dKDM4B protein over a period of time between 1:15 and 2:25 h after UV irradiation in wild type flies (Fig. 3B). In the case of Dmp53−ns/Dmp53−ns organisms, this response was not observed (Fig. 3B). Taken together, these results indicate that the transcription of the dKDM4B gene is stimulated in response to DNA damage, suggesting that the increase in dKDM4B demethylase may be linked to the reduction of H3K9me3 levels after UV irradiation. These results also suggest that the induction of dKDM4B after DNA damage requires functional Dmp53.
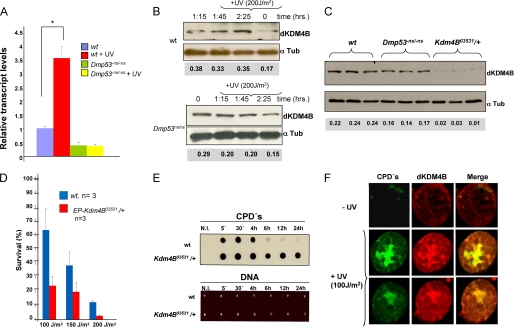
FIGURE 3.
Levels of the Kdm4B mRNA and protein in basal conditions after UV irradiation, UV sensitivity of a Kdm4B mutant fly, and dKDM4B nuclear localization after DNA damage. A, the mRNA levels were quantified by RT-qPCR from three independent experiments (*, p value = 0.002). The conditions and phenotypes are indicated in the figure. B, dKDM4B protein levels before and after UV irradiation in wild type and Dmp53−ns/Dmp53−ns third instar salivary gland cells. Note that there is an increase of ∼2.5 times of the dKDM4B protein in wild type cells, but not in the Dmp53 mutant cells after UV irradiation. C, basal dKDM4B protein levels in wild type, Dmp53−ns/Dmp53−ns, and EP-Kdm4B 03531/+ cells. D, survival of WT and Kdm4B heterozygous mutant (EP-Kdm4B 03531/+) flies after several UV doses. Calculation of the survival rate is reported under “Experimental Procedures.” The phenotypes and the irradiation doses are indicated in the figure. E, kinetic removal of CPDs after UV irradiation in wild type and dKdm4B mutant organisms. The third instar larvae were irradiated at 200 J/m2, and then DNA was purified at different times. A specific anti-CPD antibody was used to determine the presence of CPDs. The amount of loaded DNA in the membrane is shown. F, immunolocalization of CPDs (green) and dKDM4B (red) in nuclei before and after UV irradiation following the smuch protocol (44). The images were captured using a confocal microscope from isolated nuclei prepared from salivary glands either without UV irradiation or after UV irradiation after 2 h of recovery.
We next investigated the survival of a dKdm4B mutant fly after UV irradiation. We used the P-element insertion within the dKdm4B transcription unit (EP-Kdm4B03531; insertion located at nucleotide 359 of the dKdm4B 5′-UTR that disrupts the dKdm4B gene (supplemental Fig. S3; flybase). This allele is recessive, homozygous, lethal. Initially, we evaluated the levels of the dKDM4B demethylase in protein extracts from third instar larvae salivary glands of EP-Kdm4B03531/+ flies (Fig. 3C) and found a substantial reduction in the levels of dKDM4B protein in these flies cells, indicating that the dKDM4B gene is indeed affected in this mutant. We also observed that the basal levels of the dKDM4B protein are reduced in Dmp53−ns/Dmp53−ns organisms, compared with wild type flies, in agreement with the transcript measurements (Fig. 3, A and C). Based on these results, we investigated whether heterozygous flies were more sensitive to UV irradiation than wild type individuals. In general, heterozygous flies carrying a mutation in genes involved in NER are more sensitive to UV irradiation than wild type flies (25, 31). Thus, third instar larvae were irradiated at different doses of UV, and the viability of the wild type and EP-Kdm4B03531/+ organisms was evaluated by quantification of the survival organisms that develop to adulthood. Fig. 3D shows that the dKdm4B heterozygous mutant flies are significantly more sensitive to different doses of UV than wild type flies. In addition, dKdm4B mutant flies are not able to efficiently remove CPDs after UV irradiation (Fig. 3E). We previously observed that the kinetics of CPD removal in third instar larvae can be detected 4 h after irradiation and occurs faster than in mammalian cells (32). Interestingly, the H3K9me3 demethylated state is maintained in heterochromatin for several hours until the H3K9me3 levels rise again (supplemental Fig. S4). This situation may allow the NER machinery to function in these chromatin sites.
For this reason, we decided to analyze the distribution of the dKDM4B enzyme in the nuclei of third instar salivary glands cells after UV irradiation. Salivary glands were directly irradiated at 100 J/m2, and after 2 h of recovery, the tissue was fixed, and the nuclei were isolated by the smuch procedure (see “Experimental Procedures”) and inmunostained with an anti-CPD antibody and an anti-dKDM4B antibody to visualize the presence of CPDs and dKDM4B. Accordingly, we observed an increase in dKDM4B signal intensity in UV-irradiated nuclei (Fig. 3F). Interestingly, there is also a colocalization in the nuclear regions of CPDs (Fig. 3F), suggesting a recruitment of dKDM4B at damaged chromatin.
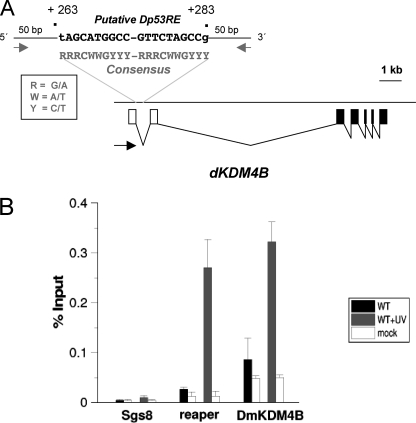
To have an insight of the role of Dmp53 in the transcription inductions of the dKdm4B gene, we searched for possible Dmp53 response elements (Dp53RE) in the region around the possible dKdm4B transcription initiation site based in the flybase gene annotation. The organization of the dKdm4B gene is complex because it is possible that it contains several promoter elements (supplemental Fig. S3). Nevertheless, we were able to identify a putative Dp53RE that matches with the p53 response element consensus sequence and has high identity with the Dp53RE present in the reaper gene (33). This sequence is just downstream from the transcription initiation site in the first intron after the 5′-UTR exon (Fig. 4A). We performed ChIP experiments to determine whether Dmp53 binds this element in the dKdm4B gene before and after UV irradiation of third instar larvae. As a positive control we used the Dp53RE present in the reaper gene (33), and as a negative DNA sequence we used the sgs8 promoter. Before UV irradiation there is little occupancy of Dmp53 in both the repear and the putative dKdm4B Dp53REs. However, after UV irradiation there is a significant increase of Dmp53 occupancy in both sequences but not in the negative control, suggesting that after UV-induced DNA damage Dmp53 can bind the Dp53RE present near the dKdmB promoter. Taken together, the results presented in this section suggest that dKDM4B is required for efficient DNA repair after UV irradiation in a Dmp53-dependent manner.
FIGURE 4.
Dmp53 binds a p53 response element in the dKdm4B gene after UV induced DNA damage. A, localization of a Dmp53 response element (Dmp53RE) in the first intron of the dKDM4B gene, near the transcription initiation site. The identified sequence of the Dmp53RE is shown in the figure. The numbers (+263 to +283) indicate the position with the putative transcription initiation site (+1) of the larger dKDM4B transcript reported in the flybase. A p53RE consensus sequence is also indicated. The black arrow indicates the transcription orientation. The Dmp53RE was identified by using the Math-inspector program from Genomatix. B, ChIP experiment to determine the Dmp53 occupancy in the dKdm4B gene-Dmp53RE before and after UV irradiation. Third instar larvae were irradiated at 200 J/m2, and after 30–60 min of recovery the salivary glands were fixed with formaldehyde and used for ChIP. The position of the primers used for dKdm4B Dmp53RE are indicated as gray arrows in A, and their sequences are listed under “Experimental Procedures.” As a positive control we used the reaper Dmp53RE (33), and as a negative control we used the sgs8 promoter used in this work.
H3K9me3 Demethylation after UV-induced DNA Damage Requires dKDM4B
We next asked whether there was a correlation between the decrease of H3K9me3 levels after UV-induced DNA damage and the dKDM4B enzyme. To investigate this question, we irradiated wild type and dKdm4B heterozygous mutant (EP-Kdm4B03531/+) third instar larvae with 200 J/m2 and evaluated the global levels of H3K9me3 in polytene chromosomes. As expected, the levels of H3K9me3 were reduced in wild type flies after UV irradiation (Fig. 5A). Interestingly, the levels of H3K9me3 levels were higher in the nonirradiated heterozygous dKDM4B mutant flies than in the wild type flies, and these levels increased after UV irradiation, despite the fact that one copy of the wild type allele is present in these heterozygous organisms (Fig. 5A). In addition, we performed ChIP experiments to measure the levels of H3K9me3 before and after UV irradiation. Initially, we analyzed the rover and AL1 loci, because a clear reduction of the levels of H3K9me3 was observed after UV-induced DNA damage. In agreement with the global analysis, the reduction of the levels of H3K9me3 at the rover and AL1 loci in response to UV irradiation was not diminished in the dKdm4B heterozygous flies when compared with wild type organisms (Fig. 5, B and C). We next performed similar ChIP assays to investigate the levels of H3K9me3 at the Nipped and Plc21 promoters. Similar to the constitutive heterochromatic regions, we observed that although one dKdm4B wild type allele is present in EP-Kdm4B03531/+ flies, no reduction in the levels of H3K9me3 occurs after UV irradiation (Fig. 5, D and E). Based on these results, it is possible that in EP-Kdm4B03531/+ organisms, the induction of dKDM4B is affected after UV treatment. On the other hand, preliminary observations also indicate that the levels of Su(var)3-9 transcript are also increased after UV irradiation in the fly,5 suggesting that a balance between methylation and demethylation of H3K9me3 may occur after severe UV irradiation. Related to this point, recent reports have shown that chromate exposure, which produces severe DNA damage in human cancer cells, increased global levels of di- and trimethylated H3K9, together with an increase in the levels of G9a histone methyl transferase (34, 35). Nevertheless, the data presented in this section support the hypothesis that the reduction of the levels of H3K9me3 after UV irradiation requires the dKDM4B demethylase.
FIGURE 5.
H3K9me3 global and specific levels in the dKdm4B mutant before and after UV irradiation. A, global H3K9me3 levels in WT and heterozygous dKdm4B mutant allele (+/Ep-Kdm4B 03531). H3K9me3 levels were quantified from at least 10 genomes (30 polytene chromosomes) for each phenotype and condition as described under “Experimental Procedures” and “Results.” The phenotypes and conditions are indicated in the figure. n = 10, p < 0.05. B–E, quantification of the H3K9me3 levels in WT and EP-Kdm4B 03531/+ flies before and after UV irradiation in the rover transposon, the AL1 sequence, the Nipped promoter region, and the Plc2 promoter region. Note that the H3K9me3 levels do not change in rover, AL1, Nipped, and Plc2 after UV irradiation in the Kdm4B heterozygous mutant. The graphs show results from independent immunoprecipitation reactions (n = 3). ChIP signals quantified by means of quantitative polymerase chain reaction are presented as mean percentages of input chromatin precipitated at each region; the error bars indicate ± S.D. Mock is a ChIP assay from the same salivary glands chromatin using an irrelevant IgG.
DISCUSSION
In this study, we found that there is a general reduction in the levels of H3K9me3 in Drosophila salivary gland cells chromosomes in response to UV irradiation. Demethylation of H3K9me3 occurs preferentially in heterochromatic regions, because the levels of H3K9me3 are low in euchromatin. This phenomenon requires the presence of a functional Dmp53, which further enhances the expression of the dKDM4B demethylase in response to UV irradiation. This finding suggests that Dmp53 can directly regulate the expression of enzymes that modify histones in response to genotoxic insults.
Dynamics of Histone Modifications in Response to UV-induced DNA Damage
Chromatin modifications are dynamic across the processes of regulation of gene expression and DNA repair. In the case of DNA damage by UV irradiation, previous studies have demonstrated that histone hyperacetylation is required for NER. In particular, levels of H3K9Ac are increased after UV irradiation in yeast, Drosophila, and mammalian cells (10, 11, 35). Additionally, this response to DNA damage is affected in p53 mutant cancer cells, as well as in Dmp53 null mutants in Drosophila (10, 11), indicating a direct role for this tumor suppressor gene in the chromatin modifications involved in NER. In the case of heterochromatic regions enriched for H3K9me3, the demethylation of H3K9me3 may be required for the increase of H3K9Ac, a hypothesis supported by our results.
Intriguingly, the basal levels of H3K9me3 are reduced in Dmp53 mutants, as indicated by global chromosome confocal analysis and ChIP experiments. A similar situation occurs with basal levels of H3K9Ac in p53 mutant human cells and basal levels of H3K14Ac in Drosophila p53 mutants (10, 11). Together, this information suggests that the regulation of histone modification is altered in p53-deficient cells, even if the lack of p53 in Drosophila and mice permits the survival of the organism. However, homozygous Dmp53−ns organisms have shorter life spans and are more sensitive to UV irradiation than wild type flies. It is also interesting to note that after UV irradiation, global levels of H3K9me3 are increased in the Dmp53−ns flies, although our ChIP results suggest that this increase is only present at the facultative heterochromatic regions. The increase in the level of H3K9me3 may be due to a general response to compact the chromatin in gene-encoding regions when the UV-induced DNA damage response is deficient in the Dmp53 mutant flies. Altogether, these data indicate that the absence of p53 alters the mechanisms that maintain a specific histone modification status under normal conditions and after genotoxic stress. Interestingly, it was recently demonstrated that heterochromatic genome stability requires the maintenance of the levels of methylated H3K9 through the histone methyltransferase Su(var)3-9 (36). Thus, alterations in these modifications caused by the absence of p53 might directly affect chromosomal integrity under basal conditions, generating a condition of chromosomal instability.
It is important to note that after NER, new H3.1 histone is deposited in the regions of chromatin that suffered the DNA damage. This deposition occurs outside of the S phase and acts to restore new nucleosomes at the damaged chromatin (37). Therefore, it is possible that in heterochromatic regions, demethylation of H3K9me3 is the initial step required to allow access of the NER machinery and also for the transient removal of histone H3, as has been proposed in the access-repair-restore model (37–40).
Histone H3K9me3 Demethylation after UV Irradiation Requires dKDM4B and Dmp53
Histone methylation and demethylation are dynamic events that control gene expression during development. Only recently has the status of H3K79 methylation been linked to NER (22). Prior to this work, H3K9 demethylases had only been linked to transcriptional control, not to UV-induced DNA damage. Heterozygous dKdm4B mutant flies are more sensitive to UV irradiation, do not remove CPDs efficiently, display higher basal levels of H3K9me than wild type flies, and exhibit deficiencies in demethylation of H3K9me3 after UV irradiation. These results suggest that demethylation of H3K9 by dKDM4B plays an important role in NER. It is also possible that other histone modifications may be altered after UV irradiation. Thus, it will be relevant to obtain a complete landscape of the dynamics of the methylated residues of H3 and H4 after DNA damage to test whether the corresponding methyl-transferases and demethylases are also involved in NER. ChIP-seq experiments are required to answer these questions.
Genotoxic stress activates p53 (among other factors) to trigger the mechanisms of DNA repair in animal cells (41). In particular, p53 modulates the transcription of multiple factors involved in DNA repair, apoptosis, and cell cycle arrest (41). In this work, we found that UV irradiation results in an increase in the mRNA and protein levels of the H3K9me3 demethylase dKDM4B. However, this response is affected in homozygous mutant Dmp53 organisms, suggesting a role for Dmp53 in the enhancement of the expression of dKDM4B after UV irradiation. In addition, a Dmp53 response element is present near the dKdm4B promoter, and Dmp53 binds this sequence after UV irradiation, suggesting a direct role for Dmp53 in dKdm4B gene regulation as a response to genotoxic stress. Indeed, a recent transcriptional analysis of p53-depleted human cells revealed that the expression of the JMJD2B/hKDM4B gene is reduced after drug-induced DNA damage, as compared with nondepleted p53 cells (43). This suggests that JMJD2B/hKDM4B is a putative target of p53, because other genes are involved in NER, such as DDB1 and XPC (42). In addition, it has been reported that overexpression of p53 generates a reduction in levels of H3K9me2 in cultured human cells (41). Our results are compatible with these reports, because dKDM4B may also demethylate H3K9me2, suggesting that demethylation of H3K9me3 following UV irradiation requires the activation of the dKDM4B gene through Dmp53. An important point in this study is that our observations were performed in a specific cell tissue (third instar salivary gland), and it will be interesting to investigate whether in other tissues and developmental stages the same response to UV irradiation is also present.
Based on our results in the fly, it will be important to determine whether human cells also show a JMJD2B/hKDM4B-dependent decrease in the level of H3K9me3 after UV-induced DNA damage and to investigate whether p53 is involved in this response. The information obtained from these types of studies will facilitate better understanding of the role of the dynamics of histone methylation in genotoxic stress and cancer.
Supplementary Material
Acknowledgments
We thank Prof. Jerry Workman and Chia Hui Lin for the anti-dKDM4B antibody and for sharing unpublished information. We also thank Andrés Saralegui for help with the confocal analysis and Ignacio López and Rene Hernandez Vargas for technical support.
This work was supported by funds from PAPIIT/UNAM grant no. IN 20109-3, Consejo Nacional de Ciencia y Tecnologia Grant 48550, and funds from the Howard Hughes Medical Institute (to M. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
L. Miller, personal communication.
Z. Palomera-Sanchez, A. Bucio-Mendez, V. Valadez-Graham, E. Reynaud, and M. Zurita, unpublished results.
- NER
- nucleotide excision repair
- CPD
- cyclobutane-pyrimidine dimer
- dKDM4B
- Drosophila lysine demethylase 4B
- Dmp53
- Drosophila p53
- H3K9me3
- histone H3 lysine 9 trimethylated
- H3K9Ac
- histone H3 lysine 9 acetylated
- Hsp70
- heat shock protein 70
- Dp53RE
- Drosophila p53 response element
- DDB
- DNA-binding protein complex
- qPCR
- quantitative PCR.
REFERENCES
- 1.Suganuma T., Workman J. L. (2008) Cell 135, 604–607 [DOI] [PubMed] [Google Scholar]
- 2.Kim T., Buratowski S. (2009) Cell 137, 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lainé J. P., Egly J. M. (2006) Trends Genet. 22, 430–436 [DOI] [PubMed] [Google Scholar]
- 4.Feuerhahn S., Egly J. M. (2008) Trends Genet. 24, 467–474 [DOI] [PubMed] [Google Scholar]
- 5.Nishi R., Alekseev S., Dinant C., Hoogstraten D., Houtsmuller A. B., Hoeijmakers J. H., Vermeulen W., Hanaoka F., Sugasawa K. (2009) DNA Repair 8, 767–776 [DOI] [PubMed] [Google Scholar]
- 6.Sugasawa K. (2009) DNA Repair 8, 969–972 [DOI] [PubMed] [Google Scholar]
- 7.Guerrero-Santoro J., Kapetanaki M. G., Hsieh C. L., Gorbachinsky I., Levine A. S., Rapić-Otrin V. (2008) Cancer Res. 68, 5014–5022 [DOI] [PubMed] [Google Scholar]
- 8.Wang H., Zhai L., Xu J., Joo H. Y., Jackson S., Erdjument-Bromage H., Tempst P., Xiong Y., Zhang Y. (2006) Mol. Cell 22, 383–394 [DOI] [PubMed] [Google Scholar]
- 9.Rubbi C. P., Milner J. (2003) EMBO J. 22, 975–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allison S. J., Milner J. (2003) Cancer Res. 63, 6674–6679 [PubMed] [Google Scholar]
- 11.Rebollar E., Valadez-Graham V., Vázquez M., Reynaud E., Zurita M. (2006) FEBS Lett. 580, 642–648 [DOI] [PubMed] [Google Scholar]
- 12.Sims R. J., 3rd, Reinberg D. (2008) Nat. Rev. Mol. Cell Biol. 9, 815–820 [DOI] [PubMed] [Google Scholar]
- 13.Probst A. V., Dunleavy E., Almouzni G. (2009) Nat. Rev. Mol. Cell Biol. 10, 192–206 [DOI] [PubMed] [Google Scholar]
- 14.Shukla A., Chaurasia P., Bhaumik S. R. (2009) Cell. Mol. Life Sci. 66, 1419–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li F., Huarte M., Zaratiegui M., Vaughn M. W., Shi Y., Martienssen R., Cande W. Z. (2008) Cell 135, 272–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicholson T. B., Chen T. (2009) Epigenetics 4, 129–132 [DOI] [PubMed] [Google Scholar]
- 17.Klose R. J., Kallin E. M., Zhang Y. (2006) Nat. Rev. Genet. 7, 715–727 [DOI] [PubMed] [Google Scholar]
- 18.Klose R. J., Zhang Y. (2007) Nat. Rev. Mol. Cell Biol. 8, 307–318 [DOI] [PubMed] [Google Scholar]
- 19.Horton J. R., Upadhyay A. K., Qi H. H., Zhang X., Shi Y., Cheng X. (2010) Nat. Struct. Mol. Biol. 17, 38–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bostelman L. J., Keller A. M., Albrecht A. M., Arat A., Thompson J. S. (2007) DNA Repair 6, 383–395 [DOI] [PubMed] [Google Scholar]
- 21.Nag R., Smerdon M. J. (2009) Mutat. Res. 682, 13–20 [DOI] [PubMed] [Google Scholar]
- 22.Chaudhuri S., Wyrick J. J., Smerdon M. J. (2009) Nucleic Acids Res. 37, 1690–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sogame N., Kim M., Abrams J. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4696–4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castro J., Merino C., Zurita M. (2002) DNA Repair 1, 359–368 [DOI] [PubMed] [Google Scholar]
- 25.Merino C., Reynaud E., Vázquez M., Zurita M. (2002) Mol. Biol. Cell 13, 3246–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reynaud E., Lomelí H., Vázquez M., Zurita M. (1999) Mol. Biol. Cell 10, 1191–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tjeertes J. V., Miller K. M., Jackson S. P. (2009) EMBO J. 28, 1878–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petesch S. J., Lis J. T. (2008) Cell 134, 74–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin C. H., Li B., Swanson S., Zhang Y., Florens L., Washburn M. P., Abmayr S. M., Workman J. L. (2008) Mol. Cell 32, 696–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lloret-Llinares M., Carré C., Vaquero A., de Olano N., Azorín F. (2008) Nucleic Acids Res. 36, 2852–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fregoso M., Lainé J. P., Aguilar-Fuentes J., Mocquet V., Reynaud E., Coin F., Egly J. M., Zurita M. (2007) Mol. Cell. Biol. 27, 3640–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aguilar-Fuentes J., Fregoso M., Herrera M., Reynaud E., Braun C., Egly J. M., Zurita M. (2008) PLoS. Genet. 4, e1000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brodsky M. H., Nordstrom W., Tsang G., Kwan E., Rubin G. M., Abrams J. M. (2000) Cell 101, 103–113 [DOI] [PubMed] [Google Scholar]
- 34.Sun H., Zhou X., Chen H., Li Q., Costa M. (2009) Toxicol. Appl. Pharmacol. 237, 258–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teng Y., Yu Y., Waters R. J. (2002) J. Mol. Biol. 316, 489–499 [DOI] [PubMed] [Google Scholar]
- 36.Peng J. C., Karpen G. H. (2009) PLoS Genet. 5, e1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polo S. E., Roche D., Almouzni G. (2006) Cell 127, 481–493 [DOI] [PubMed] [Google Scholar]
- 38.Smerdon M. J. (1983) Biochemistry 22, 3516–3525 [DOI] [PubMed] [Google Scholar]
- 39.Green C. M., Almouzni G. (2003) EMBO J. 22, 5163–5174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kruse J. P., Gu W. (2009) Cell 137, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warnock L. J., Adamson R., Lynch C. J., Milner J. (2008) Oncogene 27, 1639–1644 [DOI] [PubMed] [Google Scholar]
- 42.Adimoolam S., Ford J. M. (2003) DNA Repair 2, 947–954 [DOI] [PubMed] [Google Scholar]
- 43.Adamsen B. L., Kravik K. L., Clausen O. P., De Angelis P. M. (2007) Int. J. Oncol. 31, 1491–1500 [PubMed] [Google Scholar]
- 44.Wang Y., Zhang W., Jin Y., Johansen J., Johansen K. M. (2001) Cell 105, 433–443 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.