Abstract
Integrin-growth factor receptor cross-talk plays a role in growth factor signaling, but the specifics are unclear. In a current model, integrins and growth factor receptors independently bind to their ligands (extracellular matrix and growth factors, respectively). We discovered that neuregulin-1 (NRG1), either as an isolated EGF-like domain or as a native multi-domain form, binds to integrins αvβ3 (with a KD of 1.36 × 10−7 m) and α6β4. Docking simulation predicted that three Lys residues at positions 180, 184, and 186 of the EGF-like domain are involved in integrin binding. Mutating these residues to Glu individually or in combination markedly suppressed integrin binding and ErbB3 phosphorylation. Mutating all three Lys residues to Glu (the 3KE mutation) did not affect the ability of NRG1 to bind to ErbB3 but markedly reduced the ability of NRG1 to induce ErbB3 phosphorylation and AKT and Erk1/2 activation in MCF-7 and T47D human breast cancer cells. This suggests that direct integrin binding to NRG1 is critical for NRG1/ErbB signaling. Notably, stimulation of cells with WT NRG1 induced co-precipitation of ErbB3 with α6β4 and with αvβ3 to a much lower extent. This suggests that WT NRG1 induces integrin-NRG1-ErbB3 ternary complex formation. In contrast, the 3KE mutant was much less effective in inducing ternary complex formation than WT NRG1, suggesting that this process depends on the ability of NRG1 to bind to integrins. These results suggest that direct NRG1-integrin interaction mediates integrin-ErbB cross-talk and that α6β4 plays a major role in NRG-ErbB signaling in these cancer cells.
Keywords: Growth Factors, Integrin, Mutant, Receptor-tyrosine Kinase, Signal Transduction, Site-directed Mutagenesis, ErbB, Neuregulin
Introduction
The neuregulins (NRGs)2 are a family of four structurally related proteins that are part of the EGF family of proteins (NRG1–4) (1–4). Transmembrane NRGs typically function as precursor molecules that are cleaved by metalloproteases. This results in the release of the extracellular domain that may subsequently bind to nearby receptors (autocrine/paracrine action). NRGs contain an epidermal growth factor (EGF)-like motif that binds and activates receptor-tyrosine kinases in the EGF receptor (ErbBs) family. Neuregulin-1 (NRG1) binds to ErbB3 and ErbB4. NRG1 has 11 isoforms (5). NRG1 plays essential roles in the nervous system, heart, and breast. NRG1 signaling is involved in the development and functions of several other organ systems and human diseases, including schizophrenia (6), coronary heart diseases (7), and cancer (8). Targeted deletion of ErbB2, ErbB3, ErbB4, or NRG1 in mice leads to developmental abnormalities that are severe in the nervous system and lethal in the cardiovascular system (9–11). In cancer the interaction between ErbB receptors and ligands such as NRGs plays an important role in tumor growth. The EGF-like motif of NRGs is essential and sufficient for receptor binding and activation as well as promoting tumorigenesis (12). The presence of the autocrine loop is one of the causes that induces aberrant ErbB receptor activation and has been correlated with cancer development and progression. Disrupting this autocrine loop may provide an important therapeutic measure to control cancer cell growth (13).
Integrins have been shown to cross-talk with receptor-tyrosine kinase in growth factor signaling (14). Integrins are a family of cell adhesion receptors that recognize extracellular matrix ligands and cell surface ligands (15). Integrins are transmembrane αβ heterodimers, and at least 18 α and 8 β subunits are known (16). Integrins are involved in signal transduction upon ligand binding, and their functions are in turn regulated by signals from within the cell (15). It has been reported that there is a positive correlation between αvβ3 integrin levels and overexpression of NRG associated with melanoma tumor progression and metastasis (17–19). It has been proposed that NRG1 may play a key role in the regulation of αvβ3 integrin expression and in its signaling functions (20).
Integrin α6β4 is a receptor for the laminin family of extracellular matrix proteins, and its expression is associated with poor patient prognosis and reduced survival in a variety of human cancers (21). The β4 integrin subunit was originally identified as a tumor-related antigen expressed in metastatic cancer (22). In contrast with its function in regulating stable adhesion through the formation of hemidesmosomes in normal epithelial cells, α6β4 promotes motility and invasion in carcinoma cells (23). Moreover, suppression of α6β4 expression by siRNA diminishes invasive potential (24).
Cross-talk between growth factor receptors and integrins plays a role in growth factor signaling. In current models of NRG-ErbB signaling, integrins and ErbB receptors transduce signals independently upon binding to their ligands (e.g. extracellular matrix ligands to integrins and NRGs to ErbB receptors), and their signals are merged inside the cells. In the present study we discovered that the EGF-like domain of NRG1 binds directly to integrins. Docking simulation between the EGF-like domain of NRG1 and the headpiece of integrin αvβ3 located the integrin-binding site in NRG1. We generated a mutant NRG1 that is defective in integrin binding. This mutant was also defective in inducing ErbB3 phosphorylation, AKT, and Erk1/2 activation, whereas the mutant still binds to ErbB3 at levels comparable with that of WT NRG1. This suggests that direct integrin binding is critical for NRG1/ErbB signaling. Interestingly, WT NRG1 induced co-precipitation of ErbB3 and integrins αvβ3 and α6β4 in human breast cancer cells, suggesting that these integrins are involved in ErbB-NRG1 signaling in cancer cells. Interestingly, the integrin-binding-defective NRG1 mutant was defective in inducing co-precipitation of ErbB3 and integrins, suggesting that this process was dependent on the ability of NRG1 to bind to integrins. We propose that the binding of NRG1 to integrins is involved in NRG1/ErbB signaling.
EXPERIMENTAL PROCEDURES
Materials
Antibodies against phospho-ErbB3 (Tyr-1289), phospho-Erk1/2 (Thr-202 and Tyr-204), phosphor-Akt (Thr-308), Erk1/2, Akt, and integrins β3 and β4 were purchased from Cell Signaling Technology, Inc. (Danvers, MA). Antibody against ErbB3 was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Horseradish peroxidase-conjugated anti-rabbit IgG was purchased from Bio-Rad. We obtained recombinant human NRG1α EGF-like domain peptide (residues Ser-177—Lys-241, synthesized in Escherichia coli, >97% purity), recombinant human NRG1 isoform SMDF (296 amino acids, Spodoptera frugiperda Sf21(baculovirus)-derived), and recombinant human ErbB3 Fc chimera from R&D Systems (Minneapolis, MN). Recombinant soluble αvβ3 and K562 cells that express human αvβ3 (αvβ3-K562) have been described (25). Recombinant soluble α6β4 was synthesized as described (26). Chinese hamster ovary (CHO) cells that express WT β1 or the β1-3-1 mutant have been described (27). CHO cells that express human α6β4 (α6β4-CHO) were prepared by transfecting human β4 cDNA in pBJ-1 vector together with hygromycin gene into CHO cells that clonally express human α6 (28), selected for stable transfectants using hygromycin, and then cloned again for high expressers by cell sorting. The β1-4-1 mutant and CHO cells that express the mutant will be described elsewhere.
Methods
Plasmid Construction
The GST-NRG1 fusion protein used has the (GST)-GTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQEKAEELYQK sequence, which includes the entire EGF-like motif and the α domain. The cDNA fragment encoding the EGF-like domain was amplified using PCR with human NRG1 (the SMDF variant) cDNA (MGC-743, ATCC) as a template and further extended to include the α domain by overlap extension PCR to include the entire sequence described above. A BamHI restriction site was introduced at the 5′ end, and an EcoRI site was introduced at the 3′ end of the cDNA fragment. The resulting fragments were digested with BamHI and EcoRI and then subcloned into the BamHI/EcoRI sites of the pGEX-2T (Amersham Biosciences) vector. Site-directed mutagenesis was performed using the QuikChange method (29). The presence of the mutations was verified by DNA sequencing.
Protein Expression and Purification of the WT and 3KE Mutant NRG1
The WT NRG1 and its mutants were synthesized in E. coli BL21 (DE3) pLysS Rosetta gami 2 (Novagen) by inducing with 0.2 mm isopropyl β-d-thiogalactopyranoside for 2 h at room temperature. GST-NRG1 was purified by glutathione affinity chromatography from bacterial extracts as described in the manufacturer's instructions (GE Healthcare). To remove endotoxin, glutathione-agarose column was extensively washed with 1% Triton X-114 in PBS before eluting proteins with 5 mm glutathione. The purified GST fusion NRG1 preparations were more than 90% homogeneous in SDS-PAGE and were kept in 1 mm reduced glutathione, 2 mm oxidized glutathione in PBS at 4 °C to maintain disulfide bonds.
Cell Culture
MCF-7 and T47D human breast cancer cells and CHO cells were cultivated in DMEM (Invitrogen) supplemented with 10% (v/v) fetal bovine serum, 100 IU/ml penicillin, 100 mg/ml streptomycin, 0.25 mg/ml amphotericin B, and nonessential amino acids. K562 human erythroleukemia cells were cultivated in RPMI 1640 medium (Invitrogen) supplemented with 10% (v/v) fetal bovine serum, 100 IU/ml penicillin, 100 mg/ml streptomycin, 0.25 mg/ml amphotericin B, and nonessential amino acids.
Binding of Soluble αvβ3 and α6β4
Cell adhesion and soluble integrin binding assays were performed as described previously (30). NRG1 was immobilized to wells of 96-well microtiter plate overnight at 4 °C in 0.1 m carbonate buffer, pH 9.4. Remaining protein-binding sites were blocked by incubating with 200 μl of 0.1% BSA in PBS for 60 min at room temperature. Wells were then incubated with soluble integrin αvβ3 in 50 μl in Hepes-Tyrode's buffer supplemented with 1 mm Mn2+ at room temperature for 60 min. After rinsing the wells with the same buffer, bound integrins were determined by horseradish peroxidase (HRP)-conjugated anti-His tag mouse IgG and substrate 3,3′,5,5′-tetramethylbenzidine of HRP. We performed soluble α6β4 binding assays as described above, except that we used HRP-conjugated anti-Velcro antibody instead of anti-His tag antibody.
Competitive Binding Assay
GST fusion WT NRG1 was biotinylated by using EZ-Link Sulfo-NHS-LC-Biotin (Pierce) as described in the manufacturer's instructions. Briefly, GST -fusion WT NRG1 was incubated with sulfo-NHS-LC-Biotin for 1 h on ice, and the remaining free sulfo-NHS-LC-Biotin was quenched with Tris-HCl buffer, pH 8.0. Recombinant human ErbB3 Fc chimera (R&D Systems) was immobilized to wells of 96-well microtiter plates at 1 μg/ml coating concentration in 0.1 m NaHCO3, pH 9.4, overnight at 4 °C, and the remaining protein-binding sites were blocked by incubating with 0.1% BSA. Wells were then incubated with biotinylated GST fusion NRG1 in the presence of non-labeled GST, GST-WT NRG1, or GST-3KE for 3 h at room temperature. Bound biotinylated GST fusion NRG1 WT to wells was determined with streptavidin HRP conjugate and HRP substrate at 490 nm.
Proliferation Assay
MCF-7 cells (1 × 103 cells/well) were serum-starved overnight in serum-free DMEM and then stimulated with WT or mutant NRG1 for 48 h. Cell proliferation was measured using MTS assays.
Western Blot Analysis
MCF-7 cells grown to confluence were serum-starved in serum-free medium overnight and then treated with WT or 3KE mutant NRG1 (10 nm) for 5 or 30 min at 37 °C. Cells were washed with ice-cold PBS once and lysed with the lysis buffer (20 mm HEPES, pH 7.4, 100 mm NaCl, 10% glycerol, 1% Nonidet P-40, 1 mm MgCl2, 5 mm EDTA, 0.5% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, 20 mm NaF, 1 mm Na3VO4, protease inhibitor mixture (Sigma)). Protein concentrations in the cell lysates were determined using the BCA protein assay (Thermo Scientific). Equal amounts of cell proteins were analyzed by SDS-PAGE and transferred onto 0.45-mm pore-size polyvinylidene fluoride membrane (Millipore, Birellica, MA). The membrane was incubated with primary antibodies then HRP-conjugated secondary antibody and enhanced chemiluminescence detection reagents (Thermo Scientific). We detected and analyzed the luminescent signals on the blots within the linear range of detection using Fuji LAS 4000mini luminescent image analyzer (Fujifilm, Tokyo, Japan).
Co-immunoprecipitation
Five minutes after treatment with 10 nm WT or 3KE mutant NRG1, cells were washed with ice-cold PBS and lysed with lysis buffer. The cell lysate was incubated with anti-ErbB3 overnight at 4 °C. The immune complex was recovered by incubating with protein A-Sepharose (GE Healthcare) for 1 h at 4 °C and washed 3 times with wash buffer (20 mm HEPES, pH 7.4, 100 mm NaCl, 10% glycerol, 0.5% Nonidet P-40, 1 mm MgCl2, 1 mm phenylmethylsulfonyl fluoride, 20 mm NaF, 1 mm Na3VO4, protease inhibitor mixture (Sigma). The immunoprecipitates were analyzed by Western blotting with antibodies specific to integrin β3 or β4.
Other Methods
Docking simulation was performed as previously described (25, 30) using AutoDock3 and ADT (31). Surface plasmon resonance studies were performed as described (25, 30). We used PMV 1.54 (31) for graphics and Swiss-pdb viewer 4.01 (Swissprot) for superposing TGFα and NRG1. Statistical analysis was performed using GraphPad Prism 5.
RESULTS
Direct Binding of Integrin αvβ3 to NRG1
We recently reported that FGF1 directly interacts with αvβ3 and blocking of this interaction by mutating the integrin binding-site of FGF1 suppresses FGF1 signaling (30). These findings suggest that FGF receptor and integrins cross-talk through direct binding of FGF1 to integrins. It has been reported that integrin αvβ3 is involved in NRG1/ErbB signaling through cross-talk with ErbB (17–19). However, the specifics of this cross-talk are unclear. We hypothesized that integrins and ErbB receptors cross-talk through direct binding of NRG1 to integrins. To address this hypothesis we first tested if soluble recombinant integrin αvβ3 binds to the authentic EGF-like domain peptide (NRG1α, residues 177–241, synthesized in E. coli, R&D Systems) and GST fusion protein of NRG1, which has been widely used for studying neuregulin/ErbB signaling, in an ELISA-type assay. We found that soluble αvβ3 integrin bound to both NRG1 EGF-like domain peptide and GST-NRG1 fusion protein to the similar extent in a dose-dependent manner (Fig. 1a). Soluble αvβ3 only weakly bound to control GST. It is still possible that the integrin binding to the isolated EGF-like domain synthesized in E. coli may not represent the property of the EGF-like domain of multidomain NRG1 synthesized in eukaryotic cells. We, thus, used sensory and motor neuron-derived factor (SMDF), an isoform of NRG1 (296 amino acids, synthesized in baculovirus, R&D systems). Soluble αvβ3 bound to SMDF in a manner similar to those of isolated EGF-like domain and GST-NRG1 fusion protein (Fig. 1b). There was no appreciable difference in integrin binding function among the isolated EGF-like domain synthesized in bacteria and that from native NRG1 synthesized in eukaryotic cells and GST fusion protein of the EGF-like domain synthesized in bacteria. It is, thus, likely that the integrin binding function is the property of the EGF-like domain of NRG1. We used GST-NRG1 throughout the rest of this paper.
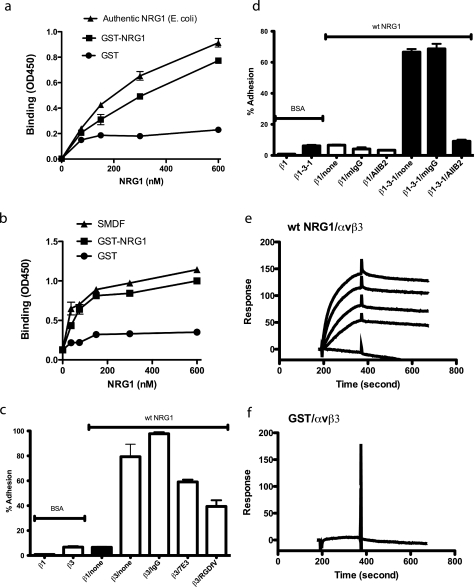
FIGURE 1.
Direct binding of the EGF-like domain of NRG1 to integrin αvβ3. a and b, the EGF-like domain of NRG1 bound to recombinant soluble αvβ3 in a dose-dependent manner in ELISA-type assays. The EGF-like domain peptide of NRG1 (NRG1α, synthesized in E. coli, R&D systems) (a), SMDF, an isoform of NRG1 (synthesized in eukaryotic cells, R&D systems) (b), GST fusion protein of NRG1, or control GST was immobilized to wells of 96-well microtiter plates. The concentrations of the coating solution are shown. Soluble recombinant integrin αvβ3 (5 μg/ml) was added to the wells in the presence of 1 mm Mn2+ and incubated for 2 h at room temperature. After washing the wells, bound αvβ3 was determined by using anti-β3 antibody and HRP-conjugated anti-mouse IgG. The data are shown as the means ± S.E. of triplicate experiments. c, specific adhesion of CHO cells that express human β3 (β3-CHO) to NRG1 is shown. Wells of 96-wellmicrotiter plate were coated with NRG1 (580 nm) or BSA, and the remaining protein-binding sites were blocked with BSA. Wells were incubated with β3-CHO cells or β1-CHO cells for 1 h at 37 °C in Tyrode's-HEPES buffer with 1 mm MgCl2. Bound cells were quantified. mAb 7E3 (to human β3, 10 μg/ml) and cyclic RGDfV (specific antagonist to αvβ3, 10 μm) blocked the adhesion of β3-CHO cells to WT NRG1. IgG represents purified mouse IgG used as a control. The data are shown as the means ± S.E. of triplicate experiments. d, adhesion of CHO cells that express human β1 (β1-CHO) or β1-3-1 (β1-3-1-CHO) to NRG1 is shown. The β1-3-1 mutation changes the specificity of β1 integrins to that of β3 integrins. Cell adhesion was performed as described above. β1-3-1-CHO cells adhered to WT NRG1, and this binding was blocked by anti-human β1 antibody AIIB2 (10 μg/ml) but not by purified mouse IgG (mIgG). BSA as a control was coated instead of NRG1. The data are shown as the means ± S.E. of triplicate experiments. The results suggest that NRG1 specifically binds to β1-3-1 (as αvβ1-3-1) but not to WT β1 (as αvβ1). e and f, surface plasmon resonance studies of NRG1-αvβ3 interaction are shown. Soluble integrin αvβ3 was immobilized to a sensor chip, and the binding of WT NRG1 and control GST (concentrations at 1000, 500, 250, 125, and 0 nm) was analyzed in the presence of 1 mm MnCl2. KD was calculated as 1.36 × 10−7 m for WT NRG-1. Control GST did not show significant binding.
We next tested if cell-surface integrins bind to immobilized NRG1. CHO cells that express recombinant β3 (β3-CHO) adhered to NRG1, whereas CHO cells that express recombinant human β1 (β1-CHO) did not (Fig. 1c). K562 erythroleukemic cells expressing recombinant αvβ3 (αvβ3-K562) adhered in a dose-dependent manner, but mock-transfected K562 cells did not (see below, Fig. 3). These results suggest that the EGF-like domain of NRG1 directly interacts with integrin αvβ3. Furthermore, anti-β3 mAb (7E3) and cyclic RGDfV (an-αvβ3-specific antagonist) reduced the adhesion (Fig. 1c). These results suggest that binding of NRG1 to αvβ3 is specific.
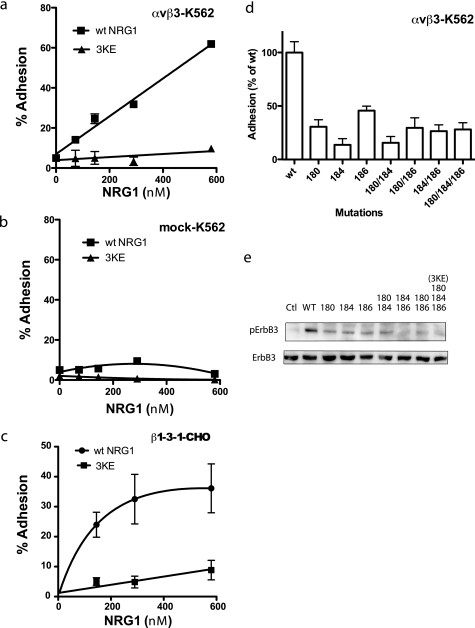
FIGURE 3.
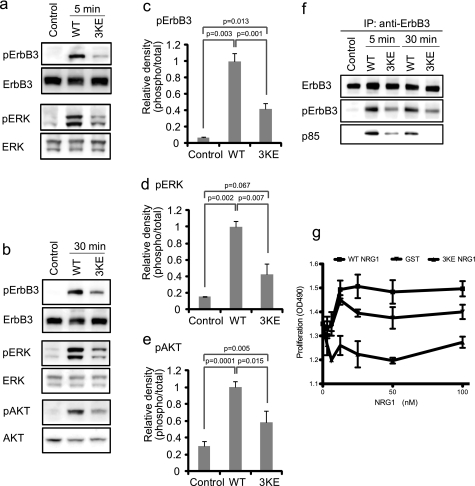
The 3KE mutant is defective in integrin binding. a and b, the 3KE mutant is defective in binding to αvβ3-K562 cells. Adhesion assays was performed using αvβ3-K562 (a) or mock-transfected cells (b) as described in Fig. 1, except that 1 mm MnCl2 was used. The data are shown as means ± S.E. of triplicate experiments. We obtained similar results using β3-CHO cells. c, the 3KE mutant is defective in binding to β1-3-1 integrin. Adhesion assays was performed using β1-3-1-CHO cells as described above. The data are shown as the means ± S.E. of triplicate experiments. The results suggest that the 3KE mutant is defective in binding to αvβ1-3-1, which mimics αvβ3 in ligand binding. d, mutating the Lys residues alone or in combination suppresses integrin binding. Adhesion assays were performed as described above. We coated wells with GST fusion proteins of NRG1 EGF-like domain (580 nm) and used αvβ3-K562 cells. The numbers represent the positions of Lys residues mutated. e, shown is the signaling function of single and double mutants. We determined the ability of the mutants to induce ErbB3 phosphorylation. MCF-7 cells were serum-starved overnight and stimulated with NRG1(10 nm) for 30 min. Cell lysates were analyzed by Western blotting with antibodies specific to phospho-ErbB3 or ErbB3. Ctl, control.
The integrin β subunit possesses an I-like domain that plays a critical role in ligand binding (32). We have shown that when a disulfide-linked five-residue sequence of β1 I-like domain (residues 177–183) of αvβ1 is switched with a corresponding sequence in β3 integrin (designated the β1-3-1 mutant), ligand binding specificity of the mutated integrin αvβ1-3-1 is altered to that of αvβ3 (27). Hence, the loop was designated “the specificity loop.” The β1-3-1 mutant (as αvβ1-3-1) bound to vitronectin and fibrinogen, but WT β1 (as αvβ1) did not (27). The crystal structure of αvβ3 showed that the specificity loop is located in the ligand binding (RGD binding) site and undergoes marked conformational changes (1 angstrom shift) upon RGD binding to αvβ3 (33). To determine whether NRG1 binds to the ligand-binding site of αvβ3 common to other known αvβ3 ligands (e.g. vitronectin and fibrinogen), we used the β1-3-1 mutant (27). CHO cells that express β1-3-1 (designated β1-3-1-CHO cells) bound to NRG1, but WT β1-CHO did not (Fig. 1d). The adhesion of β1-3-1-CHO cells to NRG1 was blocked by anti-β1 antibody AIIB2 (Fig. 1d) (note: the β1-3-1mutant is still more than 99% human β1, and therefore, its function is blocked by anti-human β1 mAb such as AIIB2 (27)). These findings suggest that NRG1 binds to the ligand-binding site of αvβ3 and that the specificity loop plays a role in NRG1 binding to αvβ3.
In surface plasmon resonance studies NRG1 GST fusion protein in solution bound to immobilized soluble αvβ3 with an apparent KD of 1.36 × 10−7 m, whereas control GST did not show significant binding (Fig. 1, e and f). This KD is similar to those of other known integrin ligands. These results suggest that the EGF-like domain of NRG1 interacts with integrin αvβ3 in a manner similar to that of other integrin ligands.
Integrin Binding-defective Mutants of NRG1
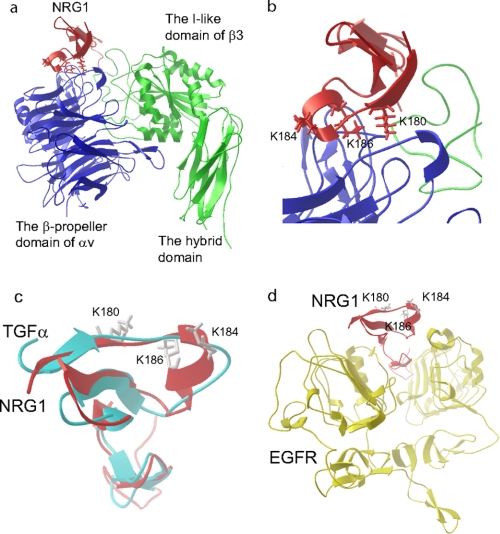
To locate the integrin-binding site in NRG1 and to generate integrin-binding-defective mutants of NRG1, we used docking simulation and site-directed mutagenesis. Docking simulation of the interaction between integrin αvβ3 and the EGF-like domain of NRG1 (PDB code 1HAF) predicts that NRG1 binds to integrin αvβ3 (PDB code 1LG5) with a high affinity (docking energy −23.5 kcal/mol) (Fig. 2a), which is consistent with our binding results. The predicted integrin binding interface includes the Lys residues at positions 180, 184, and 186 (Fig. 2b), which are conserved in NRG1α and NRG1β. Several amino acid residues from both αv and β3 subunits are predicted to bind to NRG1 (Table 1). Most amino acid residues are involved in binding to other αvβ3 ligands (34–38), suggesting that the predicted NRG1-binding site may be common to that for other ligands. To assess the position of the Lys residues in the ERG1-ErbB complex, we used the TGFα-EGFR complex, PDB code 1MOX, because NRG1 and TGFα are homologous (Fig. 2c). We replaced the TGFα with NRG1 by superposing them (Fig. 2d). The model predicted that the Lys residues at positions 180, 184, and 186 are not in the EGFR-binding site of NRG1.
FIGURE 2.
Docking simulation of αvβ3-NRG1 interaction. a, a model of NRG1-integrin αvβ3 interaction predicted by docking simulation by using AutoDock3 is shown. The headpiece of integrin αvβ3 (PDB code 1LG5) was used as a target. The model predicts that the EGF-like domain of NRG1 (PDB code 1HAF, blue) binds to the RGD-binding site of the integrin αvβ3 headpiece (green and red). b, the Lys residues at positions 180, 184, and 186 of NRG1α are located at the interface between NRG1 and αvβ3 and were selected for mutagenesis studies. c, superposition of TGFα and NRG1 is shown. d, the Lys residues at positions 180, 184, and 186 of NRG1 are not located in the binding site for EGFR. We replaced TGFα in the TGFα-EGFR complex (PDB code 1MOX) with NRG1 (PDB code 1HAF) by superposing. ErbB3 or ErbB4 is homologous to EGFR.
TABLE 1.
Amino acid residues in integrin αvβ3 that are involved in NRG1 binding
Amino acid residues in integrin αvβ3 within 6 Å of NRG1 in the docking model were identified using Swiss-pdb viewer.
| αv | β3 |
|---|---|
| Asp-84, Arg-115, Met-118, Lys-119, Gln-120 | Pro-169, Pro-170, Glu-171, Ala-172, Leu-173, Tyr-178, Asp-179, Met-180, Lys-181 |
| Gln-145, Asp-146, Ile-147, Asp-148, Ala-149, Asp-150, Gly-151, Gly-151, Gln-152 | |
| Phe-177, Tyr-178, Gln-190, Gln-182 | |
| Gln-207, Ala-209, Thr-210, Arg-211, Thr-212, Ala-213, Gln-214, Ala-215, Asp-218 |
We mutated the Lys residues simultaneously to Glu (designated the Lys-180/Lys-184/Lys-186 to Glu (3KE) mutation). We tested if the 3KE mutant is defective in binding to integrin αvβ3 in cell adhesion assays using αvβ3-K562 (Fig. 3a), control mock-transfected K562 cells (Fig. 3b), and β1-3-1-CHO cells (Fig. 3c). We found that αvβ3 and αvβ1-3-1 mutant integrins bound to WT NRG1. But there was little or no adhesion to the 3KE mutant, suggesting that the 3KE mutant is defective in binding to αvβ3. We obtained similar results using β3-CHO cells (data not shown). We were not able to obtain a KD value for 3KE to integrin αvβ3 due to low binding in surface plasmon resonance (data not shown).
To obtain more insight into which of the three Lys residues is critical for integrin binding we generated several NRG1 mutants in which one or two of the Lys residues is mutated to Glu. We determined their ability to bind to αvβ3 (Fig. 3d). We found that mutating each of the Lys residues markedly reduced integrin binding, and mutating more Lys residues did not further enhance the inhibition in integrin binding. These findings suggest that each of the three Lys residues is involved in integrin binding. We tested if these mutants can induce ErbB3 phosphorylation in MCF-7 human breast cancer cells. MCF-7 was chosen because NRG1 stimulation of MCF-7 cells induces intracellular signaling via signaling of ErbB2-ErbB3 heterodimers (39). We found that the all mutants were defective in inducing ErbB3 phosphorylation (Fig. 3e). We analyzed 3KE as a representative NRG1 mutant in more detail.
The 3KE Mutant Binds to ErbB3
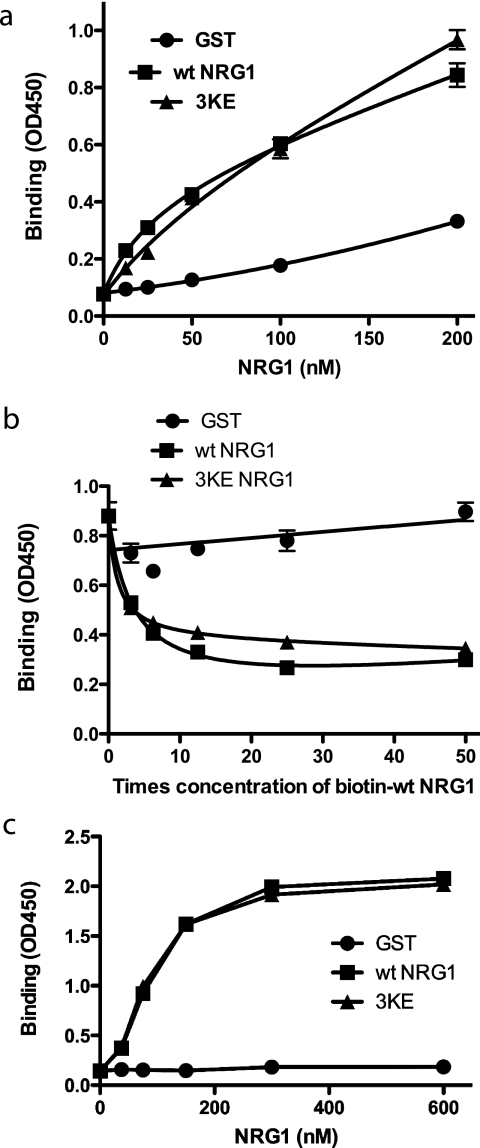
The docking simulation and our model (Fig. 2d) predict that the integrin-binding site in NRG1 is distinct from the ErbB-binding site. We tested if the 3KE mutation affects the binding of NRG1 to ErbB3 using recombinant soluble ErbB3. WT and 3KE mutant NRG1 bound to immobilized soluble ErbB3 comparably in an ELISA-type assay (Fig. 4a). Also, WT and 3KE NRG1 competed for binding of biotinylated WT NRG1 to immobilized soluble ErbB3 at comparable levels in competitive binding assays (Fig. 4b). In a reciprocal experiment soluble ErbB3 bound to immobilized NRG1 (Fig. 4c). There was no difference between immobilized WT and 3KE NRG1 in binding to soluble ErbB3. These findings suggest that the 3KE mutation has minimal effects on NRG1-ErbB3 interaction.
FIGURE 4.
The 3KE mutant of NRG1 binds to ErbB3. a, binding of the 3KE mutant of NRG1 to recombinant ErbB3 is shown. Recombinant soluble ErbB3 Fc fusion protein (R&D system) was coated onto wells of a 96-well microtiter plate (1 μg/ml). NRG1 WT or 3KE mutant was added to the wells and incubated for 1 h at room temperature. GST was used as a control. After washing the wells, bound GST NRG1 was determined by using anti-GST antibody HRP conjugate. The results suggest that the 3KE mutant of NRG1 binds to ErbB3 at levels nearly comparable with WT NRG1. The data are shown as the means ± S.E. of triplicate experiments. b, shown is a competitive binding assay. Recombinant soluble ErbB3 Fc fusion protein was coated onto wells of 96-well microtiter plate (1 μg/ml). Binding of biotin-labeled NRG1 WT (20 nm) in the presence of increasing concentrations of NRG1 WT, 3KE, or GST is shown. After washing the wells, bound biotin-labeled NRG1 WT was determined by using streptavidin HRP conjugate. The data are shown as the means ± S.E. of triplicate experiments. The results suggest that the 3KE mutant of NRG1 binds to ErbB3 at levels comparable with WT NRG1. c, binding of soluble ErbB3-Fc to immobilized NRG1 is shown. We immobilized NRG1 WT and 3KE proteins to the wells of the 96-well microtiter plate at the indicated coating concentrations and incubated with soluble ErbB3-Fc (1 μg/ml) as described above. Bound ErbB3 was detected using HRP-conjugated anti-His6 tag antibodies and peroxidase substrate. ErbB3-Fc has a His6 tag.
The 3KE Mutant Is Defective in Inducing Intracellular Signaling
We studied the ability of the 3KE mutant to induce intracellular signaling in MCF-7 cells. WT NRG1 induced phosphorylation of ErbB3, AKT, and ERK1/2 in MCF-7 cells, but the 3KE mutant induced phosphorylation of these proteins at much lower levels than WT NRG1 (Fig. 5, a–e). Also, WT NRG1 induced recruitment of the p85 subunit of phosphatidylinositol 3-kinase (PI3K) to ErbB3, whereas the 3KE mutant was defective in this function (Fig. 5f). We obtained essentially the same results using another human breast cancer cell line T47D (data not shown). These results suggest that the 3KE mutant is defective in inducing NRG1/ErbB intracellular signaling, whereas 3KE still binds to ErbB3.
FIGURE 5.
Effect of the 3KE mutation on NRG1 signaling. a and b, the 3KE mutant of NRG1 is defective in inducing ErbB3 phosphorylation, AKT activation, and ERK1/2 activation. MCF-7 cells were serum-starved overnight and stimulated with 10 nm WT and the 3KE mutant of NRG1 for 5 min (a) or 30 min (b). Cell lysates were analyzed by Western blotting. Data are representative of three independent experiments. c–e, levels of phosphorylation were quantified using a luminescence analyzer from triplicate experiments. Data were normalized using WT NRG1 as 1. f, recruitment of p85 of PI3K is shown. Cells were stimulated with WT NRG1 or 3KE, ErbB3 was immuno-purified (IP) from lysates using anti-ErbB3, and immuno-purified materials were analyzed by Western blotting. The p85 subunit of PI3K was detected in lysates of cells stimulated with WT NRG1. Much lower levels of p85 were detected in cells stimulated with 3KE. Data shown are representative of three independent experiments. g, shown is the effect of WT and 3KE NRG1 on the proliferation of MCF-7 cells. Human MCF-7 breast cancer cells were serum-starved overnight and cultured for 48 h with WT or 3KE mutant NRG1. GST was used as a control. Cell number was measured by MTS assays (OD490). The data are shown as the means ± S.E. (n = 3). p < 0.05 by 2-way ANOVA in each case. Data are representative of three independent experiments performed.
NRG1 Induces Co-precipitation of Integrins and ErbB3, Whereas 3KE Is Defective in This Function
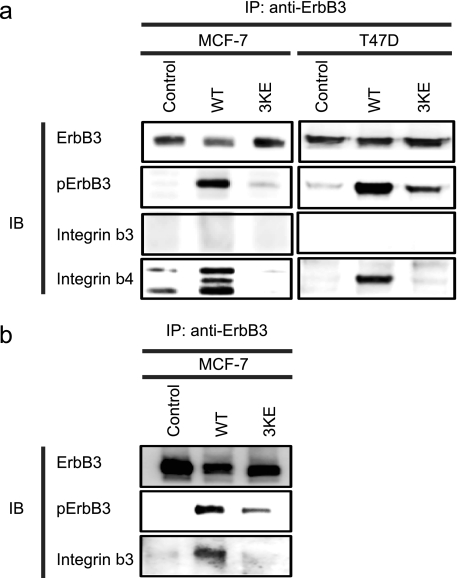
Our results so far suggest that direct binding of NRG1 to integrins is involved in NRG1 intracellular signal transduction. We demonstrated that NRG1 and αvβ3 interact in β3-CHO cells (Figs. 1–3), but it is unclear which integrins are involved in NRG1 signaling in MCF-7 cells. It is predicted that, if NRG1 binds to both ErbB3 and integrins, NRG1 mediates ternary complex (integrin-NRG1-ErbB3) formation. To address this prediction, we stimulated serum-starved MCF-7 cells with WT or 3KE NRG1 and immuno-purified ErbB3 from cell lysates and analyzed the immuno-purified materials by Western blotting using antibodies specific to different integrin β subunit. Unexpectedly, we discovered that stimulation with WT NRG1 markedly induced co-precipitation of integrin β4 with ErbB3 in 5 min in MCF-7 cells, whereas we detected only weak signals of β3 under the same conditions. But we detected co-precipitation of β3 when we used 5 times more lysates (Fig. 6b). This is consistent with the report that the expression of β3 in MCF-7 cells is very low (40). Our results suggest that α6β4 integrin plays a major role in NRG signaling in MCF-7 cells. We did not detect co-precipitation of β1 with ErbB3 under the conditions used in MCF-7 cells treated with WT NRG1 (data not shown). The middle band in MCF-7 is β4, based on its size. The higher and lower bands are nonspecific bands, because they are present in immuno-purified materials from non-treated cells (control). Likewise only β4 was detected using T47D human breast cancer cells (Fig. 6a). Notably, 3KE was defective in co-precipitation of β3 and β4 (Fig. 6, a and b). This indicates that the integrin-NRG1-ErbB3 ternary complex formation is dependent on the ability of NRG1 to bind to integrins. Taken together our results suggest that NRG1 mediates ternary complex formation between ErbB3 and integrin β3 or β4, but 3KE is defective in this function. This is consistent with a model in which NRG1 directly binds to ErbB3 and integrins, and this interaction is critically involved in NRG1/ErbB3 signaling.
FIGURE 6.
WT NRG1 induced co-precipitation of integrin β3 and β4 with ErbB3, whereas 3KE is defective in this function. a, MCF-7 cells were for serum-starved 24 h and stimulated with 10 nm NRG1 WT or 3KE for 5 min. We used 0.7 mg of protein of cell lysate for immunoprecipitation with anti-ErbB3 antibody. Immunoprecipitated materials were analyzed by Western blotting (IB). The levels of ErbB3 phosphorylation were less with the 3KE mutant. Integrin β4 was co-immunoprecipitated with the ErbB3 upon stimulation with WT NRG1, whereas the 3KE mutant was defective in this function. Integrin β3 was not detected under the conditions used. The three bands in co-precipitated β4 in MCF-7 are considered to be (from the top) α6β4 heterodimer, intact β4, and a fragment of β4 based on size. Data are representative of three independent experiments. b, we detected co-precipitation (IP) of β3 with ErbB3 when we used 5× more MCF-7 lysate for co-precipitation experiments using WT NRG1. Data are representative of three independent experiments.
NRG1 Binds to Integrin α6β4
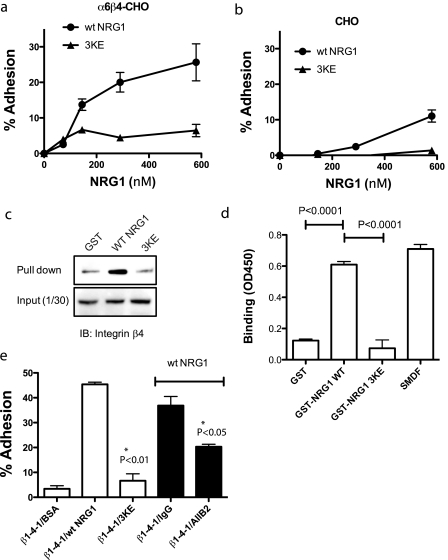
To test if integrin α6β4 binds to NRG1, we used CHO cells that express recombinant human α6β4 (α6β4-CHO cells). α6β4-CHO cells clonally express human α6 and β4 (data not shown). α6β4-CHO cells adhered well to WT NRG1 but adhered to 3KE NRG1 at much lower levels (Fig. 7a). As a control, mock-transfected CHO cells did not adhere to either WT or mutant NRG1. Monoclonal antibodies specific to human α6 (135–13c) or β4 (439–9B) did not affect adhesion of α6β4-CHO cells to WT NRG1 (data not shown), but the specificity of the NRG1 binding to β4 is shown in co-precipitation experiments (Fig. 7b). We incubated WT NRG1 or 3KE with a detergent lysate of α6β4-CHO cells and analyzed the bound proteins by Western blotting. β4 bound to WT NRG1 markedly more than to 3KE or control GST, indicating that integrin α6β4 bound to NRG1 (Fig. 7c). It is, however, still unclear if NRG1 directly interacts with α6β4. To address this question, we performed ELISA-type binding assays using recombinant soluble α6β4, which lacks transmembrane and cytoplasmic domains of α6β4 (26). We found that soluble α6β4 bound to WT NRG1 and SMDF to a similar extent but did not show significant binding to 3KE (Fig. 7d). GST was used as a negative control. These results suggest that α6β4 directly binds to the EGF-like domain of NRG1 and that this binding is not affected by whether it is synthesized in bacteria or in eukaryotic cells or whether it is synthesized as a fusion protein or as a native multidomain form.
FIGURE 7.
α6β4 binds to WT NRG1, but not to 3KE, through the WPNSDP sequence of β4. WT NRG1 was immobilized to wells of 96-well microtiter plates at 0–580 nm coating concentrations. The remaining protein-binding sites were blocked with BSA. CHO cells that express recombinant α6β4 (α6β4-CHO) (a) or control mock-transfected CHO cells (b) were incubated in the wells for 1 h at 37 °C. Bound cells were measured after gently rinsing the wells. The results suggest that NRG1 binds to integrin α6β4. 3KE NRG1 only weakly interacts with α6β4 (*, p < 0.05.). Data are shown as the means ± S.E. of triplicate experiments. c, pulldown assays are shown. To determine the specificity of binding to α6β4, we incubated GST, GST-WT NRG1, or GST-3KE NRG1 (10 μg) with lysate of α6β4-CHO cells overnight at 4 °C and recovered the material bound to GST proteins using glutathione-Sepharose. We found that WT NRG1 pulled down much more β4 than control GST, and 3KE did not. Data are representative of three independent experiments performed. IB, immunoblot. d, binding of recombinant soluble α6β4 to NRG1 is shown. WT and mutant GST-NRG1 and SMDF were immobilized as described above and incubated with recombinant soluble α6β4 (2 μg/ml), and bound α6β4 was detected using HRP-conjugated anti-Velcro antibody as described under “Experimental Procedures.” e, the β1-4-1 mutant, in which the specificity loop of β1 is replaced with the corresponding amino acid residues of β4, specifically binds to WT NRG1 but not to 3KE. We performed adhesion assays as described above. We coated wells with GST fusion proteins of WT NRG1 and 3KE (580 nm). We used CHO cells that express the β1-4-1 mutant and DMEM for adhesion assays. The data are shown as the means ± S.E. (n = 3).
Does the disulfide-linked loop in the β subunit play a role in recognizing NRG1 like β3? The disulfide-linked specificity loop is not present in β4 and is replaced with remnant residues (27). To test the role of the region corresponding to the specific loop in β4, we generated a β1 mutant in which the CTSEQNCTT sequence that contains the specificity loop was replaced with the WPNSDP sequence of β4 (designated the β1-4-1 mutant). The β1-4-1 mutant was stably expressed in CHO cells (designated β1-4-1-CHO cells), and stable transfectants were further cloned for high expressers.3 We found that β1-4-1-CHO cells adhered to WT NRG1, and inhibitory anti-human β1 mAb AIIB2 suppressed the adhesion of β1-4-1-CHO cells to WT NRG1 (Fig. 7e). However, β1-4-1-CHO cells only weakly bound to 3KE. These results suggest that the region of β4 that corresponds to the specificity loop is involved in α6β4 binding to WT NRG1, although the disulfide-linkage is not present, and that the conserved Lys residues of NRG1 are involved in α6β4 binding, as in αvβ3 binding.
Taken together, our results suggest that NRG1 binds to αvβ3 and α6β4 and that α6β4 plays a major role in NRG1/ErbB signaling in MCF-7 and T47D cells. These integrins make a ternary complex (ErbB3-NRG1-integrin) when cells were stimulated with WT NRG1. Because 3KE is defective in inducing NRG1/ErbB signaling (although 3KE still binds to ErbB3) and in inducing ternary complex formation, we propose that direct integrin binding to NRG1 and subsequent ternary complex formation is critical for NRG1/ErbB signaling.
DISCUSSION
In the present study we establish that the EGF-like domain of NRG1 directly binds to integrin αvβ3 and α6β4. Because the isolated EGF-like domain synthesized in E. coli, multidomain NRG1 isoform SMDF synthesized in eukaryotic cells, and GST fusion protein of the EGF-like domain bound to integrin αvβ3 to the similar extent, the integrin binding is a property of the EGF-like domain of NRG1. KD of NRG1 for αvβ3 (1.36 × 10−7 m) is similar to those for known integrin ligands. The NRG1 binding to αvβ3 was blocked by antibody 7E3 specific to β3, which has been mapped in the specific loop of β3 (41), and the RGDfV peptide, a specific inhibitor of αvβ3. We also demonstrated that NRG1 bound to the β1-3-1 mutant of β1 integrin, suggesting that the specificity loop of β3 is involved in αvβ3 binding. It is highly likely that NRG1 binds to a binding site common to other known αvβ3 ligands. The predicted NRG1-binding site, however, does not include the metal ion-dependent activation site (MIDAS) region of the β3 subunit (Table 1) that is commonly involved in integrin-ligand interaction. It is unclear if this is a unique property of NRG1-integrin interaction at this point. It would be interesting to address this prediction in future experiments.
The EGF-like domain of the NRGs is known to be sufficient to specifically activate ErbB receptors and induce cellular responses in culture through binding to ErbB receptors (2–4, 12, 42–44). The EGF-like domain is generally synthesized in E. coli mostly as GST fusion protein and, thus, does not undergo posttranslational modification. There is one predicted glycosylation site in the EGF-like domain of NRG1 (Asn-204). Interestingly, this site is outside of the integrin-binding site and ErbB-binding site of the domain. It is, thus, unlikely that glycosylation at Asn-204 in eukaryotic cells affects integrin or ErbB binding of the EGF-like domain. We identified Lys residues at positions 180, 184, and 186 at the N terminus of the EGF-like domain that are critical for integrin binding. It appears that all these residues are involved in integrin binding, as mutating individual residues suppressed integrin binding. Notably, the integrin binding-defective NRG1 mutants tested were all defective in inducing ErbB3 phosphorylation, suggesting that the direct binding of NRG1 to integrins may be involved in NRG signaling. To address this question, we extensively characterized the 3KE NRG1 mutant, in which the three Lys residues in the predicted integrin-binding site were mutated to Glu. Although the 3KE mutant was defective in integrin binding, it bound to ErbB3 at the level comparable with that of WT NRG1. These findings are consistent with the fact that the region that includes the three Lys residues is not involved in ErbB binding based on the crystal structure of the TGFα-EGFR complex (PDB code 1MOX) (Fig. 2, c and d). Notably, WT NRG1 induced ErbB3 phosphorylation and ERK1/2 and AKT activation in MCF-7 cells, whereas the 3KE mutant of NRG1 induced ErbB3 phosphorylation and ERK1/2 and AKT activation at much lower levels. These results suggest that ErbB3 and integrins cross-talk through direct binding to NRG1 and that this process is required for NRG1/ErbB signaling.
How does the ability of NRG1 to bind to integrins relate to the signaling function of NRG1? The present finding predicts that integrins and ErbB3 can simultaneously bind to NRG1. Consistently, we demonstrated that WT NRG1 induced co-precipitation of ErbB3 and α6β4, whereas co-precipitation of αvβ3 was at a much lower level in MCF-7 and T47D cells. We suspect that the lower levels of co-precipitation of αvβ3 with ErbB3 probably is a reflection of the low expression levels of this integrin in these cell types. We propose a model of integrin-ErbB cross-talk in which NRG1 directly interacts with both integrins and ErbB3 and mediates ternary complex formation on the cell surface. Notably, the integrin binding-defective 3KE mutant was defective in inducing ternary complex formation, suggesting that 3KE cannot recruit integrins to the ErbB-NRG1 complex and the intracellular signaling through integrin signaling pathways may be disrupted. Although NRG1 at 10 nm in solution can induce intracellular signals, how does NRG1 at such a low concentration bind to integrins that have much lower affinity to NRG1? One possibility is that NRG1 (e.g. 10 nm in solution) binds to high affinity ErbB receptors (e.g. ErbB3) and is concentrated on the cell surface. Therefore, integrins may be able to bind to the receptor-bound NRG1 on the cell surface, as NRG1 in the NRG-ErbB complex still exposes the integrin-binding site.
It has been reported that α6β4 promotes tumor cell motility and invasiveness by activating the PI3K/AKT pathway or small GTPase Rac1/nuclear factor κB (45, 46). α6β4 is distinct from other integrin receptors because the β4 subunit contains a 1000-amino acid cytoplasmic domain (15). This cytoplasmic domain is essential for coupling α6β4 to the cytoskeleton and for its ability to activate intracellular signaling pathways (47). It has been proposed that α6β4 becomes competent for signaling in response to signals that disrupt hemidesmosomes, resulting in the release of α6β4 from interactions with the cytokeratin cytoskeleton and de novo interaction with the actin cytoskeleton and signaling molecules in the apical region of the cells (48). In this signaling-competent state, α6β4 cooperates with growth factor receptors and other surface molecules to amplify intracellular signaling pathways through the β4 cytoplasmic domain (49–51). α6β4 integrin associates with ErbB2 in mammary cells and cooperates with ErbB2 to promote PI3K-dependent invasion and survival (49). In mouse mammary tumor virus-Neu mice, the introduction of a targeted deletion of the β4 cytoplasmic domain revealed that β4 integrin signaling plays a role in mammary tumor progression (52). The present study provides evidence that NRG1 (in addition to laminins) is a ligand for α6β4 and that this interaction plays a role in NRG1/ErbB signaling in cancer cells through making the α6β4-NRG1-ErbB3 ternary complex. The present results demonstrated that α6β4 in the apical region induces intracellular signaling through direct binding to NRG1. This is a drastic change in biological function of this integrin in cancer cells and migrating cells and is highly relevant to cancer initiation and progression.
The present study for the first time describes that NRG1 directly binds to integrins and this interaction is involved in NRG1/ErbB signaling using a mutant of NRG1 that cannot bind to integrins. It would be interesting to test if other members of the EGF family directly interact with integrins and thereby mediate cross-talk between integrins and EGFRs in future studies. Several other growth factors have been shown to directly interact with integrins (16), whereas the significance of these interactions has not been fully established. We have demonstrated that FGF1 directly binds to αvβ3 and the ability of FGF1 to bind to αvβ3 is required for FGF signaling (30). It is possible that other members of the FGF family bind to integrins and mediate cross-talk between integrins and FGF receptors. Likewise, we have demonstrated that insulin-like growth factor-1 directly interacts with integrins and this interaction is required for insulin-like growth factor 1 receptor activation (53). It would be interesting to test if the direct interaction between growth factors and integrins can be a general mechanism for growth factor-integrin cross-talk.
This work was supported, in whole or in part, by National Institutes of Health Grants CA131015, CA113298, CA093373, and U19 NCDDG grant (to Yoshikazu Takada). This work was also supported by Department of Defense Grant W81XWH-10-1-0312 (to Yoshikazu Takada) and Grant-in-aid for Scientific Research 20370046 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to K. S.).
J. Saegusa, M. Fujita, K. Ieguchi, M. Wu, Y. K. Takada, and Y. Takada, in preparation.
- NRG1
- neuregulin-1
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- SMDF
- sensory and motor neuron-derived factor
- EGFR
- EGF receptor.
REFERENCES
- 1.Peles E., Bacus S. S., Koski R. A., Lu H. S., Wen D., Ogden S. G., Levy R. B., Yarden Y. (1992) Cell 69, 205–216 [DOI] [PubMed] [Google Scholar]
- 2.Carraway K. L., 3rd, Weber J. L., Unger M. J., Ledesma J., Yu N., Gassmann M., Lai C. (1997) Nature 387, 512–516 [DOI] [PubMed] [Google Scholar]
- 3.Chang H., Riese D. J., 2nd, Gilbert W., Stern D. F., McMahan U. J. (1997) Nature 387, 509–512 [DOI] [PubMed] [Google Scholar]
- 4.Zhang D., Sliwkowski M. X., Mark M., Frantz G., Akita R., Sun Y., Hillan K., Crowley C., Brush J., Godowski P. J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 9562–9567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falls D. L. (2003) Exp. Cell Res. 284, 14–30 [DOI] [PubMed] [Google Scholar]
- 6.Schmitt A., Parlapani E., Gruber O., Wobrock T., Falkai P. (2008) Eur. Arch. Psychiatry Clin. Neurosci. 258, 35–39 [DOI] [PubMed] [Google Scholar]
- 7.Lemmens K., Doggen K., De Keulenaer G. W. (2007) Circulation 116, 954–960 [DOI] [PubMed] [Google Scholar]
- 8.Montero J. C., Rodríguez-Barrueco R., Ocaña A., Díaz-Rodríguez E., Esparís-Ogando A., Pandiella A. (2008) Clin. Cancer Res. 14, 3237–3241 [DOI] [PubMed] [Google Scholar]
- 9.Meyer D., Birchmeier C. (1995) Nature 378, 386–390 [DOI] [PubMed] [Google Scholar]
- 10.Meyer D., Yamaai T., Garratt A., Riethmacher-Sonnenberg E., Kane D., Theill L. E., Birchmeier C. (1997) Development 124, 3575–3586 [DOI] [PubMed] [Google Scholar]
- 11.Erickson S. L., O'Shea K. S., Ghaboosi N., Loverro L., Frantz G., Bauer M., Lu L. H., Moore M. W. (1997) Development 124, 4999–5011 [DOI] [PubMed] [Google Scholar]
- 12.Breuleux M. (2007) Cell. Mol. Life Sci. 64, 2358–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q., Ahmed S., Loeb J. A. (2004) Cancer Res. 64, 7078–7085 [DOI] [PubMed] [Google Scholar]
- 14.Miyamoto S., Teramoto H., Gutkind J. S., Yamada K. M. (1996) J. Cell Biol. 135, 1633–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hynes R. O. (2002) Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 16.Takada Y., Ye X., Simon S. (2007) Genome Biol. 8, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang C. K., Perez C., Grunt T., Waibel C., Cho C., Lupu R. (1996) Cancer Res. 56, 3350–3358 [PubMed] [Google Scholar]
- 18.Tsai M. S., Shamon-Taylor L. A., Mehmi I., Tang C. K., Lupu R. (2003) Oncogene 22, 761–768 [DOI] [PubMed] [Google Scholar]
- 19.Atlas E., Cardillo M., Mehmi I., Zahedkargaran H., Tang C., Lupu R. (2003) Mol. Cancer Res. 1, 165–175 [PubMed] [Google Scholar]
- 20.Vellon L., Menendez J. A., Lupu R. (2005) Oncogene 24, 3759–3773 [DOI] [PubMed] [Google Scholar]
- 21.Lu S., Simin K., Khan A., Mercurio A. M. (2008) Clin. Cancer Res. 14, 1050–1058 [DOI] [PubMed] [Google Scholar]
- 22.Falcioni R., Kennel S. J., Giacomini P., Zupi G., Sacchi A. (1986) Cancer Res. 46, 5772–5778 [PubMed] [Google Scholar]
- 23.Borradori L., Sonnenberg A. (1999) J. Invest. Dermatol. 112, 411–418 [DOI] [PubMed] [Google Scholar]
- 24.Lipscomb E. A., Dugan A. S., Rabinovitz I., Mercurio A. M. (2003) Clin. Exp. Metastasis 20, 569–576 [DOI] [PubMed] [Google Scholar]
- 25.Saegusa J., Akakura N., Wu C. Y., Hoogland C., Ma Z., Lam K. S., Liu F. T., Takada Y. K., Takada Y. (2008) J. Biol. Chem. 283, 26107–26115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishiuchi R., Takagi J., Hayashi M., Ido H., Yagi Y., Sanzen N., Tsuji T., Yamada M., Sekiguchi K. (2006) Matrix Biol. 25, 189–197 [DOI] [PubMed] [Google Scholar]
- 27.Takagi J., Kamata T., Meredith J., Puzon-McLaughlin W., Takada Y. (1997) J. Biol. Chem. 272, 19794–19800 [DOI] [PubMed] [Google Scholar]
- 28.Eto K., Puzon-McLaughlin W., Sheppard D., Sehara-Fujisawa A., Zhang X. P., Takada Y. (2000) J. Biol. Chem. 275, 34922–34930 [DOI] [PubMed] [Google Scholar]
- 29.Wang W., Malcolm B. A. (1999) Biotechniques 26, 680–682 [DOI] [PubMed] [Google Scholar]
- 30.Mori S., Wu C. Y., Yamaji S., Saegusa J., Shi B., Ma Z., Kuwabara Y., Lam K. S., Isseroff R. R., Takada Y. K., Takada Y. (2008) J. Biol. Chem. 283, 18066–18075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanner M. F. (1999) J. Mol. Graph. Model. 17, 57–61 [PubMed] [Google Scholar]
- 32.Luo B. H., Carman C. V., Springer T. A. (2007) Annu. Rev. Immunol. 25, 619–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiong J. P., Stehle T., Zhang R., Joachimiak A., Frech M., Goodman S. L., Arnaout M. A. (2002) Science 296, 151–155 [DOI] [PubMed] [Google Scholar]
- 34.Humphries M. J., Symonds E. J., Mould A. P. (2003) Curr. Opin. Struct. Biol. 13, 236–243 [DOI] [PubMed] [Google Scholar]
- 35.Irie A., Kamata T., Puzon-McLaughlin W., Takada Y. (1995) EMBO J. 14, 5550–5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X. P., Puzon-McLaughlin W., Irie A., Kovach N., Prokopishyn N. L., Laferté S., Takeuchi K., Tsuji T., Takada Y. (1999) Biochemistry 38, 14424–14431 [DOI] [PubMed] [Google Scholar]
- 37.Kamata T., Takada Y. (2001) Int. J. Hematol. 74, 382–389 [DOI] [PubMed] [Google Scholar]
- 38.Mould A. P., Koper E. J., Byron A., Zahn G., Humphries M. J. (2009) Biochem. J. 424, 179–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu W., Li J., Roth R. A. (1999) Biochem. Biophys. Res. Commun. 261, 897–903 [DOI] [PubMed] [Google Scholar]
- 40.Wong N. C., Mueller B. M., Barbas C. F., Ruminski P., Quaranta V., Lin E. C., Smith J. W. (1998) Clin. Exp. Metastasis 16, 50–61 [DOI] [PubMed] [Google Scholar]
- 41.Puzon-McLaughlin W., Kamata T., Takada Y. (2000) J. Biol. Chem. 275, 7795–7802 [DOI] [PubMed] [Google Scholar]
- 42.Wen D., Suggs S. V., Karunagaran D., Liu N., Cupples R. L., Luo Y., Janssen A. M., Ben-Baruch N., Trollinger D. B., Jacobsen V. L. (1994) Mol. Cell. Biol. 14, 1909–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Busfield S. J., Michnick D. A., Chickering T. W., Revett T. L., Ma J., Woolf E. A., Comrack C. A., Dussault B. J., Woolf J., Goodearl A. D., Gearing D. P. (1997) Mol. Cell. Biol. 17, 4007–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pinkas-Kramarski R., Shelly M., Glathe S., Ratzkin B. J., Yarden Y. (1996) J. Biol. Chem. 271, 19029–19032 [DOI] [PubMed] [Google Scholar]
- 45.Shaw L. M., Rabinovitz I., Wang H. H., Toker A., Mercurio A. M. (1997) Cell 91, 949–960 [DOI] [PubMed] [Google Scholar]
- 46.Zahir N., Lakins J. N., Russell A., Ming W., Chatterjee C., Rozenberg G. I., Marinkovich M. P., Weaver V. M. (2003) J. Cell Biol. 163, 1397–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murgia C., Blaikie P., Kim N., Dans M., Petrie H. T., Giancotti F. G. (1998) EMBO J. 17, 3940–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lipscomb E. A., Simpson K. J., Lyle S. R., Ring J. E., Dugan A. S., Mercurio A. M. (2005) Cancer Res. 65, 10970–10976 [DOI] [PubMed] [Google Scholar]
- 49.Gambaletta D., Marchetti A., Benedetti L., Mercurio A. M., Sacchi A., Falcioni R. (2000) J. Biol. Chem. 275, 10604–10610 [DOI] [PubMed] [Google Scholar]
- 50.Chung J., Yoon S. O., Lipscomb E. A., Mercurio A. M. (2004) J. Biol. Chem. 279, 32287–32293 [DOI] [PubMed] [Google Scholar]
- 51.Yang X., Kovalenko O. V., Tang W., Claas C., Stipp C. S., Hemler M. E. (2004) J. Cell Biol. 167, 1231–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo W., Pylayeva Y., Pepe A., Yoshioka T., Muller W. J., Inghirami G., Giancotti F. G. (2006) Cell 126, 489–502 [DOI] [PubMed] [Google Scholar]
- 53.Saegusa J., Yamaji S., Ieguchi K., Wu C. Y., Lam K. S., Liu F. T., Takada Y. K., Takada Y. (2009) J. Biol. Chem. 284, 24106–24114 [DOI] [PMC free article] [PubMed] [Google Scholar]