Abstract
Mammalian glutamate dehydrogenase (GDH) is a housekeeping enzyme central to the metabolism of glutamate. Its activity is potently inhibited by GTP (IC50 = 0.1–0.3 μm) and thought to be controlled by the need of the cell in ATP. Estrogens are also known to inhibit mammalian GDH, but at relatively high concentrations. Because, in addition to this housekeeping human (h) GDH1, humans have acquired via a duplication event an hGDH2 isoform expressed in human cortical astrocytes, we tested here the interaction of estrogens with the two human isoenzymes. The results showed that, under base-line conditions, diethylstilbestrol potently inhibited hGDH2 (IC50 = 0.08 ± 0.01 μm) and with ∼18-fold lower affinity hGDH1 (IC50 = 1.67 ± 0.06 μm; p < 0.001). Similarly, 17β-estradiol showed a ∼18-fold higher affinity for hGDH2 (IC50 = 1.53 ± 0.24 μm) than for hGDH1 (IC50 = 26.94 ± 1.07 μm; p < 0.001). Also, estriol and progesterone were more potent inhibitors of hGDH2 than hGDH1. Structure/function analyses revealed that the evolutionary R443S substitution, which confers low basal activity, was largely responsible for sensitivity of hGDH2 to estrogens. Inhibition of both human GDHs by estrogens was inversely related to their state of activation induced by ADP, with the slope of this correlation being steeper for hGDH2 than for hGDH1. Also, the study of hGDH1 and hGDH2 mutants displaying different states of activation revealed that the affinity of estrogen for these enzymes correlated inversely (R = 0.99; p = 0.0001) with basal catalytic activity. Because astrocytes are known to synthesize estrogens, these hormones, by interacting potently with hGDH2 in its closed state, may contribute to regulation of glutamate metabolism in brain.
Keywords: Enzyme Inhibitors, Enzyme Kinetics, Enzyme Mechanisms, Enzyme Mutation, Estrogen, Glutamate Dehydrogenase
Introduction
Female hormones are known to exert multiple physiological actions by binding to the estrogen receptors present in many cells (1). This binding triggers downstream signaling cascades, initiating gene transcriptions that regulate cell growth, migration, and other functions (1, 2). However, in addition to affecting nuclear DNA processes, estrogens can also act via nongenomic mechanisms (3–5), including direct actions on metabolic enzymes (6, 7). Of these, mammalian glutamate dehydrogenase (GDH)2 (EC 1.4.1.3) was originally shown to be modified by estrogens (8, 9). The enzyme catalyzes the reversible interconversion of glutamate to α-ketoglutarate and ammonia using NAD(H) or NADP(H) as cofactors, thus linking amino acid and carbohydrate metabolism. Because inhibition of mammalian GDH was obtained at estrogen concentrations (e.g. IC50 = ∼30.0 μm for 17β-estradiol) higher than those present in mammalian tissues (1, 10), the physiological relevance of these effects remains uncertain.
Whereas previous investigations employed bovine liver GDH that is encoded by the single functional GLUD gene present in all mammals (11), humans and the great apes possess two GDH-specific genes of distinct molecular biologic origin: an intron-containing GLUD1 gene (located on the 10th chromosome) that encodes the housekeeping hGDH1 and an X-linked intronless GLUD2 gene that encodes the hGDH2 isoform expressed in neural and testicular tissues (12). There is evolutionary evidence that the GLUD1 gene was retro-posed <23,000,000 years ago to the X chromosome, where it gave rise to the GLUD2 gene via random mutations and natural selection (13). As a result, the hGDH1 and hGDH2 are highly homologous, sharing in their mature form all but 15 of their 505 amino acid residues.
Despite their homology, hGDH1 and hGDH2 show distinct regulatory properties and relative resistance to thermal inactivation. Although hGDH1 is potently inhibited by GTP (IC50 = 0.1–0.3 μm), hGDH2 has dissociated its function from GTP control (IC50 = ∼100 μm) because of evolutionary replacement of Gly456 by Ala (14). Instead, regulation of hGDH2 is achieved via a novel molecular mechanism that mainly resulted from evolutionary substitution of Ser for Arg443 (15). In the absence of allosteric effectors, hGDH2 assumes a closed state associated with little catalytic activity while remaining remarkably responsive to activation by ADP and/or l-leucine (16).
Although the teleological need for duplication of human GDH has not been fully understood, there is evidence that the two human isoenzymes serve distinct cellular functions, with hGDH1 being involved in cellular energetics and hGDH2 being involved in metabolic recycling processes (17). Specifically, hGDH1, which is mainly activated by ADP and inhibited by GTP, is thought to be controlled by the need of the cell for ATP (18). This view has been supported by the discovery of gain-of-function mutations in the GLUD1 gene that attenuate GTP inhibition (19). In patients harboring these mutations, the overactive hGDH1 leads to excessive insulin release as a result of enhanced ATP synthesis in pancreatic β cells (20). On the other hand, hGDH2, which is specifically expressed in human cerebral cortical astrocytes and in testicular Sertoli cells, is thought to be involved in the production of metabolites essential to the supportive role of cells (21). Previous studies have shown that cultured astrocytes provide neurons with lactate that largely derives from the tricarboxylic acid cycle via conversion of glutamate to α-ketoglutarate (GDH reaction) (22). Dissociation of hGDH2 function from GTP control may permit the enzyme to metabolize glutamate even when an enhanced tricarboxylic acid cycle generates GTP levels sufficient to completely inactivate the housekeeping hGDH1 (23).
In light of these considerations, we sought here to investigate whether the interaction of steroid hormones with hGDH2 is distinct from that of hGDH1. For this, we obtained wild-type hGDH1 and hGDH2 in recombinant forms by expression of the GLUD1 and GLUD2 cDNA in Sf21 cells, respectively, and tested the effect of female hormones on these enzymes. Functional analyses, using either crude cell extracts or highly purified GDH reparations obtained from these extracts, unexpectedly revealed that estrogens interacted more potently with the neural and testicular tissue-specific isoenzyme (hGDH2) than with the widely expressed hGDH1. To elucidate the molecular mechanisms that render hGDH2 sensitive to estrogens, we performed site-directed mutagenesis of the GLUD1 and the GLUD2 cDNAs. The results obtained and their implications for the biological function of these compounds are presented below.
EXPERIMENTAL PROCEDURES
Materials
Sf21 cells and the baculovirus expression vectors were obtained from Invitrogen. The media for the Sf21 insect cells and fetal calf serum were from Invitrogen. Modified baculovirus (BaculoGold) was obtained from BD Pharmigen (San Diego, CA). NADPH, ADP, and GTP (lithium salt) were from Roche Applied Science. Phenylsepharose high performance was from Amersham Biosciences/GE Healthcare, and hydroxyapatite Bio-Gel HT was from Bio-Rad. The steroid hormones used in these studies were from Sigma-Aldrich.
Expression of Recombinant Proteins
Wild-type GLUD1 and GLUD2 cDNAs, cloned in pVL1393 vector, were expressed in Sf21 cells using the baculovirus expression system, as previously described (12). The Sf21 cells (of the insect Spodoptera frugiperda) were cotransfected with the plasmid DNA (pVL1393 vector containing the GLUD1 or GLUD2 cDNA insert) and modified baculovirus DNA and incubated at 27 °C for 5 days. The cultured cells were harvested 5 days post-infection and used for extracting the recombinant GDH proteins. For this, the cultured cells were homogenized in a buffer containing 0.05 m Tris-HCl, pH 7.4, 1% Triton X-100, 0.1 mm phenylmethylsulfonyl fluoride, and 0.5 m NaCl. The resulting whole homogenate was centrifuged at 7000 × g at 4 °C for 10 min, and the supernatant was used for studies employing crude extracts. Protein determination was done using the Lowry method.
Enzyme Purification
GDH was purified from Sf21 cell extracts using a previously described method (24), which involves ammonium sulfate fractionation and hydrophobic interaction and hydroxyapatite chromatography. Peak GDH activity fractions from the hydroxyapatite column were used for studying the purity of the GDH protein using SDS-PAGE according to the Laemmli procedure. GDH preparations purified to homogeneity were used for enzyme assays. Protein concentration of samples was determined densitometrically on stained SDS-PAGE gels, using bovine serum albumin as a standard.
Enzyme Assays and Regulation Studies
Enzyme activity was assayed spectrophotometrically (at 340 nm) in the direction of reductive amination of α-ketoglutarate. The reaction mixture of 1 ml contained 50 mm triethanolamine (TRA) buffer, pH 8.0, 100 mm ammonium acetate, 100 μm NADPH (except as indicated), and 2.6 mm EDTA. Enzyme reaction was initiated by adding α-ketoglutarate to 8 mm (except as indicated). Initial rates (enzyme velocity during the first 30 s after initiation of the reaction) were recorded. Wild-type and mutant hGDH1 proteins were studied in parallel. Kinetic analyses were performed to determine the Michaelis-Menten constant (Km) for α-ketoglutarate and NADPH. Several sets of experiments were performed for each purified enzyme. In each of these experiments, α-ketoglutarate varied from 0.4 to 8.0 mm, whereas ADP concentration was kept constant at 0 (base line), 25, or 250 μm. NADPH varied from 10 to 400 μm, whereas the ADP concentration was kept constant at 1.0 mm. Regulation of the human recombinant GDHs by estrogens was studied by adding each hormone into the reaction mixture while keeping the other substrates constant. The concentration of diethylstilbestrol (DES) ranged from 0 to 50 μm, whereas that of 17β-estradiol and estriol was varied from 0 to 500 μm. Progesterone was varied from 0 to 2000 μm.
Site-directed Mutagenesis of the GLUD1 cDNA
A GLUD1 DNA, cloned in pBSKII+ vector, was mutagenized at sites differing between hGDH1 and hGDH2 using the Gene Editor Mutagenesis system according to the manufacturer's protocol (Promega, Madison, WI). In each of these sites, the amino acid residue present in hGDH2 replaced the corresponding amino acid of hGDH1. To create the double mutant R443S/G456A, a GLUD1 cDNA with the G456A mutation underwent a second mutagenesis step in which the Arg at position 443 was replaced by Ser. To obtain the hGDH2 mutant proteins, a GLUD2 cDNA cloned in pBSKII+ vector was mutagenized at selected sites. All of the plasmids containing the GLUD1 or GLUD2 mutants were then amplified by T4 DNA polymerase (nicks were ligated by T4 DNA ligase) and used to transform the BMH 71–18 mutS strain of Escherichia coli. The cells were grown in the presence of an appropriate antibiotic selection mix; plasmid DNA was isolated and used to transform the JM109 strain of E. coli. Subsequently, the mutated cDNAs were subcloned in pVL1393 vector, which was used for the expression in Sf21 cells, as described above. The obtained mutant GDH isoproteins were studied in crude extracts. Selected mutants were also further purified and studied with respect to their kinetic and regulatory characteristics. Mutated and wild-type human enzymes were studied in parallel.
Statistical Analyses
All of the statistical analyses on the obtained data and plotting were performed using the Origin Program (MicroCal Software, Northampton, MA). The differences in kinetic and allosteric behavior were evaluated using Student's t test. The IC50 and SC50 values were determined graphically.
RESULTS
Production of Recombinant Wild-type and Mutant hGDH1 and hGDH2 and Their Modification by Estrogens in Crude Extracts
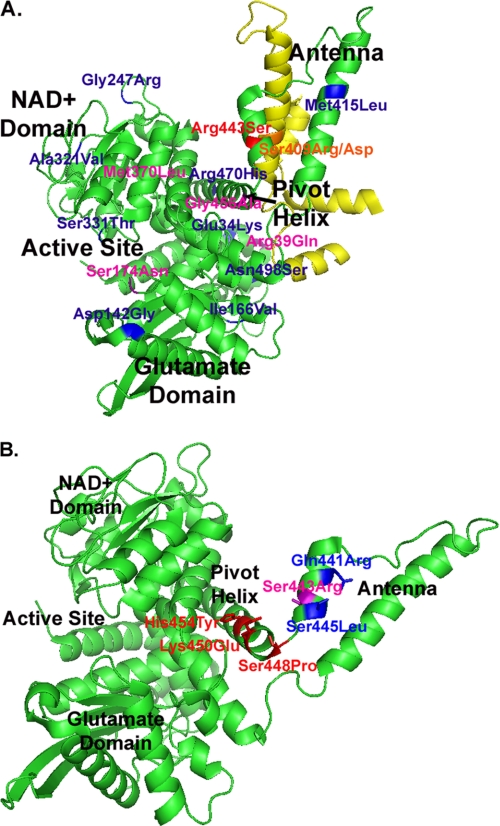
Expression of the wild-type GLUD1 and GLUD2 cDNAs in Sf21 cells produced catalytically active hGDH1 and hGDH2 as previously described (12). Functional assays (performed at 1.0 mm ADP) using crude homogenates of the cultured cells revealed that estrogens, in a concentration-dependent manner, inhibited the wild-type hGDH2 more potently than the wild-type hGDH1 (Table 1). To determine the amino acid residue(s) responsible for the sensitivity of hGDH2 to estrogens, we mutagenized the wild-type hGDH1 at residues that differ from those of hGDH2 and studied the obtained recombinant mutant proteins. As shown in Table 1 and Fig. 1, a total of 14 single amino mutants of hGDH1 were obtained. Functional analyses of the recombinant mutants, obtained by expression of the mutant DNAs in Sf21 cells, revealed that substitution of Ser for Arg443 was the single amino acid change that conferred a marked sensitivity to estrogens (Table 1); however, the sensitivity of the R443S mutant to estrogens exceeded that of the wild-type hGDH2 (Table 1). On the other hand, the substitution of Gln for Arg39, Asn for Ser174, Leu for Met370, and Ala for Gly456 in hGDH1 had a moderate opposite effect, rendering the enzyme somewhat less sensitive to estrogens (Table 1). Nine other amino acid substitutions in hGDH1 had no significant effects on the estrogen sensitivity of this enzyme (Table 1). That substitution of Ser for Arg443 is responsible for the sensitivity of the wild-type hGDH2 to estrogens was confirmed by reverse mutagenesis experiments (using the GLUD2 instead of the GLUD1 gene as template) in which the Ser443 of hGDH2 was replaced by Arg. Functional analyses of the S443R-hGDH2 mutant showed that the introduced Arg443 not only abrogated the sensitivity of hGDH2 to estrogens (Table 1 and supplemental Fig. S1) but also increased its basal activity from 4 to ∼60% of maximal. As such, the basal activity of the S443R-hGDH2 mutant became even higher than that of the wild-type hGDH1 (35–40% of maximal). These data provide additional evidence that replacement of Ser for Arg443 was the key evolutionary change that diminished the basal activity of hGDH2, thus providing this enzyme with a novel molecular mechanism for regulating its activity.
TABLE 1.
Inhibition of wild-type and mutant hGDH1 and hGDH2 by DES in crude extracts
For obtaining the hGDH1 mutants, the GLUD1 cDNA was mutagenized at sites at which hGDH1 differs from hGDH2. In each of these sites, the amino acid residue present in the GLUD2 GDH replaced the corresponding amino acid of the GLUD1 enzyme. For obtaining the S443R-hGDH2 reverse mutant, the GLUD2 cDNA was used as template. Wild-type and mutated cDNAs were expressed in Sf21 cells. Crude homogenates of these cells were used for assaying GDH activity (in the direction of reductive amination of α-ketoglutarate in TRA buffer, pH 8.0) in the absence or in the presence of increasing concentrations of DES. ADP concentration was kept constant at 1 mm. The DES IC50 (± S.E.) values were calculated from representative inhibitory curves for each enzyme using the Origin Program. The number in parentheses represents the number of experimental determinations. p values refer to comparison of the DES IC50 of the wild-type and S443R mutant hGDH2 and of each hGDH1 mutant with that of the wild-type hGDH1 using Student's t test. Mutations in hGDH1 that significantly increased the DES IC50, as well as the wild-type hGDH2 and its S443R mutant, are shown in bold type, whereas the R443S hGDH1 mutation that decreased the DES IC50 is shown in bold and italic type. For each enzyme, two to six additional independent experiments were performed, yielding similar results.
| Enzyme | DES IC50 at 1 mm ADP |
|---|---|
| μm | |
| WT hGDH1 | 32.40 ± 1.77 (27) |
| E34K-hGDH1 | 24.22 ± 3.24 (24) |
| R39E-hGDH1 | 51.52 ± 4.32 (27); <0.01 |
| D142E-hGDH1 | 29.87 ± 3.15 (24) |
| I166V-hGDH1 | 40.92 ± 2.60 (27) |
| S174N-hGDH1 | 61.44 ± 3.82 (27); <0.0001 |
| G247R-hGDH1 | 41.89 ± 3.21 (24) |
| A321V-hGDH1 | 44.12 ± 2.90 (24) |
| S331T-hGDH1 | 34.85 ± 4.15 (21) |
| M370L-hGDH1 | 52.90 ± 3.39 (30); <0.001 |
| M415L-hGDH1 | 24.29 ± 2.50 (30) |
| R443S-hGDH1 | 1.89 ± 0.11 (24); <0.0001 |
| G456A-hGDH1 | 60.67 ± 3.44 (21); <0.0001 |
| R470H-hGDH1 | 39.01 ± 2.39 (24) |
| N498S-hGDH1 | 26.45 ± 2.35 (26) |
| WT hGDH2 | 10.06 ± 1.27 (36); <0.0001 |
| S443R-hGDH2 | 91.33 ± 9.05 (33); <0.0001 |
FIGURE 1.
Location of the introduced mutations in hGDH1 (A) and in hGDH2 (B). Shown is a cartoon diagram of the apo form of human GDH1 (Protein Data Bank entry 1LIF). For simplicity, only one of six subunits that compose the GDH hexamer are shown (in green), including its NAD+-binding domain, the glutamate-binding domain, the active site, the pivot helix, and the antenna. In A, the evolutionary mutation (R443S) that renders the enzyme markedly sensitive to estrogens is shown in red, whereas mutations that decrease estrogen sensitivity are shown in purple. Residues, mutation of which has no significant effect on estrogen sensitivity, are shown in blue. The antenna of an adjacent subunit, on the ascending helix of which Ser409 (orange) is located, is shown in yellow. As described in the text, Arg443 from one subunit is connected with hydrogen bonds with Ser409 of the adjacent subunit. In B, residues in the pivot helix and the junction of the pivot helix with the antenna, mutation of which in hGDH2 decreases basal activity (K450E, H454Y, and S448P) are shown in red. Residues in the antenna, mutation of which in hGDH2 increases basal activity (Q441R and S445L), are shown in blue. The reverse residue 443 mutation in hGDH2 (S443R) that markedly increases basal activity is shown in purple.
Modification of Purified Wild-type hGDH1 and hGDH2 by Estrogens
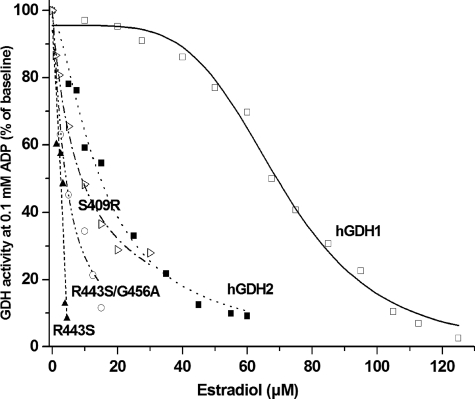
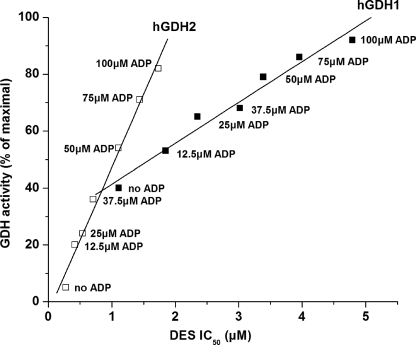
The R443S hGDH1 mutant, the wild-type hGDH1, and the wild-type hGDH2 were purified to homogeneity and used for additional studies. Functional analyses of the purified enzymes confirmed the data obtained on crude extracts by showing that estrogens interacted more potently with the wild-type hGDH2 than with the wild-type hGDH1 (Table 2 and Fig. 2). Thus, in the absence of ADP, the wild-type hGDH2 was ∼18-fold more sensitive to DES (IC50 = 0.08 ± 0.01 μm) than the wild-type hGDH1 (IC50 = 1.67 ± 0.06 μm; p < 0.001) (Table 2). However, in the presence of 0.1 or 1.0 mm ADP, the wild-type hGDH2 was ∼7- and ∼4-fold more sensitive to DES, respectively, than the wild-type hGDH1 (Table 2). Similarly, 17β-estradiol showed a ∼18-fold higher affinity for hGDH2 (IC50 = 1.53 ± 0.24 μm) than for hGDH1 (IC50 = 26.94 ± 1.07 μm; p < 0.001) (Table 2). As with DES, this differential sensitivity to 17β-estradiol was less pronounced when inhibitory assays were performed in the presence of 0.1 or 1.0 mm ADP (Fig. 2 and Table 2). Also, estriol and progesterone inhibited the wild-type hGDH2 more strongly than wild-type hGDH1, but at higher concentrations than those required for DES or for 17β-estradiol (Table 2). Study of the inhibitory curves of the wild-type hGDH1 and hGDH2 at specific ADP concentrations (Fig. 3) revealed that modification of these enzyme by estrogens correlated inversely with their catalytic activity levels (measured prior to the addition of the female hormones) (R = 0.9845 for the wild-type hGDH1 and R = 0.9935 for hGDH2). As shown in Fig. 3, the obtained regression line for the wild-type hGDH2 was significantly steeper than that for the wild-type hGDH1, indicating that the estrogen effect is more strongly linked to the state of activation of hGDH2 than that of hGDH1. Because hGDH1 activity is subject to negative modulation by GTP, we sought to determine whether the presence of inhibitory concentrations of GTP alters the sensitivity of the enzyme to estrogens. The results revealed that, in the absence of ADP, inhibitory concentrations of GTP had no effect on the modulation of hGDH1 activity by estrogens (supplemental Fig. S2A). However, at 1.0 mm ADP, the presence of inhibitory concentrations of GTP potentiated the interaction of estrogens with the wild-type hGDH1 (supplemental Fig. S2B).
TABLE 2.
Inhibition of purified wild-type hGDH1 and hGDH2 and mutant R443S and R443S/G456A-hGDH1 by female steroidal hormones
The IC50 (± S.E.) values were calculated from the inhibitory curves for each enzyme using the Origin Program. Highly purified enzyme preparations were used for these experiments as described under “Experimental Procedures.” GDH activity was determined in the direction of reductive amination of α-ketoglutarate in TRA buffer, pH 8.0, in the presence of increasing concentrations of female steroidal hormones. DES was varied from 0 to 50 μm, 17β-estradiol and estriol from 0 to 500 μm, and progesterone from 0 to 2000 μm. Estrogen inhibitory curves for the recombinant enzymes were obtained either in the absence of ADP (No ADP) or in the presence of 0.1 and 1.0 mm ADP. The R443S single mutant and the R443S/G456A double mutant were only tested at 0.1 and 1.0 mm ADP, because these enzymes were relatively unstable when assayed without ADP.
| IC50 (μm) |
||||
|---|---|---|---|---|
| DES | 17β-Estradiol | Estriol | Progesterone | |
| μm | ||||
| No ADP | ||||
| Wild-type hGDH1 | 1.67 ± 0.06 | 26.94 ± 1.07 | 144.77 ± 18.87 | 118.78 ± 3.63 |
| Wild-type hGDH2 | 0.08 ± 0.01a | 1.53 ± 0.24a | 11.34 ± 0.74a | 12.31 ± 2.64a |
| 0.1 mm ADP | ||||
| Wild-type hGDH1 | 7.06 ± 0.41 | 69.23 ± 1.31 | 315.53 ± 26.19 | 596.39 ± 50.87 |
| R443S-hGDH1 | 0.49 ± 0.03a | 2.22 ± 0.76a | 2.27 ± 0.09a | 8.61 ± 1.23a |
| R443S/G456A-hGDH1 | 4.36 ± 0.36a | |||
| Wild-type hGDH2 | 1.05 ± 0.09a | 15.10 ± 1.22a | 188.72 ± 17.92b | 58.86 ± 24.52a |
| 1 mm ADP | ||||
| Wild-type hGDH1 | 26.50 ± 2.24 | 127.42 ± 10.38 | 398.39 ± 9.66 | 743.54 ± 145.98 |
| R443S-hGDH1 | 2.19 ± 0.14a | 14.81 ± 1.05a | 42.71 ± 6.98a | 78.57 ± 9.19a |
| R443S/G456A-hGDH1 | 6.35 ± 0.69a | 68.75 ± 7.40a | ||
| Wild-type hGDH2 | 8.38 ± 0.86a | 104.40 ± 32.55 | 274.90 ± 9.19a | 392.21 ± 8.92c |
a p < 0.001 compared with the wild-type hGDH1 studied in the presence of the same ADP concentration.
b p < 0.01 compared with the wild-type hGDH1 studied in the presence of the same ADP concentration.
c p < 0.05 compared with the wild-type hGDH1 studied in the presence of the same ADP concentration.
FIGURE 2.
Inhibition of purified recombinant wild-type hGDH1 and hGDH2 and the R443S, R443S/G456A, and S409R hGDH1 mutants by 17β-estradiol in the presence of 0.1 mm ADP. GDH activity was measured in the direction of reductive amination of α-ketoglutarate in the presence of increasing concentrations of 17β-estradiol (0–125 μm). The data points represent the mean values of three experimental determinations and are expressed as percentages of GDH activity measured at 0.1 mm ADP in the absence of 17β-estradiol.
FIGURE 3.
Estrogen inhibition of wild-type hGDH1 and hGDH2 at different activation states induced by ADP. The y axis shows the catalytic activity (expressed as percentages of maximal obtained at 1.0 mm ADP) displayed by each recombinant enzyme at the specific concentration of ADP prior to the addition of DES. The x axis shows the DES IC50 (± S.E.) values for each of the wild-type enzymes. ADP concentrations were varied from 0 to 100 μm (numbers next to the data points). The enzyme assays were performed in the direction of α-ketoglutarate reductive amination in TRA buffer, pH 8.0, as described under “Experimental Procedures.” The R correlation coefficient and P (probability that R is 0) values of the linear regression were calculated using the Origin Program. The slope of the regression line (± S.E.) is given below as the rate of change of enzyme basal activity (expressed as percentages of maximal) per increase in DES IC50: slope for hGDH1, 14.35 ± 1.14 (R = 0.9845, P < 0.0001); and slope for hGDH2, 51.16 ± 2.60 (R = 0.9935, P < 0.0001).
Modification of Kinetic Properties of Wild-type hGDH1 and hGDH2 by Estrogens
Kinetic analyses performed by varying the levels of the enzyme substrates in the presence of inhibitory concentrations of DES revealed that the Km for α-ketoglutarate, NH3, or glutamate was not altered by the inhibitor. However, kinetic analyses of NADPH binding in the presence of DES concentrations sufficient to decrease the activity of hGDH1 or hGDH2 by 50% (IC50 for each of the wild-type enzymes) revealed that the estrogen provoked enzyme inhibition by excess cofactor (NADPH > 100 μm). In contrast, in the absence of DES, we observed no enzyme inhibition by excess cofactor (studied up to 400 μm of NADPH) of either wild-type hGDH1 or hGDH2 (supplemental Fig. S3).
Substitution of Ser for Arg443 is Responsible for the Sensitivity of hGDH2 to Estrogens
Functional analyses of the purified R443S-hGDH1 revealed that this mutant was 14–32-fold more sensitive to estrogens than the wild-type hGDH1, thus confirming results obtained on crude cell extracts (Table 2 and Fig. 2). Also, the purified R443S-hGDH1 mutant was substantially more sensitive to estrogens than the purified wild-type hGDH2, thus confirming data obtained with the use of crude cell extracts (Table 1). To test whether the drastic effect of the R443S change is modified by other amino acid substitutions, as shown in crude extracts (Table 1), we studied a double mutant of hGDH1 that carries the R443S and the G456A change in the same amino acid chain. As shown in Table 1, the G456A single amino acid change has an opposite effect, rendering hGDH1 less sensitive to estrogens (by ∼2-fold). Functional assays of the recombinant R443S/G456A-hGDH1 enzyme, purified to homogeneity, revealed that the sensitivity of this double mutant to estrogens was intermediate between that of the R443S single mutant and the wild-type hGDH2 (Table 2 and Fig. 2). As such, these data are consistent with the thesis that, whereas the R443S change was largely responsible for the sensitivity of the wild-type hGDH2 to estrogens, its drastic effect is modified by the opposing moderate influences of four other amino acid substitutions acquired during the evolution of hGDH2.
Study of hGDH1 and hGDH2 Mutants Reveals that Sensitivity to Estrogens Correlates Inversely with their Specific Basal Activity
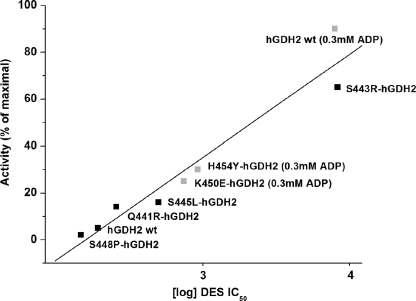
Because the R443S mutant displays a fraction of the basal activity of the wild-type hGDH1, we sought to test whether other single amino acid substitutions in hGDH1 that decrease basal activity alter sensitivity to estrogen hormones. Initially, we studied the S409(R/D) hGDH1 mutants, which, similar to the R443S hGDH1 mutant, show low basal activity.3 Introduction of Arg at residue 409 is thought to disrupt side chain bonds in the antenna region between Arg443 of one subunit and Ser409 of the adjacent subunit (15). Functional analyses of the expressed enzyme, purified to homogeneity, revealed that the S409R-hGDH1 mutant was indeed much more sensitive to estrogens than the wild-type hGDH1 (Fig. 2), thus supporting the possibility that estrogen interaction with GDH is potentiated by a closed enzyme conformation. This possibility was further tested by studying several hGDH2 mutants obtained by mutagenesis of residues in the regulatory domain of GLUD2 that affect basal activity (25) (Fig. 1). In this regard, we have previously shown that substitution of Glu for Lys450 or Tyr for His454, residues located in the pivot helix, diminished the basal activity of hGDH2 (25). On the other hand, replacement of Gln441 by Arg or Ser445 by Leu, residues located in the antenna, enhanced the basal activity of hGDH2 (25). A third mutant located in the junction of the antenna with the pivot helix (Ser448Pro) (Fig. 1) decreased basal activity. The study of estrogen interaction with these mutants revealed that all three mutations that depressed basal activity (K450E, H454Y, and S448P) made hGDH2 more sensitive to estrogens, whereas the two mutations that enhanced basal activity (Q441R and S445L) attenuated estrogen inhibition (Fig. 4). Regression analyses revealed that inhibition of mutant and wild-type hGDH2 by DES correlated inversely with their catalytic activity levels (R = 0.9660) (Fig. 4). Similarly, regression analyses involving the wild-type hGDH1 and several of its mutants revealed that modification of these enzymes by estrogens correlated inversely with their basal activity levels (R = 0.9864) (Fig. 5 and supplemental Fig. S4). In addition, the sensitivity of these enzymes to estrogens correlated significantly with their relative resistance to thermal inactivation (R = 0.9772) (supplemental Fig. S5).
FIGURE 4.
Regression analysis of DES IC50 for the wild-type hGDH2 and its mutants versus the basal catalytic activities of the recombinant enzymes. The y axis shows the catalytic activity (expressed as percentages of maximal) displayed by each recombinant enzyme prior to the addition of DES, measured either in the absence of ADP (basal activity) (Q441R, S443R, S445L, and S448P mutants and wild-type hGDH2) or in the presence of 0.3 mm ADP (K450E and H454Y mutants and wild-type hGDH2). At lower concentrations of ADP (0.1 or 0.2 mm), the K450E and H454Y mutants displayed very little measurable activity in the absence of ADP. For reasons of comparison, the DES IC50 for the wild-type hGDH2 was also obtained in the presence of 0.3 mm ADP. The x axis shows the DES IC50 values for each recombinant enzyme, in logarithmic scale. The R correlation coefficient and P (probability that R is 0) values of the linear regression were calculated using the Origin program (R = 0.9660, P < 0.0001).
FIGURE 5.
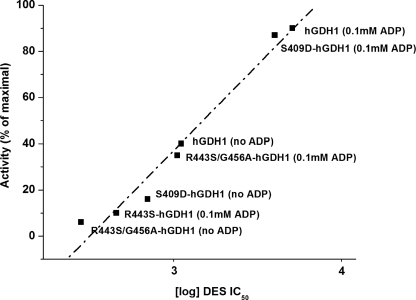
Regression analysis of DES IC50 for the wild-type hGDH1 and its mutants versus the catalytic activities of the recombinant enzymes. The y axis shows the catalytic activity displayed by each recombinant enzyme prior to the addition of DES expressed as percentages of maximal activity. The DES IC50 values were calculated from the inhibitory curves for each of the recombinant enzyme using the Origin Program and are shown on the x axis, in logarithmic scale. The wild-type hGDH1 enzyme and its S409D and R443S/G456A mutants were studied either under basal conditions (no ADP) or in the presence of 0.1 mm ADP. However, the R443S mutant, which displayed very little activity under base-line conditions, was studied only in the presence of 0.1 mm ADP. The R correlation coefficient and P (probability that R is 0) values of the linear regression were calculated using the Origin program (R = 0.9864, P < 0.0001).
DISCUSSION
Here we report that estrogens inhibit the wild-type hGDH2 with a greater potency than the wild-type GDH1. To elucidate the molecular basis of these interactions, we studied several mutants of hGDH1, obtained by amino acid substitutions at hGDH1 residues that differ from those of hGDH2. The results showed that replacement of Arg443 by Ser was the only evolutionary amino acid change that made the enzyme markedly sensitive to estrogens; however, the R443S mutant proved to be substantially more sensitive to estrogens than the wild-type hGDH2. These differences between wild-type hGDH2 and the R443S mutant were explained by the study of additional single amino acid substitutions acquired during the evolution of hGDH2, which revealed that four such changes (R39Q, S174N, M370L, and G456A) reduced the sensitivity of the enzyme to estrogens. Consistent with this possibility are data obtained here by the study of a double hGDH1 mutant that carries the R443S and G456A changes in the same amino acid chain. As described under “Results,” the sensitivity of this double mutant to estrogens was intermediate between that of the R443S single mutant and the wild-type hGDH2. Hence, whereas the R443S was the single amino acid change that rendered the wild-type hGDH2 sensitive to estrogens, its drastic effect is modified by the moderate opposing influences of the four other amino acid substitutions acquired during the evolution of hGDH2. Three of these residues are located on strategic sites of the enzyme: R39Q is located at the back of the antenna (near the ADP-binding site), S174N is located at the entrance of the catalytic cleft, and G456A is located on the pivot helix (Fig. 1).
Structural analyses of the R443S mutant have suggested that introduction of Ser443 disrupts side chain bonds in the antenna between Arg443 of one subunit and Ser409 of the adjacent subunit, leading to a closed conformation (15). This possibility is supported by additional studies3 showing that substituting Arg for Ser409, which also disrupts the above side chain bonds, diminishes basal activity. In light of this, we studied here the interaction of estrogens with the S409R-hGDH1 mutant and found that this change also made the enzyme sensitive to estrogens. We then studied additional mutations in hGDH1 and hGDH2 that affect the catalytic function of these enzymes and showed that the sensitivity of these mutants to estrogens correlated inversely with their basal activity levels. As such, whereas mutations that render the enzyme sensitive to estrogen involve different amino acid residues, the common denominator here is that these substitutions decrease catalytic activity by favoring a “closed state.” Additional observations showing that the sensitivity of the wild-type hGDH1 and hGDH2 to estrogens correlated inversely with their state of activation induced by various ADP concentrations are consistent with this model because ADP is known to activate GDH by helping the catalytic cleft to open. Moreover, our data showing that the regression line (correlating estrogen sensitivity with basal activity levels) for hGDH2 was significantly steeper than that for hGDH1 suggest that the estrogen effect is more strongly linked to the state of activation of hGDH2 than to that of hGDH1.
Previous studies have shown that GTP, a selective inhibitor of hGDH1, binds to both the closed and the open state of the enzyme, promoting a closed conformation (26). At the structural level, GTP is shown to occupy the space behind the NAD+ domain, when it rotates during catalysis to close the active site (26). This in turn prevents the NAD+ domain from moving back to open the catalytic cleft (26). To explore whether GTP binding alters the estrogen sensitivity of hGDH1, we obtained estrogen inhibitory curves in the presence of GTP. The results revealed that inhibitory concentrations of GTP had no effect on the modulation of hGDH1 activity by estrogens. Only when the GTP effect was partially counteracted by ADP did the presence of GTP modify estrogen regulation. As such, our observations suggest that estrogen inhibition of human GDHs does not require the GTP sites to be functional.
Although inhibition of mammalian GDH by estrogens was one of the earliest observations on the direct effects of these hormones on metabolic enzymes (27), modification of F0F1 ATPase (an enzyme central in ATP production or hydrolysis) by estrogens has received attention lately (6, 7). It is presently unclear, however, whether these direct effects are biologically relevant, because the concentrations required for inhibition of bovine liver GDH (hGDH1 in the human) or of F0F1 ATPase (26–145 μΜ) are above the physiological levels of these hormones. In light of these considerations, the present findings, showing that hGDH2 is much more sensitive to estrogens than hGDH1 or F0F1 ATPase, suggest that these hormones may target hGDH2 preferentially.
It has been argued (17) that hGDH2 has adapted to the unique environment that prevails in the nerve tissue in which GTP levels are higher than those found in other tissues. Moreover, resistance to GTP inhibition may allow hGDH2 to function in nerve terminals even when the tricarboxylic acid cycle generates GTP levels sufficient to inhibit hGDH1 (23). Because the sensitivity of hGDH2 to estrogens relates to the propensity of the enzyme to assume under base-line conditions a closed conformation and because ADP antagonizes estrogen inhibition by helping the catalytic cleft to open, regulation of hGDH2 may be achieved by the opposing actions of estrogens and ADP, akin to the regulation of hGDH1 by the antagonistic effects of GTP and ADP (16).
Our observations showing that the affinity of DES for hGDH2 (IC50 = 0.08 μm) is comparable with that of GTP for hGDH1 (IC50 = 0.1–0.3 μm) suggest that regulation of hGDH2 by estrogens is physiologically relevant. Tissue fractionation studies (28) have shown that progesterone levels are relatively high in the mitochondria, where hGDH2 is located (29). In females, the plasma levels of free estradiol vary markedly during their menstrual cycle (0.15–2.0 nm) (1). Only during pregnancy do these levels (0.13 μm) approach those required for inhibition of the wild-type hGDH2 under base-line conditions. Whereas the levels of circulating estrogens in nonpregnant women are below those used here to inactivate hGDH2, there is evidence for extensive local biosynthesis in the central nervous system and other tissues and that this synthesis could raise estrogen to levels higher than those of serum (30). In the brain, astrocytes, the cells that express hGDH2, are the only neural cells capable of synthesizing estrogens from androstenedione (31); they express both 17β-hydroxysteroid dehydrogenase and P450-aromatase. Astrocytes also express the enzyme glutamine synthetase (32) that catalyzes conversion of glutamate taken up by astrocytes to glutamine, which is then exported to neurons to be used as a precursor of transmitter glutamate. As such, prevention of glutamate oxidative dehydrogenation by estrogens (via inhibition of hGDH2) may permit a larger fraction of glutamate to be converted via glutamine synthetase to glutamine. Previous studies have shown that estrogens increase the expression of glutamine synthetase (33). These observations, taken together, suggest that astrocytes may modulate glutamatergic mechanisms by regulating their estrogen production. Moreover, inhibition of hGDH2 by estrogens may provide a mechanism by which these hormones protect against glutamate excitotoxicity and oxidative stress as discussed below.
Although the leads provided by the present studies need to be further pursued, there is evidence that whereas control of GDH velocity is of importance for cell functions, deregulation of this enzyme may lead to disease processes in the human. As noted above, regulatory mutations in the hGDH1 gene that attenuate GTP inhibition lead to hyperinsulinism/hyperammonemia syndrome characterized by hypoglycemic episodes that often follow intake of l-leucine (stimulates the overactive enzyme). In addition, patients harboring these mutations suffer from seizures that may be independent from their hypoglycaemic episodes (19). These observations, taken together with recent findings showing that overexpression of GDH1 in mice results in age-dependent degenerative changes in the CA1 region of the hippocampus (34), suggest that a tight regulation of GDH activity is of importance for nerve tissue function and degeneration. Regarding deregulation of hGDH2 in human disorders, recent studies revealed that patients with Parkinson disease, who were hemizygous for T1492G polymorphism in the GLUD2 gene, experienced onset of their disease 8–13 years earlier than patients with other genotypes in two populations of diverse genetic background (35). However, female Parkinson disease patients who were heterozygous for this allele were protected. The T1492G polymorphism is within the coding region of the GLUD2 gene and results in the substitution of Ala for Ser445 in the regulatory domain of hGDH2 (35). The S445A variant, obtained in recombinant form by expressing the T1492G-GLUD2 cDNA in Sf21 cells, displayed enhanced basal activity that was sensitive to inhibition by estrogens (35). As such, the protection of female heterozygotes (for T1492G polymorphism) from the gain-of-function properties of the S445A hGDH2 variant has been attributed to estrogen inhibition of the overactive enzyme (35). Also, independent studies have provided evidence that female hormones protect from Parkinson disease (36) and that apoptosis of dopaminergic neurons occurs in aromatase knock-out mice in the male (37). These observations suggest that inhibition of mitochondrial glutamate oxidative dehydrogenation by estrogens may account, at least in part, for their neuroprotective actions. Moreover, because mitochondrial glutamate oxidation is associated with increased hydroxyl-radical formation in an animal model of Parkinson disease (38), the ability of estrogens to modify the mitochondrial glutamate metabolism may in part account for their protective effects against oxidative stress. Hence, elucidating the mechanisms by which estrogens regulate GDH activity may have important implications for understanding the role of these hormones in cell biology in health and disease and for the development of novel therapeutic approaches to human degenerative disorders.
Supplementary Material
This work was supported by European Community and the Ministry of Development/General Secretariat for Research and Technology Contract Grant PENED 2003/03ED576 and by Association for the Advancement of Research and Treatment of Neurologic Disorders of Crete EY ZHN.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
V. Mastorodemos, K. Kanavouras, M. Providaki, M. Kokkinidis, and A. Plaitakis, unpublished data.
- GDH
- glutamate dehydrogenase
- DES
- diethylstilbestrol
- h
- human
- TRA
- triethanolamine.
REFERENCES
- 1.Gruber C. J., Tschugguel W., Schneeberger C., Huber J. C. (2002) N. Engl. J. Med. 346, 340–352 [DOI] [PubMed] [Google Scholar]
- 2.Green P. S., Simpkins J. W. (2000) Int. J. Dev. Neurosci. 18, 347–358 [DOI] [PubMed] [Google Scholar]
- 3.Amantea D., Russo R., Bagetta G., Corasaniti M. T. (2005) Pharmacol. Res. 52, 119–132 [DOI] [PubMed] [Google Scholar]
- 4.Ba F., Pang P. K., Benishin C. G. (2004) Neurochem. Int. 45, 31–38 [DOI] [PubMed] [Google Scholar]
- 5.Goodman Y., Bruce A. J., Cheng B., Mattson M. P. (1996) J. Neurochem. 66, 1836–1844 [DOI] [PubMed] [Google Scholar]
- 6.McEnery M. W., Pedersen P. L. (1986) J. Biol. Chem. 261, 1745–1752 [PubMed] [Google Scholar]
- 7.McEnery M. W., Hullihen J., Pedersen P. L. (1989) J. Biol. Chem. 264, 12029–12036 [PubMed] [Google Scholar]
- 8.Yielding K. L., Tomkins G. M. (1960) Proc. Natl. Acad. Sci. U.S.A. 46, 1483–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warren J. C., Carr D. O., Grisolia S. (1964) Biochem. J. 93, 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pons M., Michel F., Descomps B., Crastes de Paulet A. (1978) Eur. J. Biochem. 84, 257–266 [DOI] [PubMed] [Google Scholar]
- 11.Mavrothalassitis G., Tzimagiorgis G., Mitsialis A., Zannis V., Plaitakis A., Papamatheakis J., Moschonas N. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 3494–3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shashidharan P., Michaelidis T. M., Robakis N. K., Kresovali A., Papamatheakis J., Plaitakis A. (1994) J. Biol. Chem. 269, 16971–16976 [PubMed] [Google Scholar]
- 13.Burki F., Kaessmann H. (2004) Nat. Genet. 36, 1061–1063 [DOI] [PubMed] [Google Scholar]
- 14.Zaganas I., Plaitakis A. (2002) J. Biol. Chem. 277, 26422–26428 [DOI] [PubMed] [Google Scholar]
- 15.Zaganas I., Spanaki C., Karpusas M., Plaitakis A. (2002) J. Biol. Chem. 277, 46552–46558 [DOI] [PubMed] [Google Scholar]
- 16.Plaitakis A., Metaxari M., Shashidharan P. (2000) J. Neurochem. 75, 1862–1869 [DOI] [PubMed] [Google Scholar]
- 17.Plaitakis A., Zaganas I. (2001) J. Neurosci. Res. 66, 899–908 [DOI] [PubMed] [Google Scholar]
- 18.Smith E. (1979) Proc. Am. Phil. Soc. 123, 73–84 [Google Scholar]
- 19.Stanley C. A., Lieu Y. K., Hsu B. Y., Burlina A. B., Greenberg C. R., Hopwood N. J., Perlman K., Rich B. H., Zammarchi E., Poncz M. (1998) N. Engl. J. Med. 338, 1352–1357 [DOI] [PubMed] [Google Scholar]
- 20.Stanley C. A., Fang J., Kutyna K., Hsu B. Y., Ming J. E., Glaser B., Poncz M. (2000) Diabetes 49, 667–673 [DOI] [PubMed] [Google Scholar]
- 21.Spanaki C., Zaganas I., Kleopa K. A., Plaitakis A. (2010) J. Biol. Chem. 285, 16748–16756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sonnewald U., Westergaard N., Schousboe A. (1997) Glia 21, 56–63 [DOI] [PubMed] [Google Scholar]
- 23.Zaganas I., Kanavouras K., Mastorodemos V., Latsoudis H., Spanaki C., Plaitakis A. (2009) Neurochem. Int. 55, 52–63 [DOI] [PubMed] [Google Scholar]
- 24.Kanavouras K., Mastorodemos V., Borompokas N., Spanaki C., Plaitakis A. (2007) J. Neurosci. Res. 85, 1101–1109 [DOI] [PubMed] [Google Scholar]
- 25.Kanavouras K., Borompokas N., Latsoudis H., Stagourakis A., Zaganas I., Plaitakis A. (2009) J. Neurochem. 109, 167–173 [DOI] [PubMed] [Google Scholar]
- 26.Peterson P. E., Smith T. J. (1999) Structure 7, 769–782 [DOI] [PubMed] [Google Scholar]
- 27.Kallos J., Shaw K. P. (1971) Proc. Natl. Acad. Sci. U.S.A. 68, 916–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haukkamaa M., Wichmann K. (1977) Clin. Endocrinol. 7, 111–119 [DOI] [PubMed] [Google Scholar]
- 29.Mastorodemos V., Kotzamani D., Zaganas I., Arianoglou G., Latsoudis H., Plaitakis A. (2009) Biochem. Cell Biol. 87, 505–516 [DOI] [PubMed] [Google Scholar]
- 30.Simpson E., Rubin G., Clyne C., Robertson K., O'Donnell L., Davis S., Jones M. (1999) Endocr.-Relat. Cancer 6, 131–137 [DOI] [PubMed] [Google Scholar]
- 31.Zwain I. H., Yen S. S. (1999) Endocrinology 140, 3843–3852 [DOI] [PubMed] [Google Scholar]
- 32.Hertz L., Dringen R., Schousboe A., Robinson S. R. (1999) J. Neurosci. Res. 57, 417–428 [PubMed] [Google Scholar]
- 33.Blutstein T., Devidze N., Choleris E., Jasnow A. M., Pfaff D. W., Mong J. A. (2006) J. Neuroendocrinol. 18, 692–702 [DOI] [PubMed] [Google Scholar]
- 34.Bao X., Pal R., Hascup K. N., Wang Y., Wang W., Xu W., Hui D., Agbas A., Wang X., Michaelis M. L., Choi I., Belousov A. B., Gerhardt G. A., Michaelis E. K. (2009) J. Neurosci. 29, 13929–13944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plaitakis A., Latsoudis H., Kanavouras K., Ritz B., Bronstein J. M., Skoula I., Mastorodemos V., Papapetropoulos S., Borompokas N., Zaganas I., Xiromerisiou G., Hadjigeorgiou G. M., Spanaki C. (2010) Eur. J. Hum. Genet. 18, 336–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quesada A., Lee B. Y., Micevych P. E. (2008) Dev. Neurobiol. 68, 632–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hill R. A., Chua H. K., Jones M. E., Simpson E. R., Boon W. C. (2009) Mol. Cell Neurosci 41, 1–7 [DOI] [PubMed] [Google Scholar]
- 38.Panov A., Dikalov S., Shalbuyeva N., Taylor G., Sherer T., Greenamyre J. T. (2005) J. Biol. Chem. 280, 42026–42035 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.