Abstract
During breast cancer metastasis to bone, tumor cells home to bone marrow, likely targeting the stem cell niche, and stimulate osteoclasts, which mediate osteolysis required for tumor expansion. Although osteoblasts contribute to the regulation of the hematopoietic stem cell niche and control osteoclastogenesis through production of proresorptive cytokine RANKL (receptor activator of NF-κB ligand), their role in cancer metastases to bone is not fully understood. C57BL/6J mouse bone marrow cells were treated for 3–12 days with ascorbic acid (50 μg/ml) in the presence or absence of 10% medium conditioned by breast carcinoma cells MDA-MB-231, 4T1, or MCF7. Treatment with cancer-derived factors resulted in a sustained 40–60% decrease in osteoblast differentiation markers, compared with treatment with ascorbic acid alone, and induced an osteoclastogenic change in the RANKL/osteoprotegerin ratio. Importantly, exposure of bone cells to breast cancer-derived factors stimulated the subsequent attachment of cancer cells to immature osteoblasts. Inhibition of γ-secretase using pharmacological inhibitors DAPT and Compound E completely reversed cancer-induced osteoclastogenesis as well as cancer-induced enhancement of cancer cell attachment, identifying γ-secretase activity as a key mediator of these effects. Thus, we have uncovered osteoblasts as critical intermediary of premetastatic signaling by breast cancer cells and pinpointed γ-secretase as a robust target for developing therapeutics potentially capable of reducing both homing and progression of cancer metastases to bone.
Keywords: Bone, Breast Cancer, Cell Differentiation, Cell-Cell Interaction, Notch Pathway, Bone Metastases, γ-Secretase, Osteoblast, Osteoclast, Premetastatic Niche
Introduction
Bone is one of the most common sites for distant metastases from breast cancer (1). Once bone metastases have occurred, they cannot be cured, and the patient 5-year survival rate falls from 95% to 20% (2). Bone metastasis is associated with significant morbidity due to the disruption of bone architecture and mineral homeostasis, which leads to hypercalcemia, pathological fractures, and considerable pain burden.
To home and grow in the bone, cancer cells need to establish successful interactions with the bone microenvironment. Bone is a dynamic tissue that provides support and protection for organs and maintains body mineral homeostasis. Bone is constantly remodeled by the coordinated action of specialized bone cells—osteoclasts that destroy bone and osteoblasts that build bone (3). Osteoclasts are cells of hematopoietic origin that resorb bone by lowering the extracellular pH to dissolve hydroxyapatite crystals and release proteolytic enzymes, such as cathepsin K and matrix metalloproteinase-9 (MMP-9),4 to digest the organic matrix (4). Osteoblasts are derived from mesenchymal stem cells and secrete the extracellular matrix which later mineralizes to form bone. Major pathways controlling osteoblast differentiation include Wnt/β-catenin, Notch, and TGFβ signaling (5–7). The formation of osteoclasts is regulated by cells of osteoblastic lineage, which produce the proresorptive cytokine, receptor activator of NF-κB ligand (RANKL), as well as its negative regulator, soluble decoy receptor osteoprotegerin (OPG).
The presence of a developing tumor has been suggested to alter the microenvironment of distant sites even before the tumor cells arrive, thus forming a “premetastatic niche” that facilitates homing of tumor cells and development of metastatic lesions (8–10). With regard to the bone, tumor cells have been suggested to simulate the behavior of hematopoietic stem cells (11), which allows them to harvest resources from the hematopoietic stem cell niche to establish neoplasms (12). The importance of osteoblasts as key regulators of the hematopoietic stem cell niche has been established (13, 14); however, their role in the formation of a premetastatic niche has not been assessed. It has been shown that breast cancer cells inhibit osteoblast differentiation and induce osteoblast apoptosis (15–18). Osteoblasts are central for the osteolytic effects of breast cancer cells, which do not secrete RANKL themselves (19), but produce factors such as parathyroid hormone-related protein (20) that stimulate osteoblasts to produce RANKL while inhibiting production of OPG (21–23). In turn, RANKL stimulates osteoclast formation, often leading to catastrophic bone destruction (24, 25). In addition, we have previously shown that breast cancer-derived factors can directly induce osteoclastogenesis from late osteoclast precursors (26, 27). During bone resorption, growth factors trapped in the bone matrix, such as TGFβ and insulin-like growth factor, are released and act back on the tumor cells to stimulate their growth (28, 29). Several cytokines have been implicated in the progression of cancer metastasis, with TGFβ (30) and Wnt signaling inhibitor DKK-1 (18) being of considerable importance in the metastatic process. In addition, Notch signaling has been implicated in the control of the hematopoietic stem cell niche (31, 32) as well as in cancer development (33), suggesting that it can also play a role in formation of the premetastatic niche. Notch signaling is initiated by ligand binding, which induces γ-secretase-mediated release of the Notch intracellular domain (NICD), which translocates to the cell nucleus and alters gene expression (33).
In this study, we examined the effect of breast carcinoma cells on the bone marrow cultures that retain potential for differentiation into both osteoblasts and osteoclasts and thus better represent the complex bone microenvironment. Our data demonstrate that soluble factors produced by breast cancer cells inhibit osteoblast differentiation and stimulate osteoblast-dependent osteoclast differentiation, as a result augmenting subsequent attachment of breast cancer cells to bone cells. We also present evidence for a critical role of γ-secretase in these effects.
EXPERIMENTAL PROCEDURES
Test Compounds
l-Ascorbic acid (AA, Sigma, A5960) was freshly prepared and added to the medium on the day of medium change. LiCl (Sigma, L0505) was diluted in water, SB 216763 (Tocris Bioscience, 1616), SB 431542 (Tocris Bioscience, 1614), DAPT (Calbiochem, 565770), Compound E (CE; Calbiochem, 565790) were diluted in dimethyl sulfoxide, which was used as a vehicle (0.1%) in corresponding experiments. All inhibitors were present during the whole culture period. OPG (Sigma, 08137), and pan-specific TGFβ antibody produced in rabbit (R&D Systems, AB-100-NA) were incubated with MDA-MB-231 conditioned medium (CM) for 10 min before adding to cultures.
Cell Cultures
The MDA-MB-231 and MCF7 human breast carcinoma cell line, 4T1 murine breast carcinoma cell line, and MCF10A human mammary epithelial cells were kindly provided by Dr. P. Siegel (McGill University) and cultured as described previously (26). The MC3T3-E1 mouse preosteoblastic cell line was kindly provided by Dr. M. T. Kaartinen (McGill University). Cells were cultured to 50% confluence, except for MCF10A, which was cultured to 100% confluence, in T-75 tissue culture flasks. Conditioned medium was collected after 48 h of incubation, centrifuged at 2000 rpm for 5 min, aliquoted, and stored at −80 °C.
All animal studies were performed in accordance with the McGill University guidelines established by the Canadian Council on Animal Care. Mice (C57BL6/J, male, 6 weeks old) were purchased from Charles River. Mouse-derived bone marrow cells were collected from mouse tibia and femora under aseptic conditions as described previously (34). Bone marrow cells were plated at a density of 2.5 × 106 cells/cm2 and cultured in minimal essential medium supplemented with 1% penicillin-streptomycin (Wisent, 450-201-EL) and 10% fetal bovine serum (Hyclone, SH 30396-03). One day after plating, 50 μg/ml AA was added to induce osteoblast differentiation, and CM from the indicated cells (10%) was added to experimental cultures. All cultures were supplemented with fresh medium every other day. On the indicated days, samples were fixed with 10% formalin and stained for alkaline phosphatase (ALP; Fast Red, Sigma, F4381), tartrate-resistant acid phosphatase (TRAP; Sigma, 387A) and analyzed using BioQuant software. Mineralization was assessed using Von Kossa staining (Sigma, S6506). Pit resorption assay was performed as described previously (35).
Cell Proliferation Assays
Bone marrow cells were plated in 96-well flat-bottom plates at a density of 2.5 × 106/cm2 and cultured as described. Proliferation assay was performed after 9 days of culture using the BrdU CHEMICON Cell Proliferation Assay kit (Millipore, 2750) according to the manufacturer's instructions. Absorbance was measured at 450 nm using a microplate reader (Beckman Coulter AD340), with a higher optical density indicating a higher BrdU concentration in the sample.
RNA Isolation and RT-PCR
Total RNA was isolated from primary cultures using the RNeasy mini kit and QIAshredder columns (Qiagen, 74104 and 79654). For real-time PCR, 2 μg of total RNA was reverse transcribed using a cDNA archive kit (Applied Biosystems, 74322171). Real-time PCR was performed using 7500 Applied Biosystems instrument, with TaqMan Universal PCR Master Mix (Applied Biosystems, 4304437) and the following TaqMan gene expression assays: TRAP (Mm00475698_m1), MMP-9 (Mm00600163_m1), cathepsin K (Mm00484036_ m1), osterix (Mm00504574_m1), collagen-1a1 (Mm00801666_g1), β-actin (Mm00607939_s1), RANKL (Mm00441908_m1), OPG (Mm01205928 _m1), Hey1 (Mm00468865_m1), and Hes1 (Mm 01342805_m1). Real-time PCR for Runx2, Delta1, JAG2, Cyclin A, Cyclin D1, p53, and GAPDH was performed using SYBR Green Universal PCR Master Mix (Applied Biosystems, 4367659) and the following primers: Runx2 forward, TGGCTTGGGTTTCAGGTTAG, and reverse, TCGGTTTCTTAGGGTCTTGGA; Delta1 forward, TTGGGCTTCTCTGGCTTCAAC, and reverse, CCACACACTTGGCACCGTTAG; JAG2 forward, CAAGTTCTGTGACGAGTGTGTCCC, and reverse, TTGCCCAAGTAGCCATCTGG; Cyclin A forward, CTGCCTTCCACTTAGCTCTC, and reverse, GAGGTAGGTCTGGTGAAGGT; Cyclin D1 forward, CAGAAGTGCGAAGAGGAGGTC, and reverse, TCATCTTAGAGGCCACGAACAT; p53 forward, CACAGCGTGGTGGTACCTTA, and reverse, GCACAAACACGAACCTCAAA; GAPDH forward, TTCCGTGTTCCTACCCCCAA, and reverse, GATGCCTGCTTCACCACCTT.
Immunofluorescence and Apoptosis Assay
Cells plated on glass coverslips were fixed with 10% formalin and immunostained as described previously (36). We used monoclonal antibody for β-catenin (Cell Signaling, 9587) and NICD (Santa Cruz Biotechnology, sc-6014). Staining was completed with biotinylated goat anti-mouse IgG (Invitrogen, A10519) and Alexa Fluor 488-conjugated streptavidin (Invitrogen, S11223). Nuclei were counterstained using DAPI dihydrochloride (Invitrogen, D1306). Ten random images/experimental condition were collected in each experiment, each image containing 8–25 precursors. Cell counts were performed by counting DAPI-labeled nuclei. Nuclear fluorescence intensity was evaluated using Volocity software, by first circling DAPI-labeled nuclei and then assessing the average fluorescence of the protein of interest within that area. For evaluation of apoptosis, nuclear morphology was examined and rated positive for apoptosis if it exhibited nuclear condensation and a loss of membrane integrity. The rate of apoptosis was estimated as a proportion of cells demonstrating nuclear fragmentation from the total number of cells analyzed. In addition, the fluorescent-tagged annexin-V was used to detect apoptotic cells (Santa Cruz Biotechnology, sc-4252-AK). Live cultures were rinsed with PBS and incubated with the FITC-annexin in the supplied incubation buffer for 15 min at room temperature. Cultures were then fixed in 10% formalin, stained with DAPI, and immediately examined. 182–487 cells/experimental condition were scored.
Immunoblotting
For protein isolation, cells were treated with lysis buffer containing 50 mm Tris, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 2 mm EDTA, and protease inhibitor mixture. Nuclear extraction was conducted using lysis buffers, first of 10 mm Tris, pH 8, 1.5 mm MgCl2, 5 mm KCl, 0.5 mm DTT, 0.1 m PMSF, 0.5% Nonidet P-40, and second of 20 mm Tris, pH 8, 25% glycerol, 1.5 mm MgCl2, 0.5 mm DTT, 0.1 m PMSF, 0.2 mm EDTA, and 0.4 mm NaCl. Immunoblotting was performed as described previously (26) using anti-RANKL (Santa Cruz Biotechnology, sc-52950) and anti-NICD (Santa Cruz Biotechnology, sc-6014) followed by horseradish peroxidase-conjugated secondary antibodies (Jackson Laboratories, 705-065-003) and chemiluminescent substrate (Supersignal West Pico; Pierce, 34080). Blots were reprobed with α-tubulin antibody (Sigma-Aldrich, T9026) as a loading control.
Cell Attachment Assay
Bone marrow cultures were treated as indicated for 9 days. MDA-MB-231 cells were loaded with Cell Tracker Green (5 μm; Invitrogen, C2925) in serum-free DMEM for 1 h, washed, incubated in serum-free DMEM for an additional 1 h, washed, trypsinized, centrifuged, resuspended in serum-free DMEM at a cell density of 4 × 104/ml, and applied to bone cell cultures. After 40 min of incubation, cultures were washed three times with serum-free DMEM and fixed with 10% formalin for 10 min before imaging.
Statistical Analysis
Data are presented as representative images, representative experiments, or as means ± S.E., with n indicating the number of independent experiments. Differences were assessed by Student t test and accepted as statistically significant at p < 0.05.
RESULTS
Breast Cancer Cells Inhibit Differentiation of Osteoblasts and Stimulate Differentiation of Osteoclasts
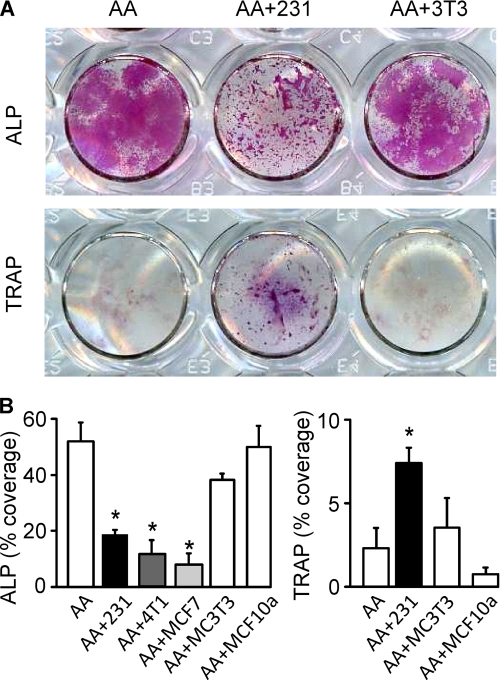
We examined the effects of soluble factors produced by human metastatic breast cancer cells MDA-MB-231, human metastatic breast cancer cells MCF7, or mouse metastatic breast cancer cells 4T1 on the differentiation of osteoblasts and osteoclasts from precursors derived from mouse bone marrow. Bone marrow cells were treated with AA in the presence or absence of medium conditioned by MDA-MB-231 cells, MCF7 cells, or 4T1 cells for 9 days and examined for the expression of osteoblast differentiation marker ALP (Fig. 1A, upper) and osteoclast differentiation marker TRAP (Fig. 1A, lower). Medium conditioned by the MCF10A human breast epithelial cell line and MC3T3-E1 mouse preosteoblastic cell line were used as controls. Cultures treated with AA displayed robust ALP staining in osteoblastic nodules. In contrast, in cultures treated with AA in the presence of MDA-MB-231, 4T1, or MCF7 CM, the ALP-positive area was significantly reduced (Fig. 1B, left), and the staining exhibited punctuated pattern localized in small clusters (Fig. 1A, and supplemental Fig. 1A). Cultures treated with AA alone rarely contained visible osteoclasts. In contrast, treatment with MDA-MB-231, 4T1, or MCF7 CM induced the formation of four to nine large multinucleated osteoclasts/cm2. Because the numbers of visible osteoclasts were relatively low, we examined changes in area covered by TRAP-positive cells (Fig. 1A and 1B, right) and found a significant increase in TRAP-positive area in MDA-MB-231 CM-treated cultures. Close investigation confirmed the identity of large, multinucleated TRAP-positive osteoclasts, typically located under a layer of osteoblastic cells (supplemental Fig. 1C). Both the inhibition of osteoblast differentiation and stimulation of osteoclast formation by MDA-MB-231 CM were sustained for 6–15 days of culture (supplemental Fig. 1). Addition of MCF10A or MC3T3 CM did not affect osteoblast or osteoclast differentiation (Fig. 1).
FIGURE 1.
Breast cancer cells inhibit osteoblasts and stimulate osteoclasts. Mouse bone marrow cells were grown for 3–15 days with AA (50 μg/ml) without additions (open bars) or in the presence of MDA-MB-231, 4T1, or MCF7 CM (10%, shaded bars) or controls MC3T3-E1 CM (10%) and MCF10A CM (10%). A, representative images of cultures treated with AA only (AA, left), with AA and MDA-MB-231 CM (AA+231, center), or with AA and MC3T3-E1 CM (AA+3T3, right), fixed on day 6–9, and stained for ALP (red, upper) or TRAP (purple, lower). Scanned are the wells of a 24-well plate. B, average area covered on day 9 by ALP-positive cells (left) and on day 6 by TRAP-positive cells (right). Treatment with MDA-MB-231, 4T1, or MCF7 CM significantly reduced ALP-positive osteoblast staining (left). Treatment with MDA-MB-231 CM significantly increased TRAP-positive osteoclast staining (right). Supplementation of cultures with AA and conditioned medium from MC3T3 or MCF10A did not produce significantly different results from treatment with AA alone. Data are means ± S.E. (error bars), n = 2–6 independent experiments, p < 0.05.
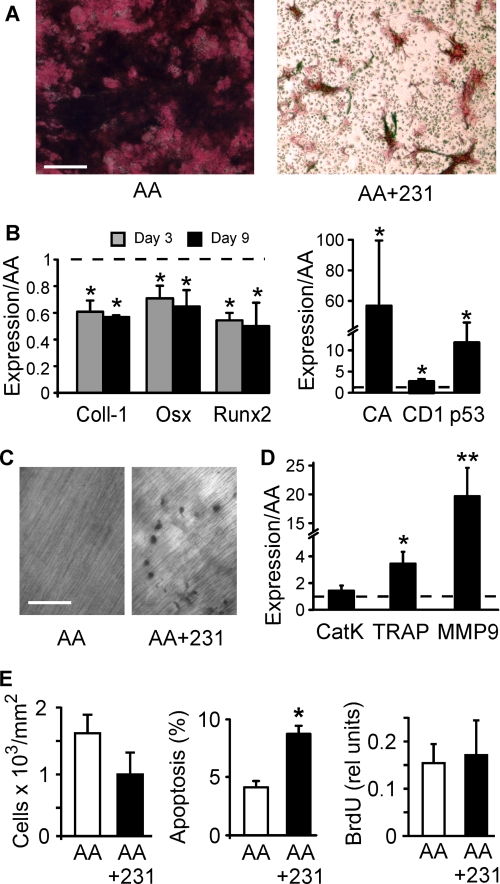
We next assessed whether the functional activity of osteoblasts and osteoclasts reflects their observed differentiation status. When AA-treated cultures were provided with a source of inorganic phosphate to induce mineralization, they developed mineralized nodules easily identified by Von Kossa staining (Fig. 2A, left). Addition of MDA-MB-231 CM to these cultures prevented mineralization (Fig. 2A, right) and induced significant and sustained decrease in expression of osteoblast differentiation markers, collagen-1, osterix, and Runx2 (Fig. 2B, left), confirming inhibition of osteoblast differentiation by soluble factors produced by breast cancer cells. Moreover, treatment with 4T1 CM led to significant increases in the expression of cell proliferation markers, Cyclin A, Cyclin D1, and p53 (Fig. 2B, right). To assess osteoclast functional activity, bone marrow cells were plated on dentin slices and treated for 9 days with either AA alone or a combination of AA and MDA-MB-231 CM. Whereas cultures treated with AA alone did not exhibit osteoclastic resorption (Fig. 2C, left), numerous resorption pits were identified in cultures treated with AA and MDA-MB-231 CM (Fig. 2C, right). We quantified the gene expression of osteoclast differentiation markers cathepsin K, TRAP, and MMP-9 by real-time PCR. Treatment with MDA-MB-231 CM induced a significant and marked increase in expression of TRAP and especially MMP-9, whereas expression of cathepsin K was increased but did not reach a change of statistical significance (Fig. 2D).
FIGURE 2.
Breast cancer cells maintain osteoblasts in an immature state and induce differentiation of functional osteoclasts. A, bone marrow cells were grown for 12 days with AA (50 μg/ml) and β-glycerophosphate (10 mm) in the absence (left) or presence of MDA-MB-231 CM (10%, right). The cultures were fixed and stained for ALP (red) and mineralized deposits (black). Scale bar is 100 μm. B, bone marrow cells were grown for 3–9 days with AA (50 μg/ml) in the absence or presence of MDA-MB-231 CM (10%, left) or 4T1 CM (10%, right). Expression of Collagen-1 (Coll-1), osterix (Osx), and Runx2 was analyzed on day 3 (gray) or 9 (black). Expression of Cyclin A (CA), Cyclin D1 (CD1), and p53 was analyzed on day 9. Data are means ± S.E. (error bars), normalized to expression of β-actin (left) or GAPDH (right), and presented relative to levels observed in AA only samples (dashed line), n = 3–5 independent experiments, p < 0.05. C, bone marrow cells were grown for 9 days on dentin slices with AA (50 μg/ml) in the absence (left) or presence of MDA-MB-231 CM (10%, right), then the cells were removed, and dentin was stained with toluidine blue to reveal resorption pits. Scale bars represent 100 μm. D, expression of Cathepsin K (Cat K), TRAP, and MMP-9 was analyzed on day 9. Data are means ± S.E., normalized to expression of β-actin, and presented relative to levels observed in AA only samples (dashed line), n = 4–6 independent experiments, p < 0.05. E, bone marrow cells were grown for 9 days with AA (50 μg/ml) in the absence or presence of MDA-MB-231 CM (10%). The parallel samples were fixed, stained with DAPI nuclear stain, and the cell density was estimated (left). The rate of apoptosis was estimated as a proportion of cells demonstrating nuclear fragmentation from the total number of cells analyzed (center). Cell proliferation was measured by BrdU incorporation (right). Data are means ± S.E., n = 3–5 independent experiments, p < 0.05.
It is conceivable that the effect of breast cancer cells on osteoblast may be due to induction of cell death rather than inhibition of differentiation. We have found that the average cell density was not significantly different in cultures treated with AA alone or a combination of AA and MDA-MB-231 CM (Fig. 2E, left). Nevertheless, cultures treated with AA and MDA-MB-231 CM exhibited significant increase in nuclear fragmentation and loss of membrane integrity compared with cells treated with AA alone (Fig. 2E, middle). In addition, cultures treated with MDA-MB-231 CM demonstrated an increase in the number of cells positive for early apoptosis marker, annexin-5 (from 6 ± 4% in control cultures, to 14 ± 2% in MDA-MB-231 CM-treated cultures, n = 2 independent experiments). Cell proliferation, as assessed using a BrdU incorporation assay, was not significantly different in cultures treated with AA and MDA-MB-231 CM compared with treated with AA alone (Fig. 2E, right). Thus, although MDA-MB-231 CM induced higher apoptosis rates in bone cells, it resulted only in relatively small changes in the total numbers of cells in culture.
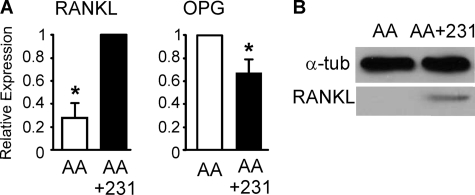
Because the cultures were not treated with exogenous osteoclastogenic factors, we assessed whether breast cancer-derived factors affect the expression of RANKL and OPG by osteoblasts. We have found that MDA-MB-231 CM induced a 3-fold increase in RANKL expression level and a 30% decrease in OPG expression level (Fig. 3A). Immunoblotting further confirmed significant increase in the protein levels of RANKL in MDA-MB-231 CM-treated cultures (Fig. 3B).
FIGURE 3.
Breast cancer cells induce osteoclastogenic change in RANKL/OPG expression. Bone marrow cells were grown for 9 days with AA (50 μg/ml) in the absence (AA, open bars) or presence of MDA-MB-231 CM (10%, AA+231, filled bars). A, expression of RANKL and OPG normalized to expression of β-actin and presented relative to levels observed in cells grown with AA+231 for RANKL and AA only for OPG. Data are means ± S.E. (error bars), n = 5 independent experiments, p < 0.05. B, RANKL protein level assessed by immunoblotting in whole cell lysates. Shown is a representative immunoblot with α-tubulin as a loading control.
Notch Signaling Is Stimulated in Bone Cells by Breast Cancer-derived Factors
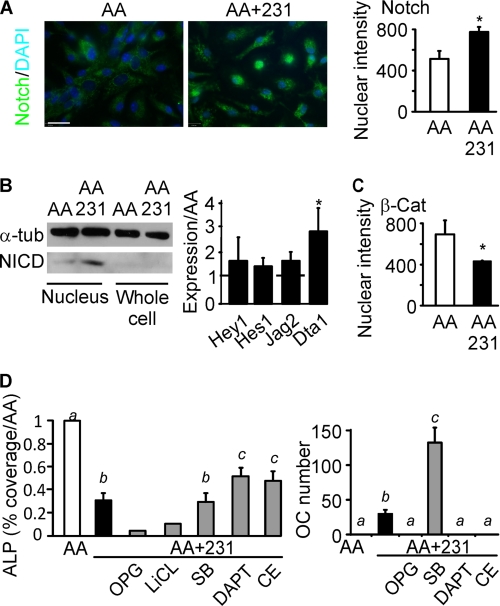
The role of Notch signaling in the inhibition of osteoblast differentiation has been firmly established (37). We assessed the status of this signaling pathway in osteoblastic cultures exposed to breast cancer-derived factors. Localization of the cleaved NICD was examined by immunofluorescence. Cultures treated only with AA exhibited little nuclear staining (Fig. 4A, left), whereas treatment with MDA-MB-231 CM resulted in the appearance of the nuclear staining of the NICD (Fig. 4A, center). Quantification of the intensity of nuclear staining for NICD demonstrated a significant increase in nuclear localization of the NICD in MDA-MB-231 CM-treated cultures compared with cultures treated with AA alone (Fig. 4A, right). The increase in nuclear NICD was confirmed by immunoblotting (Fig. 4B, left). NICD direct transcriptional targets Hey-1 and Hes-1, and Notch ligand Jag-2 exhibited a trend toward higher expression in cultures treated with breast cancer-derived factors, whereas Notch ligand Delta-1 was significantly higher in cultures treated with breast cancer-derived factors (Fig. 4B, right). Wnt and Notch signaling are known to cross-talk; therefore, we assessed the activation of β-catenin using immunofluorescence. MDA-MB-231 CM-treated cultures exhibited significantly less nuclear intensity for β-catenin compared with control cultures (Fig. 4C), confirming the inhibition of Wnt signaling by breast cancer factors.
FIGURE 4.
Notch signaling pathway is stimulated in bone cells by breast cancer-derived factors. Bone marrow cells were grown for 9 days with AA (50 μg/ml) in the absence (AA, open bars) or presence of MDA-MB-231 CM (10%, AA+231, black bars). A, NICD localization was assessed by immunofluorescence (green), and nuclei were stained using DAPI (blue). Left and center, representative images of negative (left) and positive (center) nuclear staining for NICD are shown. Scale bar is 20 μm. Right, nuclear intensity of NICD is quantified. Data are means ± S.E. (error bars), n = 3 independent experiments, p < 0.05. B, left, NICD level was assessed by immunoblotting in nuclear extracts and whole cell lysates. Shown is a representative immunoblot with α-tubulin as a loading control. Right, expression of the transcriptional targets of the NICD, Hey-1 and Hes-1, and Notch ligands Jag-2 and Delta1 (Dta1) was analyzed on day 9. Data are means ± S.E., normalized to expression of β-actin for Hey-1 and Hes-1 or GAPDH for Jag-2 and Delta1 and presented relative to levels observed in cells grown with AA only (dashed line), n = 3 independent experiments. C, nuclear intensity of β-catenin (β-Cat) is shown. Data are means ± S.E., n = 3 independent experiments, p < 0.05. D, bone marrow cells were grown for 9 days with AA (50 μg/ml), MDA-MB-231 CM (10%), and the following inhibitors (gray bars): OPG (500 ng/ml), LiCl (10 mm), SB216763 (SB, 10 μm), DAPT (100 nm), or CE (100 nm). The parallel samples were fixed and stained for ALP or TRAP. Left, area covered by ALP-positive cells was normalized to the samples grown with AA only. Right, number of TRAP-positive osteoclastic cells was counted in the same experiments. Data are means ± S.E., n = 3–6 independent experiments except for OPG and LiCl data, where n = 3 replicates; different letters indicate significant difference at p < 0.05.
To investigate the roles of NOTCH and Wnt pathways in the effects of breast cancer-derived factors on osteoblast and osteoclast differentiation, we employed pharmacological inhibitors of both pathways. We treated bone marrow cells for 9 days with AA alone or with AA and MDA-MB-231 CM (10%) together with glycogen synthase kinase inhibitors LiCl or SB216763, or γ-secretase inhibitors DAPT or CE, and examined the expression of osteoblast marker ALP and osteoclast marker TRAP. Glycogen synthase kinase inhibitors were unable to rescue osteoblast differentiation (Fig. 4D, left). Moreover, treatment with SB216763 induced an additional increase in osteoclast numbers (Fig. 4D, right). Although inhibitors of γ-secretase partially rescued MDA-MB-231 CM-induced osteoblast inhibition (Fig. 4D, left), their effect was relatively minor, and it was not observed in cultures treated with 4T1 and MCF7 CM (supplemental Fig. 2). However, both γ-secretase inhibitors completely prevented the stimulation of osteoclast formation by breast cancer-derived factors (Fig. 4D, right). Addition of exogenous OPG blocked osteoclast formation but further stimulated the inhibition of osteoblast differentiation (Fig. 4F). Moreover, antagonizing the TGFβ pathway, which was previously suggested as a mediator of antiosteoblastic effects of breast cancer cells (15), was ineffective in preventing MDA-MB-231 CM-induced osteoblast inhibition (supplemental Fig. 3).
Exposure to Breast Cancer-derived Factors Enhances Subsequent Breast Cancer Cell Attachment to Immature Osteoblasts
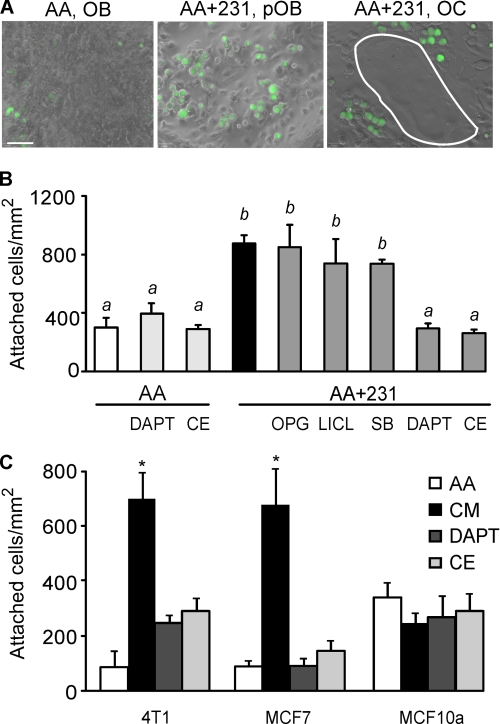
Because bone cells have been shown to be critical players in mediating stem cell attachment to the hematopoietic bone marrow niche (13, 38, 39), we investigated how exposure to soluble factors produced by breast cancer cells may affect the direct interaction of breast cancer cells with bone cells. Bone marrow cultures were treated for 9 days with AA alone or with a combination of AA and MDA-MB-231 CM, and then MDA-MB-231 cells labeled with Cell Tracker Green were incubated for 40 min on top of bone cells. Although some breast cancer cells adhered to mature osteoblasts in cultures treated with AA alone (Fig. 5A, left), markedly more breast cancer cells attached to immature osteoblast precursors in AA and MDA-MB-231 CM-treated cultures (Fig. 5A, center). Notably, the breast cancer cells did not attach to osteoclasts (Fig. 5A, right). Quantification confirmed that the treatment with MDA-MB-231 CM significantly increased subsequent attachment of breast cancer cell to bone cells (Fig. 5B). Treatment with OPG or glycogen synthase kinase inhibitors LiCl or SB216763 did not interfere with cancer factor-induced breast cancer cell attachment to bone cells (Fig. 5B). In contrast, treatment with γ-secretase inhibitor DAPT or CE fully reversed the MDA-MB-231 CM-induced breast cancer cell attachment to osteoblasts (Fig. 5B). In the absence of conditioned medium, neither DAPT nor CE affected cancer cell attachment to bone cells (Fig. 5B).
FIGURE 5.
Exposure to breast cancer-derived factors enhances subsequent breast cancer cell attachment to immature osteoblasts. Bone marrow cells were grown for 9 days with AA (50 μg/ml) in the absence (AA, open bars) or presence of MDA-MB-231 CM (10%), combined with vehicle (AA+231, black bars) or the following inhibitors (gray bars): OPG (500 ng/ml), LiCl (10 mm), SB216763 (SB, 10 μm), DAPT (100 nm), or Compound E (CE, 100 nm). The MDA-MB-231 cells were labeled with Cell Tracker Green and added to bone marrow cultures for 40 min, and then the cultures were washed to remove nonattached cells, fixed, and analyzed. A, representative images demonstrate attachment of breast cancer cells (green) to mature osteoblasts (OB) in cultures treated with AA only (left); to immature osteoblast precursors (pOB) in cultures treated with AA and MDA-MB-231 CM (center); or to osteoclasts (OC, white outline) in cultures treated with AA and MDA-MB-231 CM (right). Scale bar is 20 μm. B, significantly more breast cancer cells attached to bone marrow cultures treated with AA and MDA-MB-231 CM compared with cultures treated with AA alone. Inhibitors of γ-secretase DAPT and CE prevented this effect of MDA-MB-231 CM, whereas glycogen synthase kinase inhibitors and OPG were ineffective. Data are means ± S.E. (error bars), n = 2–6 independent experiments; different letters indicate significant difference at p < 0.05. C, bone marrow cells were grown for 9 days with AA (50 μg/ml) in the absence (AA, open bars) or presence of 4T1, MCF7, or MCF10A CM (10%), either alone (CM, black bars) or with γ-secretase inhibitors DAPT (100 nm) or CE (100 nm). The same cells as were used for CM treatment were labeled with Cell Tracker Green and added to bone marrow cultures for 40 min. Treatment with 4T1 or MCF7 CM significantly increased attachment of these cells to bone marrow cultures, which was inhibited by γ-secretase inhibitors. Data are means ± S.E., n = 3 independent experiments, p < 0.05.
In keeping with the effects of MDA-MB-231-derived factors, MCF7 and 4T1 cells were also found to produce soluble factors that significantly increase the subsequent attachment of these breast cancer cells to bone cells, an effect that was drastically inhibited by DAPT or CE (Fig. 5C). In contrast, the normal breast epithelial cell line MCF10A was not effective in promoting cell attachment to osteoblasts (Fig. 5C).
DISCUSSION
This study demonstrates that soluble factors produced by breast cancer cells inhibit osteoblast differentiation while stimulating osteoclast differentiation. In addition to establishing an osteolytic environment, this change in bone cells also supports the subsequent attachment of breast cancer cells to immature osteoblasts. We have identified γ-secretase as a critical mediator of these effects. Pharmacological inhibition of γ-secretase completely reversed cancer-induced osteoclastogenesis and enhancement of cancer cell attachment, providing a potential therapeutic target capable of reducing both the homing and progression of cancer metastases to bone.
Because of the usually osteolytic nature of breast cancer metastases in bone, the effects of breast cancer cells on osteoclasts have been studied extensively; however, much less attention has been given to the interactions of breast cancer cells with osteoblasts. Using osteoblastic cell lines, primary calvarial osteoblasts, or co-cultures of separately isolated osteoblastic and osteoclastic cells, it has been shown that breast cancer cells inhibit osteoblast differentiation (15, 17, 18), induce osteoblast apoptosis (15, 16), and increase the production of pro-osteoclastic factors by osteoblasts (22, 23). To model the cell types and interactions potentially encountered by tumor cells in the bone microenvironment more accurately, we have developed a unique culture system that allows for monitoring the differentiation of osteoblasts and the osteoblast-dependent differentiation of osteoclasts directly from bone marrow cells. This model has allowed us to investigate complex interactions among osteoblasts, osteoclasts, and cancer cells. We have shown that although breast cancer cells induce osteoblast apoptosis, this effect may be countered by cell proliferation, resulting in maintenance of sufficient pool of osteoblastic cells acting as a source of proresorptive factors. We have confirmed that in the presence of breast cancer cells, immature osteoblasts up-regulated the production of RANKL and down-regulated the production of OPG. This can be due to direct effect of breast cancer cells on osteoblasts, or it can be a consequence of osteoblast differentiation status because it has been shown before that immature osteoblasts produce more RANKL and less OPG compared with more mature cells (40–43).
Inhibition of osteoblastogenesis combined with a stimulation of osteoclastogenesis by the soluble factors produced by breast cancer cells induced a significant shift in the bone microenvironment toward (i) more supportive environment for the homing of arriving cancer cells and (ii) more osteolytic milieu for the further growth of tumors at the bone site. The ability of breast cancer cells to modify the distant microenvironment of the bone tissue is consistent with the notion of a premetastatic niche (9). The involvement of osteoblasts in the maintenance of the hematopoietic stem cell niche is long recognized (13, 38, 39). It has been speculated that in the metastatic niche tumor cells take advantage of the stem cell habitat in the bone marrow (14), thus equating hematopoietic stem cell niche with the metastatic niche. We have shown that not only can osteoblasts directly support the attachment of breast cancer cells, but also that breast cancer cells can augment this property of the osteoblasts while acting distantly. These findings are consistent with the role of osteoblasts as a part of a premetastatic niche permitting the attraction of tumor cells and their incorporation into the niche. Cancer cells attached preferentially to younger, more immature osteoblasts, suggesting that the increase in cancer cell attachment is likely a combination of direct effect of breast cancer cells on osteoblasts and indirect consequence of osteoblast differentiation status. Breast cancer cells never attached to osteoclasts, and inhibition of osteoclast formation using OPG did not prevent breast cancer factor-induced increase in breast cancer attachment to osteoblasts. However, osteoclasts still appear to play a role in these effects of breast cancer cells. First, soluble factors produced by breast cancer cells induced a 20-fold increase in the expression of an osteoclastic gene MMP-9, which was previously shown to be associated with the premetastatic niche (10). Second, the inhibition of a breast cancer factor-induced increase in breast cancer attachment to osteoblasts correlated with a strong inhibition of osteoclast formation, but the stimulation of osteoblastogenesis was not required, suggesting supportive action of osteoclasts in breast cancer attachment. Thus, we demonstrated that soluble factors produced by breast cancer introduce changes in osteoblasts and osteoclasts consistent with the establishment of a premetastatic niche.
To assess the potential mediators of the effect of breast cancer cells on osteoblast and osteoclast precursors, we considered the involvement of pathways known to affect osteoblast and osteoclast differentiation, including Wnt, TGFβ, and Notch. Wnt signaling is an essential pathway in osteogenesis (44), which has been shown to play a critical role in myeloma bone metastases (45) and was shown to be altered in the breast cancer bone metastases model (18). TGFβ is well known to play an important role in promoting tumor progression specifically in models of MDA-MB-231 breast cancer metastasis (46–49) and has been suggested to mediate the inhibitory effects of breast cancer cells on osteoblasts (15). Notch signaling has recently been identified as a key mediator of bone formation (6) and a mediator of osteosarcoma (32). Notch signaling is also implicated in regulation of osteoclastogenesis (50, 51). We have shown that normalizing Wnt signaling or antagonizing TGFβ signaling did not interfere with the effects produced by the breast cancer-derived factors on bone cells. In contrast, inhibition of γ-secretase, a key enzyme mediating Notch signaling, resulted in the full reversal of breast cancer-induced osteoclastogenesis and enhancement of cancer cell attachment. Although the inhibition of Notch signaling also resulted in a partial rescue of MDA-MB-231-induced inhibition of osteoblast differentiation, this effect was relatively minor and not reproduced when different breast cancer cells, MCF7 and 4T1, were employed. The ineffectiveness of inhibitors of γ-secretase in fully rescuing osteoblastic phenotype suggests that RANKL/OPG expression and the promotion of tumor cell attachment in osteoblasts may be regulated independently from differentiation. These data may also suggest an important role for osteoblast-independent interactions, such as the direct effects of breast cancer cells on osteoclast differentiation (26, 27). It is also possible that the main target of γ-secretase inhibitors is in osteoclasts, rather than osteoblasts, implicating osteoclasts in control of breast cancer cell attachment to bone, in line with the previously shown involvement of osteoclasts in the regulation of the hematopoietic stem cell niche (52).
Thus, our study suggests the critical role of γ-secretase in the homing and establishment of osteolytic bone metastases from breast cancer. Complete reversal of key prometastatic events such as cancer-induced enhancement of cancer cell attachment and osteoclastogenesis by inhibition of γ-secretase provides a robust therapeutic target to develop drugs potentially capable of reducing both the homing and progression of cancer metastases to bone.
Supplementary Material
Acknowledgments
We thank Dr. P. Siegel and Dr. M. T. Kaartinen, McGill University, for providing cell lines used in this study and Quyen Su for help in preliminary data analysis.
This work was supported by the Institute for Musculoskeletal Health and Arthritis/Canadian Institute for Health Research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- MMP-9
- matrix metalloproteinase-9
- AA
- l-ascorbic acid
- ALP
- alkaline phosphatase
- CE
- Compound E
- CM
- conditioned medium
- NICD
- notch intracellular domain
- OPG
- osteoprotegerin
- RANKL
- receptor activator of NF-κB ligand
- TRAP
- tartrate-resistant acid phosphatase.
REFERENCES
- 1.Coleman R. E. (2006) Clin. Cancer Res. 12, 6243S–6249S [DOI] [PubMed] [Google Scholar]
- 2.Kim C. J., Bland K. I., Yeatman T. J. (2004) The Breast Comprehensive Management of Benign and Malignant Disorders, W. B. Saunders, Philadelphia, PA [Google Scholar]
- 3.Hattner R., Epker B. N., Frost H. M. (1965) Nature 206, 489–490 [DOI] [PubMed] [Google Scholar]
- 4.Stenbeck G. (2002) Semin. Cell Dev. Biol. 13, 285–292 [DOI] [PubMed] [Google Scholar]
- 5.Deng Z. L., Sharff K. A., Tang N., Song W. X., Luo J., Luo X., Chen J., Bennett E., Reid R., Manning D., Xue A., Montag A. G., Luu H. H., Haydon R. C., He T. C. (2008) Front. Biosci. 13, 2001–2021 [DOI] [PubMed] [Google Scholar]
- 6.Engin F., Yao Z., Yang T., Zhou G., Bertin T., Jiang M. M., Chen Y., Wang L., Zheng H., Sutton R. E., Boyce B. F., Lee B. (2008) Nat. Med. 14, 299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hilton M. J., Tu X., Wu X., Bai S., Zhao H., Kobayashi T., Kronenberg H. M., Teitelbaum S. L., Ross F. P., Kopan R., Long F. (2008) Nat. Med. 14, 306–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendoza M., Khanna C. (2009) Int. J. Biochem. Cell Biol. 41, 1452–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaplan R. N., Riba R. D., Zacharoulis S., Bramley A. H., Vincent L., Costa C., MacDonald D. D., Jin D. K., Shido K., Kerns S. A., Zhu Z., Hicklin D., Wu Y., Port J. L., Altorki N., Port E. R., Ruggero D., Shmelkov S. V., Jensen K. K., Rafii S., Lyden D. (2005) Nature 438, 820–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaplan R. N., Psaila B., Lyden D. (2006) Cancer Metastasis Rev. 25, 521–529 [DOI] [PubMed] [Google Scholar]
- 11.Wang J., Loberg R., Taichman R. S. (2006) Cancer Metastasis Rev. 25, 573–587 [DOI] [PubMed] [Google Scholar]
- 12.Neiva K., Sun Y. X., Taichman R. S. (2005) Braz. J. Med. Biol. Res. 38, 1449–1454 [DOI] [PubMed] [Google Scholar]
- 13.Calvi L. M., Adams G. B., Weibrecht K. W., Weber J. M., Olson D. P., Knight M. C., Martin R. P., Schipani E., Divieti P., Bringhurst F. R., Milner L. A., Kronenberg H. M., Scadden D. T. (2003) Nature 425, 841–846 [DOI] [PubMed] [Google Scholar]
- 14.Shiozawa Y., Havens A. M., Pienta K. J., Taichman R. S. (2008) Leukemia 22, 941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mercer R. R., Miyasaka C., Mastro A. M. (2004) Clin. Exp. Metastasis 21, 427–435 [DOI] [PubMed] [Google Scholar]
- 16.Fromigué O., Kheddoumi N., Lomri A., Marie P. J., Body J. J. (2001) J. Bone Miner. Res. 16, 1600–1610 [DOI] [PubMed] [Google Scholar]
- 17.Phadke P. A., Mercer R. R., Harms J. F., Jia Y., Frost A. R., Jewell J. L., Bussard K. M., Nelson S., Moore C., Kappes J. C., Gay C. V., Mastro A. M., Welch D. R. (2006) Clin. Cancer Res. 12, 1431–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bu G., Lu W., Liu C. C., Selander K., Yoneda T., Hall C., Keller E. T., Li Y. (2008) Int. J. Cancer 123, 1034–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau Y. S., Danks L., Sun S. G., Fox S., Sabokbar A., Harris A., Athanasou N. A. (2007) Breast Cancer Res. Treat. 105, 7–16 [DOI] [PubMed] [Google Scholar]
- 20.Chen H. L., Demiralp B., Schneider A., Koh A. J., Silve C., Wang C. Y., McCauley L. K. (2002) J. Biol. Chem. 277, 19374–19381 [DOI] [PubMed] [Google Scholar]
- 21.Dougall W. C., Chaisson M. (2006) Cancer Metastasis Rev. 25, 541–549 [DOI] [PubMed] [Google Scholar]
- 22.Thomas R. J., Guise T. A., Yin J. J., Elliott J., Horwood N. J., Martin T. J., Gillespie M. T. (1999) Endocrinology 140, 4451–4458 [DOI] [PubMed] [Google Scholar]
- 23.Zhu J., Jia X., Xiao G., Kang Y., Partridge N. C., Qin L. (2007) J. Biol. Chem. 282, 26656–26664 [DOI] [PubMed] [Google Scholar]
- 24.Stewart A. F., Vignery A., Silverglate A., Ravin N. D., LiVolsi V., Broadus A. E., Baron R. (1982) J. Clin. Endocrinol. Metab. 55, 219–227 [DOI] [PubMed] [Google Scholar]
- 25.Taube T., Elomaa I., Blomqvist C., Beneton M. N., Kanis J. A. (1994) Bone 15, 161–166 [DOI] [PubMed] [Google Scholar]
- 26.Guo Y., Tiedemann K., Khalil J. A., Russo C., Siegel P. M., Komarova S. V. (2008) Bone 43, 386–393 [DOI] [PubMed] [Google Scholar]
- 27.Tiedemann K., Hussein O., Sadvakassova G., Guo Y., Siegel P. M., Komarova S. V. (2009) J. Biol. Chem. 284, 33662–33670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guise T. A., Mundy G. R. (1998) Endocr. Rev. 19, 18–54 [DOI] [PubMed] [Google Scholar]
- 29.Chirgwin J. M., Guise T. A. (2000) Crit. Rev. Eukaryot. Gene Expr. 10, 159–178 [PubMed] [Google Scholar]
- 30.Guise T. A., Chirgwin J. M. (2003) Clin. Orthop. Relat. Res. 415, S32–S38 [DOI] [PubMed] [Google Scholar]
- 31.Varnum-Finney B., Purton L. E., Yu M., Brashem-Stein C., Flowers D., Staats S., Moore K. A., Le Roux I., Mann R., Gray G., Artavanis-Tsakonas S., Bernstein I. D. (1998) Blood 91, 4084–4091 [PubMed] [Google Scholar]
- 32.Engin F., Bertin T., Ma O., Jiang M. M., Wang L., Sutton R. E., Donehower L. A., Lee B. (2009) Hum. Mol. Genet. 18, 1464–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shih IeM., Wang T. L. (2007) Cancer Res. 67, 1879–1882 [DOI] [PubMed] [Google Scholar]
- 34.Armstrong S., Pereverzev A., Dixon S. J., Sims S. M. (2009) J. Cell Sci. 122, 136–144 [DOI] [PubMed] [Google Scholar]
- 35.Komarova S. V., Pereverzev A., Shum J. W., Sims S. M., Dixon S. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2643–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komarova S. V., Pilkington M. F., Weidema A. F., Dixon S. J., Sims S. M. (2003) J. Biol. Chem. 278, 8286–8293 [DOI] [PubMed] [Google Scholar]
- 37.Canalis E. (2008) Sci. Signal. 1, pe17. [DOI] [PubMed] [Google Scholar]
- 38.Taichman R. S., Emerson S. G. (1998) Stem Cells 16, 7–15 [DOI] [PubMed] [Google Scholar]
- 39.Zhang J., Niu C., Ye L., Huang H., He X., Tong W. G., Ross J., Haug J., Johnson T., Feng J. Q., Harris S., Wiedemann L. M., Mishina Y., Li L. (2003) Nature 425, 836–841 [DOI] [PubMed] [Google Scholar]
- 40.Gori F., Hofbauer L. C., Dunstan C. R., Spelsberg T. C., Khosla S., Riggs B. L. (2000) Endocrinology 141, 4768–4776 [DOI] [PubMed] [Google Scholar]
- 41.Thomas G. P., Baker S. U., Eisman J. A., Gardiner E. M. (2001) J. Endocrinol. 170, 451–460 [DOI] [PubMed] [Google Scholar]
- 42.Baldock P. A., Thomas G. P., Hodge J. M., Baker S. U., Dressel U., O'Loughlin P. D., Nicholson G. C., Briffa K. H., Eisman J. A., Gardiner E. M. (2006) J. Bone Miner. Res. 21, 1618–1626 [DOI] [PubMed] [Google Scholar]
- 43.Holmen S. L., Zylstra C. R., Mukherjee A., Sigler R. E., Faugere M. C., Bouxsein M. L., Deng L., Clemens T. L., Williams B. O. (2005) J. Biol. Chem. 280, 21162–21168 [DOI] [PubMed] [Google Scholar]
- 44.Diarra D., Stolina M., Polzer K., Zwerina J., Ominsky M. S., Dwyer D., Korb A., Smolen J., Hoffmann M., Scheinecker C., van der Heide D., Landewe R., Lacey D., Richards W. G., Schett G. (2007) Nat. Med. 13, 156–163 [DOI] [PubMed] [Google Scholar]
- 45.Qiang Y. W., Chen Y., Stephens O., Brown N., Chen B., Epstein J., Barlogie B., Shaughnessy J. D., Jr. (2008) Blood 112, 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siegel P. M., Massagué J. (2003) Nat. Rev. Cancer 3, 807–821 [DOI] [PubMed] [Google Scholar]
- 47.Yin J. J., Selander K., Chirgwin J. M., Dallas M., Grubbs B. G., Wieser R., Massagué J., Mundy G. R., Guise T. A. (1999) J. Clin. Invest. 103, 197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang Y., He W., Tulley S., Gupta G. P., Serganova I., Chen C. R., Manova-Todorova K., Blasberg R., Gerald W. L., Massagué J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13909–13914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deckers M., van Dinther M., Buijs J., Que I., Löwik C., van der Pluijm G., ten Dijke P. (2006) Cancer Res. 66, 2202–2209 [DOI] [PubMed] [Google Scholar]
- 50.Yamada T., Yamazaki H., Yamane T., Yoshino M., Okuyama H., Tsuneto M., Kurino T., Hayashi S., Sakano S. (2003) Blood 101, 2227–2234 [DOI] [PubMed] [Google Scholar]
- 51.Bai S., Kopan R., Zou W., Hilton M. J., Ong C. T., Long F., Ross F. P., Teitelbaum S. L. (2008) J. Biol. Chem. 283, 6509–6518 [DOI] [PubMed] [Google Scholar]
- 52.Kollet O., Dar A., Shivtiel S., Kalinkovich A., Lapid K., Sztainberg Y., Tesio M., Samstein R. M., Goichberg P., Spiegel A., Elson A., Lapidot T. (2006) Nat. Med. 12, 657–664 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.