Abstract
The canonical heptahelical bundle architecture of seven-transmembrane domain (7TM) receptors is intertwined by three intra- and three extracellular loops, whose local conformations are important in receptor signaling. Many 7TM receptors contain a cysteine residue in the third extracellular loop (EC3) and a complementary cysteine residue on the N terminus. The functional role of such EC3-N terminus conserved cysteine pairs remains unclear. This study explores the role of the EC3-N terminus cysteine pairs on receptor conformation and G protein activation by disrupting them in the chemokine receptor CXCR4, while engineering a novel EC3-N terminus cysteine pair into the complement factor 5a receptor (C5aR), a chemo attractant receptor that lacks it. Mutated CXCR4 and C5aRs were expressed in engineered yeast. Mutation of the cysteine pair with the serine pair (C28S/C274S) in constitutively active mutant CXCR4 abrogated the receptor activation, whereas mutation with the aromatic pair (C28F–C274F) or the salt bridge pair (C28R/C274E), respectively, rescued or retained the receptor activation in response to CXCL12. In this context, the cysteine pair (Cys30 and Cys272) engineered into the EC3-N terminus (Ser30 and Ser272) of a novel constitutively active mutant of C5aR restrained the constitutive signaling without affecting the C5a-induced activation. Further mutational studies demonstrated a previously unappreciated role for Ser272 on EC3 of C5aR and its interaction with the N terminus, thus defining a new microswitch region within the C5aR. Similar results were obtained with mutated CXCR4 and C5aRs expressed in COS-7 cells. These studies demonstrate a novel role of the EC3-N terminus cysteine pairs in G protein-coupled receptor activation and signaling.
Keywords: Chemokines, Cysteine-mediated Cross-linking, G Protein-coupled Receptors (GPCR), G Proteins, Sulfhydryl, C5a Receptor, CXCR4
Introduction
G protein-coupled receptors (GPCRs),2 often referred to as seven-transmembrane domain receptors, are one of the most biologically important superfamilies of receptors (1). Phylogenetic analysis classifies the superfamily of receptors into glutamate, rhodopsin, adhesive, frizzled, and secretin families (2). The rhodopsin family alone includes ∼750 GPCRs (3). ∼60% of GPCRs respond to sensory and olfactory neurons; ∼30% respond to protein and peptide hormones, amino acids, biogenic amines, and lipids, whereas the remaining GPCRs respond to nutrients and metabolites. In response to ligands, the receptors recruit either G proteins or other intracellular effector proteins (4), which transduce the extracellular chemical signals and further stimulate intracellular signaling cascades (5). This extraordinary ability to sense such a diverse array of chemical signals with selective precision classifies GPCRs as a “druggable proteome” (6) that generates ∼10% of the global pharma revenue (7).
Given their cellular and pharmacological importance, understanding the receptor function at the molecular level is a prerequisite, and indeed the recent advancements in the high resolution crystals of rhodopsin-like GPCRs have been significantly informative (8–12). Nevertheless, a central question remains how the canonical receptor topology interconverts between several possible conformations in response to diverse chemical stimuli. Traditionally, the GPCR activation mechanism is described as “molecular switches” identified in conserved “microdomains” (13) and recently reviewed elsewhere (14, 15). Briefly, ligand/agonist binding or an activating point mutation (CAM) triggers a series of molecular macroswitches, such as the global rotamer toggling (16, 17) on TM6 or the disruption of an ionic lock (18, 19), between TM3 and TM6, to unlock the G protein-binding site in the intracellular face of the receptors leading to G protein activation. However, ∼80% of the rhodopsin family receptors do not have the putative residues to support the described “rotamer toggle” switch, the “ionic lock,” or both (20). Although a universal activation mechanism is also suggested by the ability of human receptors to activate evolutionarily distant yeast G proteins, how this is accomplished is difficult to reconcile within a framework of low sequence homology (≤20%) between receptors (21), the variable length of the peptide loops, and the diversity of N termini in receptors. Indeed, the significant variation observed in pharmacological properties of receptors (22, 23) within the same family (24) does not favor a single, unified activation mechanism model.
Within the rhodopsin family of GPCRs, the EC3 loop varies in length between 4 and 27 residues and plays a key role in the activation of the neuropeptide class of receptors (25). Minor alterations affecting its folding or structure have been shown to impair both ligand binding and signaling (26–32). This study focuses specifically on conserved cysteine residues in EC3 with respect to the NPXXY motif in TM7. Using the Ballesteros and Weinstein numbering system that designates the most conserved residue in the TM as X.50 (for example, in TM7, the proline position in the NPXXY is 7.50), C7.25 is the position at which cysteine residues are conserved in EC3. The EC3 cysteine (C7.25) in several receptors has been proposed to participate in a disulfide linkage with another conserved cysteine in the N terminus. These paired cysteine residues are highly conserved among chemokine-binding receptors and have been demonstrated to be important for both EC3 structure and perhaps high affinity ligand binding (30, 33–36). However, their exact role in structure-function relationship is still unclear.
This study focuses on the role of such cysteine pairs in ligand binding, activation, and maintaining the overall inactive/active conformation of the receptors. Two representative rhodopsin family receptors that signal through Gαi subunits have been chosen for this study as follows: CXCR4 (37), activated by chemokine CXCL12 (SDF-1), and C5aR, activated by anaphylatoxin C5a (38). We chose to express the receptors in engineered yeast (39) because this system offers several advantages for this analysis as follows: there are no other endogenous GPCRs that might interact with the mutant receptors, and there is only one G protein. We find that the EC3-N terminus cysteine pairs serve different functions in these receptors. The cysteine pair adds functionality similar to a weak agonist by increasing the receptor activation in CXCR4, although it conformationally constrains an activated receptor similar to an inverse agonist when engineered into the C5aR. Moreover, our mutational data suggest a more involved and coordinated interaction between EC3 and the N terminus that is essential in these receptors to maintain the active sites for the ligands and the overall conformation of the receptors for signaling.
EXPERIMENTAL PROCEDURES
Yeast Strains
The Saccharomyces cerevisiae strain BY1173 (39) with the following genotype [MATa His3 Leu2 Trp1 Ura3 Can1 gpa1Δ::ade2Δ::3XHA far1Δ::ura3Δ Fus1::PFus1-His3 LEU2::Leu2 PFus1LacZ SST2Δ::ura3Δ ste2Δ::G418R Trp1:: GPA1/Gαi(1/2)] has been used in this study unless otherwise specifically mentioned. This specific strain expresses a chimeric yeast-human Gα subunit (GPA1-Gαi(1/2)/(5)) in which the C-terminal five amino acids of human Gαi(1/2) replaced the C-terminal five amino acids of the yeast Gα subunit, GPA1. This strain also carries a deletion allele of SST2, which is known to down-regulate the pheromone-response pathway by accelerating the GTPase activity of GPA1. The engineered MAPK mating pathway (1) in BY1173 is coupled to the FUS1-LacZ reporter enzyme, which stimulates basal production of β-galactosidase upon ligand-dependent or -independent activation of the receptors coupling to the chimeric G protein.
Yeast Transformation
All the receptor and ligand plasmids were transformed to the yeast following the standard lithium-acetate protocol. All plasmids used for transformation are 2-μm-based and contained an REP3 element for autonomous replication in S. cerevisiae and ampicillin-R for selection in Escherichia coli. All the receptor plasmids possess an ADE2-selectable marker, which expresses the gene of interest under the control of the constitutive phosphoglycerate kinase (PGK1) promoter. All the ligand plasmids possess a URA3-selectable marker, which expresses the gene of interest under the control of the constitutive alcohol dehydrogenase (ADH) promoter. Control transformations were made using empty ADE2 and empty URA3 plasmids to control for plasmid copy number in conditions that lack receptor, ligand, or both. Positive transformants were selected by allowing the yeast to grow at 30 °C on adenine-uracil dropout solid media.
β-Galactosidase Assays
Three positive transformants were selected from each plate and subjected to liquid β-galactosidase assays. The yeast cells were grown overnight in appropriate synthetic dropout medium until confluent. The confluent culture was diluted to ∼0.15 A600 0.5 ml−1 and further grown at 30 °C in a humidified environment for 4–5 h. For dose-response studies, an appropriate concentration of the peptide agonist was added to the cells expressing the respective receptor mutants. The expression of the β-galactosidase was monitored by lysing the cells with 50 μl of buffer consisting of a 50:50 mixture of 5% (w/v) Triton X-100 (Bio-Rad) in 250 mm PIPES, pH 6.8, and 4.86 mg ml−1 chlorophenol red galactosidase (Roche Applied Science) in 25 mm PIPES, pH 6.8, and incubating in the dark at 37 °C. β-Galactosidase activity was determined by terminating the reaction after ∼1–1.5 h by adding 50 μl of 1 m Na2CO3 and measuring A570 on a Bio-Rad model 680 microplate reader. Color development for yeast cells expressing the wild type CXCR4 required overnight incubation with lysis buffer. Data were plotted and analyzed using Prism 5 (GraphPad software).
Construction of Mutant Receptor and Ligands
Point mutations were introduced into the receptor and ligand plasmids by site-directed mutagenesis using Pfu Turbo polymerase (Stratagene). The presence of the only desired point mutation was confirmed by sequencing both strands of the entire gene at the Protein and Nucleic Acid Chemistry Laboratory, Washington University School of Medicine, St. Louis, MO.
Western Blots
The expression levels of some single/double point mutants of C5aR were assessed by Western blot. Yeast cells carrying empty ADE2 plasmid or plasmids encoding wild type/mutants were grown overnight in adenine dropout liquid media. The overnight cultures were adjusted to ∼1 A600 ml−1. Yeast cells were lysed in a 50:50 mixture of 4× LDS NuPAGE buffer (Invitrogen) and 62.5 mm Tris, pH ∼7.5, supplemented with 1 μg/ml leupeptin, 1 μg/ml aprotinin, and 500 μm phenylmethylsulfonyl fluoride. ∼200 mg of 0.5-mm glass beads (Biospec Products, Bartlesville, OK) was added to the mixture, followed by vortexing at high speed for 5 min. The emulsion was heated for 10 min at 50 °C, cooled on ice, and centrifuged for 5 min. 25 μl of each supernatant was resolved on a NuPAGE® Novex 4–12% BisTris gel, transferred to nitrocellulose membrane, and immunoblotted with a rabbit polyclonal anti-C5aR antibody raised against full-length endogenous C5aR (Novus Biologicals, Littleton, CO). The blot was then washed and probed with horseradish peroxidase conjugated anti-rabbit IgG (Santa Cruz Biotechnology) at room temperature. Protein bands were detected using ECL methods.
Sequence Analysis of the GPCRs
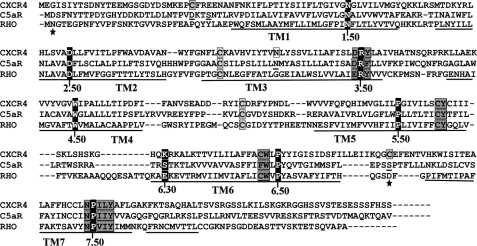
All sequences were downloaded from the GPCR data base and aligned with ClustalX-2.0.11 program. The sequence alignment presented in Fig. 1 has been further manually annotated.
FIGURE 1.
Sequence alignment of CXCR4 and C5aR with rhodopsin highlighting the most conserved residues and the known activation switches in GPCRs. Ballesteros and Weinstein nomenclature assigns X.50, to the “fingerprint residues” shown as white letters with black background in each transmembrane domain. Important transmembrane activation domains are shaded in gray. Conserved cysteine residues participating in disulfide bond formation are shown with gray boxes. Residues shown with black boxes indicate lack of complementary charged residues at 6.30 to support the ionic lock activation switch. Point mutation known to confer constitutive activity in CXCR4 is shown with white box. Residues shown as underlined have been subjected to mutagenesis for engineering C5aR. Residues marked with stars have been mutated to cysteine in other studies for engineering an extra disulfide bond between EC3 and N terminus of rhodopsin.
Mammalian Cell Culture and Transfections
Mutant C5aR receptor plasmids were constructed by site-directed mutagenesis using Pfu Turbo polymerase (Stratagene) in IRES vector (Clontech) encoding Gα16. Wild type and mutant CXCR4 plasmids were constructed by two-step restriction endonuclease digestion followed by two-step ligation to introduce receptor and Gα16 genes into the IRES vector. COS-7 cells maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum, 100 μg/ml streptomycin sulfate, 100 units/ml penicillin G were transiently transfected with wild type or mutant receptors using Lipofectamine LTX with plus reagents (Invitrogen) according to the manufacturer's protocol.
Inositol Phosphate Accumulation Assays
COS-7 cells expressing a promiscuous Gα16 subunit with either wild type or mutant receptors were supplemented with 1 μCi of [3H]inositol (PerkinElmer Life Sciences) for 24 h followed by stimulation in the absence or presence of 10–100 nm ligands at 37 °C for 1 h. Accumulated IP3 fractions were separated as described elsewhere (40) and counted in Scintiverse scintillation fluid (Fisher) until 5% significance.
Membrane Preparation and Saturation Binding Assays
Membranes of COS-7 cells expressing wild type or mutant receptors were prepared as described elsewhere (41). The membranes (1–10 μg of total protein) were thawed on ice and incubated for 1 h with 0.1 nm 125I-labeled C5a in binding buffer (Hanks' balanced salt solution, supplemented with 25 mm HEPES, pH 7.4, and 0.1% BSA). Binding reactions were performed on ice in the presence of varying concentrations of recombinant nonradioactive C5a (Sigma) and terminated by vacuum filtration through GF/C filters (Whatman) presoaked in 0.1% poly(ethyleneimine) (Sigma) with a Millipore (Temecula, CA) harvester. Filters were rapidly washed with ice-cold binding buffer and then added to scintillation tubes with 3 ml of the scintillation fluid. Same protocol was followed using 0.1 nm 125I-labeled CXCL12 binding to CXCR4 membranes in the presence of varying concentrations of recombinant nonradioactive CXCL12 (Abcam Inc.). Data were analyzed with GraphPad Prism 5.0.
Molecular Modeling
Approximately 20% overall sequence identity was shared between C5aR and rhodopsin. Besides the low level of sequence identity, the 2.8-Å resolution x-ray structure of bovine rhodopsin (Protein Data Bank code 1F88) was chosen as the starting three-dimensional template for generating homology-based model coordinates for C5aR, recruiting the program MODELLER (42). The conserved disulfide between TM3 and EC2 (Cys109–Cys188) of C5aR could be modeled successfully with MODELLER. To realize the putative engineered disulfide bond between the N terminus and the EC3 loop, small peptides corresponding to the EC3 and N terminus were modeled manually, and the hypothetical φ, ψ angles were grafted onto the MODELLER defined coordinates. The modified C5aR coordinate was then submitted to careful energy minimization in a cubic box with periodic boundary involving gromos-96 43a1 force field (43) as implemented in GROMACS 3.3.1. Placed in center of a periodic box, large enough to accommodate ∼10.5 nm solvent layer on each side, the receptor coordinate was energy minimized to 100 kJ mol−1 nm−1 tolerance with steepest descent, first in vacuum and then in the presence of simple point charge (SPC) water model, as provided in GROMACS. Numerical integrations were performed in step size of 2 fs. Bonds were constrained with SHAKE to the tolerance 0.0001. Nonbonded pair list cut-off was 1.4 nm with shift function. Solvent density was set to the value corresponding to 1 atm at 300 K. Receptor and solvent were coupled independently to Berendsen bath at 300 K, to the coupling time constant 0.1 ps. Amide bond geometry, including the φ, ψ angles of individual residues were analyzed to assess the general quality of the model. For comparison purposes, the crystal structure of wild type and engineered rhodopsin were also energy-minimized. The figures were made using the program PyMOL.
RESULTS
Sequence Alignment of Rhodopsin Family Receptors
Human GPCRs were aligned in search of conserved cysteine residues on EC3 within 25 amino acids relative to the highly conserved P7.50 of NPXXY (TM7) motif and on the N terminus within 20–25 amino acids relative to the highly conserved N1.50 (TM1). The alignment identified 48 receptors within 15 subfamilies of rhodopsin family of GPCRs (summarized in Table 1), which have such EC3-N terminus cysteine pairs. Of the 48 receptors, 24 receptors were identified as chemokine or chemokine-type receptors. However, many other receptors (Table 1) such as angiotensin, purigenic, and cysteinyl leukotriene receptors were also found to possess such EC3-N terminus cysteine pairs. Receptors with multiple cysteine residues either in EC3 or in the N terminus were excluded from this analysis; examples included the muscarinic acetylcholine, dopamine, MC4R, nicotinic, and serotonin type-7 receptors. Interestingly, most of the receptors possessed the cysteine pairs at a highly conserved position on EC3 (C7.25), and on N terminus (C1.22), although some variability was noted for EC3 and N-terminal cysteine positions in some receptors (Table 1). The minor positional difference could be due to the differences of both the N terminus and loop lengths in receptors. The CXCR4 receptor was chosen as a representative of chemokine family receptor for further study. To better understand the functional role of such cysteine pairs, a novel EC3-N terminus cysteine pair (Cys30 and Cys272) was engineered into C5aR, a chemoattractant receptor that lacks such a cysteine pair.
TABLE 1.
Summary of receptors with conserved cysteine residues on EC3 and N terminus
| Receptor subfamilies | Total no.a | Cysteine positions on EC3 and N erminus |
|---|---|---|
| GPR182 | 2 | (C7.25, C1.22) |
| Angiotensin type 1 | 1 | (C7.25, C1.22) |
| Angiotensin type 2 | 1 | (C7.25, C1.22) |
| B1 bradykinin | 1 | (C7.25, C1.22) |
| B2 bradykinin | 1 | (C7.25, C1.22) |
| Chemokine | 16 | (C7.25, C1.22) |
| Chemokine-type receptors | 8 | (C7.25, C1.22) |
| EBV-induced | 2 | (C7.25, C1.22) |
| Leukotriene type 1 | 1 | (C7.25, C1.23) |
| Leukotriene type 2 | 1 | (C7.27, C1.25) |
| Lysosphingolipid and lysophosphatidic acid | 2 | (C7.25, C1.24) |
| Lysophosphatidic acid 4 | 1 | (C7.25, C1.24) |
| Prostacyclin or prostaglandin | 3 | (C7.24, C1.24) |
| Purigenic type | 5 | (C7.26, C1.23) |
| Uncharacterized | 3 | (C7.25, C1.22) |
a Data are based on GPCR database.
Sequence Alignment of CXCR4 and C5aR with Rhodopsin
Sequence alignment to rhodopsin, presented in Fig. 1, demonstrates that CXCR4 has an extra pair of conserved cysteine residues in EC3 (C7.25) and the N terminus (C1.22), which in principle can support a putative disulfide linkage, whereas both rhodopsin and C5aR lacks this cysteine pair. Considering rhodopsin crystal structure as a template, the predicted EC3 loop is of 15 residues in CXCR4, and of 10 residues in C5aR compared with 6 residues in rhodopsin. Moreover, the alignment highlights the known activation microswitches such as W6.48 of XWXP (TM6), R3.50 of (D/E)RY (TM3), Y5.58 of XY (TM5), and Y7.53 of NPXXY (TM7) motifs that are conserved in CXCR4, C5aR, and rhodopsin. The W6.48 (TM6) represents the global “rotamer toggle” microswitch in GPCRs. R3.50 (TM3), D3.49 (TM3), and E6.30 (TM6) represent the ionic lock microswitch in GPCRs. In contrast to rhodopsin, CXCR4 and C5aR do not conserve the ionic lock because they lack a negatively charged residue at the appropriate position in TM6 (K6.30, and S6.30, respectively). Y5.58 (TM5) is recently known to latch R3.50 (TM3) in an active state of opsin (11). Y7.53 of TM7 is an allosteric activation microswitch participating in formation of complex hydrogen bond networks with conserved water molecules (44).
Signaling Profile of Mutant Human CXCR4 in Yeast
Mutations C28A, C274A, or C28A/C274A of the N-terminal (Cys28) and EC3 (Cys274) cysteine residues has been shown to significantly alter CXCR4 function as a co-receptor in HIV-1 Env-mediated cell fusion in a strain-specific manner (35). In tissue culture studies, mutation of C28A or C274A has also been shown to decrease the specific binding of CXCL12 by ∼50% amounting to an ∼6-fold increase in the Kd value for mutant receptors compared with wild type CXCR4 (30). However, the role of these cysteine residues in CXCL12-dependent or -independent CXCR4 signaling is essentially not known. We mutated C28S, C274S or C28S/C274S both in wild type and CAM of CXCR4 and expressed the receptors in yeast strain BY1173, which features a chimera of amino acids 1–467 of GPA1 followed by the last five amino acids of human Gαi2 (39). In the isolated environment of the yeast cell, a single human GPCR can be studied in the absence of other receptors competing for G protein binding, and the readout of receptor activation is clear because only a single G protein chimera is expressed. The yeast strain used in this study was engineered so that receptor signaling leads to activation of the yeast-mating pathway resulting in expression of a PFUS1-β-galactosidase reporter gene (39).
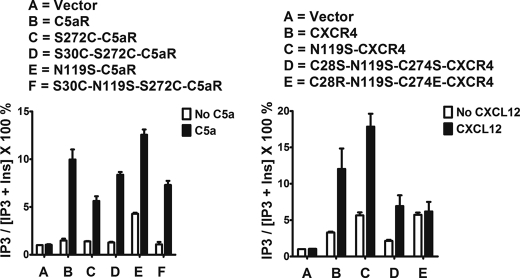
Wild type CXCR4 displayed weak signaling in response to the CXCL12 (45) that was co-expressed in the yeast (Fig. 2, right panel, lane A). Upon overnight incubation with substrates, yeast co-expressing WT-CXCR4 and CXCL12 displayed ligand-dependent signaling (1.34 ± 0.27-fold; n = 2, data not shown); however, the signal was near the basal activity of the system making it difficult to assess the effects of cysteine mutations in the context of wild type CXCR4. Thus, we chose to study the effect of cysteine mutation on CXCR4 signaling in the TM3 mutant N119S (N/S3.35) (Fig. 2, left panel), which displays robust constitutive signaling and responds to agonist both in yeast and mammalian cells (46, 47). As noted in Fig. 2, right panel, the N119S mutant CXCR4 displayed near-maximal signaling that increased in response to CXCL12 co-expression (1.35 ± 0.18-fold; n = 5). Individual C28S and C274S mutations displayed ∼50% reduced constitutive active signaling, which upon CXCL12 stimulation displayed an apparently similar fold increase in signaling (Fig. 2, right panel). In contrast, the double mutation C28S/C274S completely impaired the constitutive activity of CXCR4 and abrogated the CXCL12-mediated signaling. These data clearly suggest that the double cysteine mutations have an additive effect on the receptor activity relative to individual cysteine mutations. If the sole function of the cysteine pair were to form a disulfide bridge, one would expect that the individual mutations would give similar results when compared with the double mutation.
FIGURE 2.
Signaling profile of mutant CXCR4 receptors co-expressed with CXCL12 in yeast. Left panel, canonical receptor topology for CXCR4. The N119S mutation on TM3 is known to produce constitutive active signaling in CXCR4. Cys28 and Cys274, presumed to form a disulfide linkage between N terminus and EC3 loop, is highlighted in white with black background. Right panel illustrates the contrasting functional effect of C28S, C274S, C28S/C274S, C28F/C274F, and C28R/C274E on constitutively active mutant CXCR4 signaling. Lane A illustrates the weak signaling induced by CXCL12 when co-expressed with WT-CXCR4 in yeast. Vector represents the β-galactosidase activity of the engineered yeast in the absence of both receptor and ligand. Each bar represents the means ± S.D. of signaling activity for three independent transformants, and data are representative of at least two independent experiments. Unpaired t test suggests statistical significance with ***, p ≤ 0.0005, and *, p < 0.05 versus no CXCL12.
Rescuing the Constitutive Signaling of CXCR4 in Yeast
The complete loss of constitutive signaling in the C28S/C274S mutant CXCR4 and its failure to respond to CXCL12 (Fig. 2, lane E) could be attributed to folding defects leading to improper receptor trafficking or to the loss of overall conformational integrity due to the absence of the EC3-N terminus cysteine pair. Mutation of conserved cysteine pairs with serine has been shown to completely impair the antimicrobial effects of Mesentericin Y105 (48), a known bacteriocin. In this setting, noncovalent interactions such as hydrophobic or π-stacking could functionally replace the conserved disulfide linkage in type IIa bacteriocin antimicrobial peptides (49, 50). To examine such possibilities in CXCR4, we looked for mutant pairs that introduce aromatic-aromatic (C28F/C274F) or salt bridge interactions (C28R/C274E) between EC3 and N terminus, which can potentially rescue the function by acting as a surrogate to disulfide linkage.
As illustrated in Fig. 2, right panel, the double mutant C28F/C274F did not rescue the constitutive signaling of CXCR4, but it did display potent CXCL12-stimulated signaling (Fig. 2, right panel, compare lane E with lane F). On the other hand, the double mutant C28R/C274E rescued both the constitutive signaling as well as CXCL12-dependent signaling similar to the CXCR4-CAM (Fig. 2, right panel, compare lane B with lane G). Energetically, single exposed salt bridge interactions (0.8/1 kcal/mol) could be weaker than the hydrophobic π-stacking interactions (−1.3/−2 kcal/mol), which can provide more conformational stability to the receptor. The energetic component of the engineered interactions may account for the difference in constitutive signaling displayed by C28F/C274F and C28R/C274E mutant receptors. These data demonstrate that an interaction between EC3 and the N terminus of CXCR4 could be essential for receptor activation and that this interaction does not need to be mediated by cysteine pairs.
Engineering C5aR into a Chemokine-type Receptor
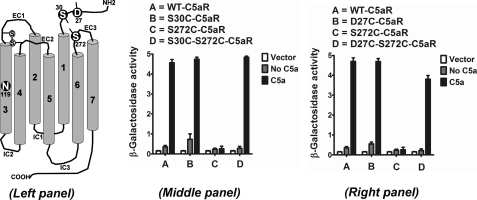
Chemokine-binding GPCRs have evolved with an extra cysteine pair in the N terminus and EC3 loop, which separates them from other GPCRs in the rhodopsin family. Except for CXCR6, the chemokine binding receptors possess a cysteine residue at a highly conserved position on EC3 (C7.25), corresponding to P7.50 of NPXXY (TM7) motif, which is complemented by another cysteine residue at a highly conserved position (C1.22), corresponding to N1.50 on the N terminus with the exception of CXCR5 (C1.19). We wanted to know the effects of introducing the conserved cysteine pair in a GPCR that lacks it. For this analysis, we chose to engineer the cysteine pair into the C5aR. As highlighted in Fig. 1, Ser272 (S7.25) of C5aR aligned with the conserved cysteine of CXCR4 on EC3 and was therefore mutated to cysteine. This position also corresponds to Asp282 in bovine rhodopsin, which has been successfully engineered to form a disulfide bond between Asp282 of EC3 and Asn2 of N terminus, as demonstrated in a crystal structure (51). To sample the flexibility of the N terminus and its probability to form the putative disulfide linkage with S272C of EC3, two residues, Asp27 (D1.22) and Ser30 (S1.25), respectively, on the N terminus of C5aR were chosen for cysteine mutation (Fig. 3, left panel).
FIGURE 3.
Engineering C5aR to a chemokine-type receptor; signaling profile of mutant C5aR receptors, co-expressed with C5a in yeast. Left panel, canonical receptor topology for C5aR. The N119S mutation on TM3 is a novel constitutively active mutant identified for C5aR in this study. Asp27, Ser30, and Ser272, highlighted in white with black background, have been mutated to cysteine for engineering a possible extra disulfide linkage between the N terminus and EC3 loop of C5aR. Middle and right panel, respectively, illustrate the effect of S30C, S272C, S30C/S272C, and D27C, S272C, D27C/S272C on C5a-stimulated C5aR signaling. Vector represents the β-galactosidase activity of the engineered yeast in the absence of both receptor and ligand. Each bar represents means ± S.D. of signaling activity for three independent transformants, and data are representative of at least two independent experiments.
Signaling Profile of Engineered C5aR in Yeast
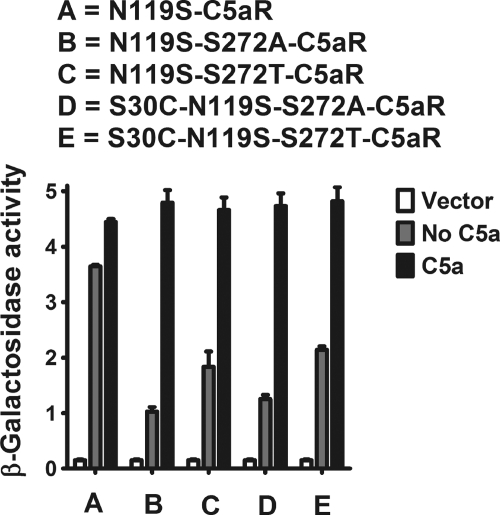
Mutation S30C (Fig. 3, middle panel) or D27C (Fig. 3, right panel) on the N terminus did not display any appreciable loss in signaling compared with wild type C5aR in response to C5a, although S30C mutants displayed more C5a-independent activity compared with both D27C and wild type C5aR. This is in accordance with the earlier observations that the N terminus tolerates many point mutations (52). On the other hand, S272C mutation on EC3 (Fig. 3) failed to demonstrate C5a-mediated signaling. The loss of function due to the S272C mutation could be rescued by the additional mutation of S30C or D27C mutation in the N terminus of C5aR (Fig. 3, middle or right panel, respectively). As noted in Fig. 4, introduction of S272A or S272T mutation on EC3 of C5aR also severely impaired C5a-mediated signaling, suggesting a strict requirement for Ser272 in EC3 of C5aR.
FIGURE 4.

Signaling profile of wild type and mutant C5aR receptors, co-expressed with wild type and C27R C5a in yeast. The nonfunctional mutants, such as S272A and S272T, have not been tested with C27R C5a. Vector represents the β-galactosidase activity of the engineered yeast in the absence of both receptor and ligand. Each bar represents means ± S.D. of signaling activity for three independent transformants, and data are representative of at least two independent experiments.
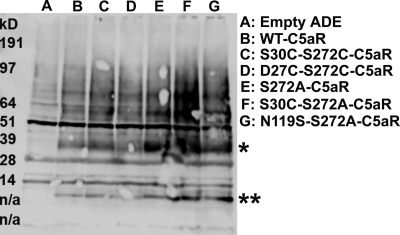
Surprisingly, the S30C mutation on the N terminus rescued S272A and S272T nonfunctional mutant C5aRs (Fig. 4). This result demonstrates that the rescue effect observed in the D27C/S272C and S30C/S272C may not be uniquely dependent on introduction of a disulfide bridge but could be due to local hydrophobic or conformational interactions between the membrane-proximal portion of the N terminus of C5aR and EC3. One possible mechanism by which the D27C or S30C might rescue activity of the mutated Ser272 C5aRs could be by binding the unpaired cysteine on the C5a ligand. In our previous work (53), saturation mutagenesis identified positions in the membrane-proximal portion of the N terminus into which cysteine mutations occurred that could apparently interact with the unpaired cysteines present in the C5a ligand. To exclude this possibility in this study, we co-expressed the “cysteine-less” C5a ligand that contains a C27R mutation with the S30C-Ser272-mutated C5aRs. Illustrated in Fig. 4, the cysteine-less C5a ligand efficiently activated the S30C-S272T and S30C-S272A C5aR, as well as the wild type C5aR. Wild type C5aR displayed maximal signaling in response to human C5a, which contains an unpaired cysteine (Cys27), and a mutated C5a in which the unpaired cysteine was substituted for a conserved residue in the rat C5a sequence (C27R). This result is consistent with previous findings from our group (53) and demonstrates that the Cys27 on C5a is not required for C5aR signaling. Moreover, this result demonstrates that the signaling observed for the single cysteine-mutated C5aR is unlikely to be due to the formation of a disulfide bond between the C5a ligand and the receptor. Western blot analyses of whole yeast lysates of these single and double mutants of C5aR (presented in Fig. 5) demonstrate that these mutants were expressed well in yeast, although at different levels. Of note, the expression level did not correlate with the strength of signaling. For example, the weak signaling mutant S272A C5aR was expressed more highly than the wild type C5aR or the strong signaling mutant D27C/S272C-C5aR (Fig. 5, compare lane E with lanes B and D). This lack of correlation between receptor expression and activity might reflect that the receptors are expressed in excess of the G proteins and are therefore not limiting.
FIGURE 5.
Effect of point mutations on expression profile of C5aR in yeast as assessed by Western blot. 25 μl of whole yeast lysates expressing the mutant or the wild type receptors were resolved on a 4–12% BisTris gel and detected as described under “Experimental Procedures.” The single and double asterisks, respectively, indicate the full-length and proteolytic fragments of the receptors. n/a, not applicable.
Previous work by several groups implicated EC3 residues for interactions with the arginine residue at the C terminus (Arg74) of the C5a ligand. Mutational studies support that Asp282 located at the EC3/TM7 junction in the C5a receptor forms a salt bridge interaction with Arg74. In support of this model, by removing Arg74 from C5a (C5a-des-Arg74, a natural metabolite of C5a generated by proteolysis), the C5a-des-Arg74 ligand activated the D282A-C5aR with similar potency compared with the full-length C5a (54). To test if the S272A mutation affects the potential interaction of Arg74 with EC3, we generated a C5a-des-Arg74 ligand that could be expressed in yeast. In our studies in yeast, C5a stimulated weak signaling in D282A-C5aR, whereas C5a-des-Arg74 induced potent signaling in D282A-C5aR (Fig. 6), consistent with the earlier studies in mammalian cells (54, 55). However, both C5a and C5a-des-Arg74 failed to stimulate signaling in S272A-C5aR (Fig. 6), suggesting that the S272A mutation in EC3 induces a conformational change that might compromise the active site and the affinity of the C5a for C5aR. To test this possibility, we assessed the ability of the 10-mer peptide agonist (YSFKPMPLaR) (54, 56) to activate wild type and mutant C5aRs. As noted in Fig. 7, left panel, ∼10 μm of the peptide was able to stimulate strong signaling both in wild type and mutant C5aRs including S272A-C5aR. In concentrations ranging from 100 pm to 10 μm (Fig. 7, right panel), the peptide displayed EC50 of 4.6 ± 0.14 nm for wild type C5aR, which increased ∼120-fold (488 ± 14 nm) for S272A-C5aR. On the other hand, both the double mutants S30C/S272A and S30C/S272C displayed moderately increased EC50 values of 38 ± 1.4 nm (∼9-fold) and 108 ± 1.4 nm (∼23-fold), respectively, for the peptide. These data demonstrate that the mutated C5a receptors were competent to transduce signals, and the lack of C5a-induced signaling in S272A/S272T/S272C mutant C5aRs could be due to decreased C5a affinity toward the mutants. These data further support that the rescue effect observed in double mutants S30C/S272A or S30C/S272C is due to the conformational interactions between the membrane-proximal portion of the N terminus of C5aR and EC3. The ∼2.5-fold difference observed in EC50 values between S30C/S272A and S30C/S272C affirms that these mutant receptors likely have similar conformations. This supports the hypothesis that GPCRs can exist in a multitude of conformational ensembles in equilibrium and can differentially modulate G protein binding with respect to their ligands (57, 58).
FIGURE 6.

Comparison of signaling profile of wild type, D282A, and S272A mutant C5aR receptors, co-expressed with C5a and metabolite C5a-des-Arg74 in yeast. C5a-des-Arg74 stimulates only ∼30% of the signaling in wild type C5aR compared with the signaling stimulated in response to C5a. D282A mutant displays reduced signaling in response to C5a consistent with other studies but displays near-maximal signaling in response to metabolite C5a-des-Arg74. The nonfunctional mutant S272A neither responds to C5a nor to the metabolite C5a-des-Arg74. Vector represents the β-galactosidase activity of the engineered yeast in the absence of both receptor and ligand. Each bar represents means ± S.D. of signaling activity for three independent transformants, and data are representative of at least two independent experiments.
FIGURE 7.
Quantifying the efficacy of the 10-mer peptide agonist toward wild type and mutant C5aRs. Left panel, maximal signaling stimulated in wild type, S272A, S30C/S272A, and S30C/S272C mutant C5aRs in response to ∼10 μm peptide agonist. Vector represents the β-galactosidase activity of the engineered yeast in the absence of both receptor and ligand. Each bar represents means ± S.D. of signaling activity for three independent transformants, and data are representative of at least two independent experiments. Right panel, dose-dependent signaling stimulated by the 10-mer peptide agonist in wild type, S272A, S30/S272A, and S30C/S272C mutant C5aRs. y axis represents β-galactosidase activity normalized to the maximum signaling, and each point represents means ± S.D. of signaling activity for three independent transformants, and data are representative of at least two independent experiments.
Identification of CAM of Human C5aR
Alignment of CXCR4 and C5aR (Fig. 1) revealed that like CXCR4 C5aR also has an asparagine in TM3 (N3.35). Sequence alignment of 736 human GPCRs of the rhodopsin family demonstrated a polar (Cys/Ser/Thr/Asn), nonpolar (Gly/Ala/Val/Ile/Leu/Met) or an aromatic (Phe/Tyr) residue in 47, 34, and 18% of the receptors at this position, respectively. We found that N3.35 (TM3) is a conserved residue among 15% of the receptors. The most abundant amino acids at N3.35 are Ser/Cys/Thr in ∼32% of the receptors, followed by Gly/Ala in ∼14% of the receptors. Mutation of N3.35 to Gly/Ala/Ser has been previously shown to confer robust constitutive active signaling in CXCR4 (47) and moderate constitutive signaling in angiotensin II AT1 receptor (59, 60). Based on this, we constructed the TM3 mutant, Asn/S3.35 of C5aR. As illustrated in Fig. 8, left panel, the N119S mutant C5aR displayed strong constitutive activity in yeast, with a modest increase in signaling upon stimulation with C5a to a level comparable with that observed with maximal wild type C5aR signaling, thus identifying a robust novel CAM of C5aR.
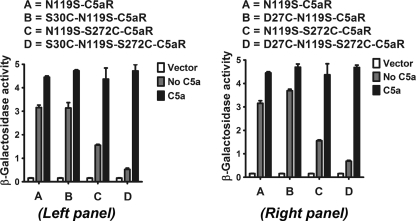
FIGURE 8.
Positional effect of N-terminal and EC3 cysteines on constitutive signaling profile of C5aR and their respective response to C5a, co-expressed in yeast. Left and right panels, respectively, illustrate the effect of S30C, S272C, S30C/S272C, and D27C, S272C, D27C/S272C on ligand-independent and -dependent signaling of constitutively active C5aR. The contrasting effect of S30C/S272C, D27C/S272C, and S272C on constitutive signaling of C5aR is noted in both panels. Vector represents the β-galactosidase activity of the engineered yeast in the absence of both receptor and ligand. Each bar represents means ± S.D. of signaling activity for three independent transformants, and data are representative of at least two independent experiments.
Effect of Engineered Cysteine Pair on Constitutive Signaling of C5aR in Yeast
As noted in Fig. 8, neither the D27C nor the S30C mutation in the N terminus affected the constitutive signaling or C5a-stimulated signaling, congruent with results observed when these mutations were introduced into the wild type C5aR (Fig. 3). The loss of function in the wild type C5aR imparted by the S272C mutant could be rescued with either an S30C or a D27C mutation (Fig. 3), suggesting that this region of the N terminus of C5aR might directly interact with EC3. Thus, the importance of Ser272 on EC3 conformation and its ability to form the putative disulfide linkage with S30C on N terminus was systematically probed by substituting Ser272, respectively, to S272T, S272A, and S272C in the CAM of C5aR.
In contrast to wild type EC3 mutants (Figs. 3 and 4), the introduction of S272A/S272C/S272T into the CAM did not abolish receptor activity; the mutated receptors retained some constitutive signaling and exhibited maximal C5a-stimulated signaling (Figs. 8 and 9). Of note, the S30C and D27C mutations in the N terminus decreased the maximal constitutive signaling displayed by N119S-S272C mutant without affecting the C5a-induced signaling (Fig. 8, compare lanes C with D in both panels). However, the presence of the S30C mutation on the N terminus did not result in a substantial change in the constitutive signaling of S272A or S272T mutants, and like the single point mutants, these double mutants also displayed maximal C5a-stimulated signaling (Fig. 9, compare lanes B with D and C with E, respectively). The apparent loss in constitutive signaling in S30C/S272C C5aR CAM (Fig. 8, compare lanes A with D in both panels) may be attributed to the conformational stability induced by the cysteine pair in C5aR CAM. This is an interesting observation as EC3-N terminus cysteine pair displays contrasting function in CXCR4 and C5aR.
FIGURE 9.
Probing the side chain effect at Ser272 of EC3 on constitutive signaling of C5aR in yeast. The constitutive signaling profile of S272T, S272A, and S272C mutants in the presence and absence of S30C at the N terminus of C5aR. Maximal signaling is noted for all the single and double mutants in response to C5a. Vector represents the β-galactosidase activity of the engineered yeast in the absence of both receptor and ligand. Each bar represents means ± S.D. of signaling activity for three independent transformants, and data are representative of at least two independent experiments.
Signal Transduction and Membrane Binding Assays of Mutated Receptors Expressed in Mammalian Cells
Although the yeast system allows for convenient testing of large numbers of mutated receptors, the yeast cell wall is not permeable to large agonists like C5a and CXCL12. Therefore, to determine binding affinities and to confirm receptor phenotypes, we selected several C5aR and CXCR4 mutant receptors to express in COS-7 cells. As noted in Fig. 10, left panel, wild type and the selected mutant C5aRs displayed robust inositol phosphate signaling in response to 10 nm C5a (a concentration that is ∼10-fold higher than the reported EC50 for C5a (61)). The S272C-mutated C5aR demonstrated slightly diminished C5a-mediated signaling relative to the wild type C5aR (6.68- versus 4.02-fold over basal, respectively). As shown in Table 2, this difference might be due to decreased binding affinity (∼5-fold less relative to wild type C5aR) because there is little difference in expression levels (Bmax 7.88 versus 8.43 pmol/mg, respectively). Introduction of the second cysteine in the N terminus partially rescued receptor activation and C5a binding affinity. Relative to S272C-C5aR, the S30C/S272C double mutant demonstrated increased IP3 production and increased binding affinity (6.52- versus 4.02-fold over basal and Kd 1.23 versus 3.61 nm, respectively). The rescue effect of S30C for S272C seen in COS-7 cells is consistent with the effects observed in yeast. Of note, the S272A/S272T/S272C mutant C5aRs displayed significantly blunted signaling phenotypes relative to that observed for S272C-C5aR expressed in COS-7 cells (Figs. 3 and 4 compared with Fig. 10). The basis for this is not clear but likely reflects an increased sensitivity to mutations in human receptors expressed in yeast. The N119S mutant C5aR displayed modest constitutive activity in COS-7 cells (Fig. 10, left panel) that was similar to that observed in yeast and was also further stimulated in response to C5a. With respect to wild type C5aR, the N119S mutant displayed slightly increased binding affinity for C5a (Table 2; Kd 0.22 versus 0.65 nm). The combined cysteine mutations S30C and S272C reduced the constitutive activity of N119S mutant (Fig. 10, left panel) and decreased the binding affinity to near wild type levels (Kd 1.11 versus 0.65 nm) with no significant reduction in C5a-mediated signaling.
FIGURE 10.
Signaling assays in COS-7 cells expressing Gα16 with wild type or mutant receptors. Left and right panels, respectively, represent C5aR and CXCR4. The cells were stimulated with 10 nm C5a (C5aR) or 100 nm CXCL12 (CXCR4). IP3 accumulation values are normalized to the mock transfections, where each bar represents means ± S.D. of three independent experiments.
TABLE 2.
Analysis of receptor binding affinities in COS-7 cell membranes
The apparent Kd and Bmax values are derived from binding assays by fitting the raw data into one site homologous binding models. Values represent means ± S.D. from three independent experiments.
| Receptors | Kd | Bmax |
|---|---|---|
| nm | pmol/mg | |
| C5aR | 0.65 ± 0.07 | 8.43 ± 0.85 |
| S272C-C5aR | 3.61 ± 0.38 | 7.88 ± 0.87 |
| S30C/S272C-C5aR | 1.23 ± 0.04 | 5.08 ± 0.15 |
| N119S-C5aR | 0.22 ± 0.07 | 4.02 ± 0.52 |
| S30C/N119S/S272C-C5aR | 1.11 ± 0.03 | 7.03 ± 0.42 |
| CXCR4 | 3.32 ± 0.24 | 9.41 ± 0.72 |
| N119S-CXCR4 | 1.63 ± 0.23 | 20.86 ± 2.55 |
| C28S/N119S/C274S-CXCR4 | 13.33 ± 1.15 | 16.42 ± 1.70 |
| C28R/N119S/C274E-CXCR4 | 7.59 ± 0.20 | 16.64 ± 0.46 |
Wild type CXCR4 expressed in COS-7 cells displayed basal signaling in IP3 assays, which further increased upon stimulation with 100 nm CXCL12 (Fig. 10, right panel, 3.63-fold stimulation over basal). Consistent with our studies in yeast, N119S-CXCR4 also displayed modest constitutive activity as assessed in IP3 assays. Some of the increased basal activity could be due to an increase in receptor expression relative to wild type CXCR4 (Table 2; Bmax 20.86 versus 9.41 pmol/mg, respectively). In favor of more active conformation, N119S-CXCR4 demonstrated a 2-fold increase in binding affinity for CXCL12 (Kd 1.63 versus 3.32 nm). Removal of the cysteine pair decreased the apparent constitutive activity in C28S/N119S/C274S CXCR4 (Fig. 10, lane D, right panel) and decreased the binding affinity for CXCL12 with no effect on receptor expression (Table 2) or its ability to be activated by 100 nm CXCL12 (Fig. 10, right panel, lane D). Similar to the studies in yeast, a potential salt bridge (C28R/C274E) substituted for the cysteine pair appreciably restored the constitutive activity in N119S mutant CXCR4 expressed in COS-7 cells. Although the putative salt bridge improved the binding affinity for CXCL12 in respect to C28S/N119S/C274S mutant (Table 2), no further stimulation was observed in presence of 100 nm CXCL12 in mammalian cells (Fig. 10, right panel). These results further support an important interaction between EC3 and the N terminus of CXCR4.
Hypothetical Molecular Model of C5aR
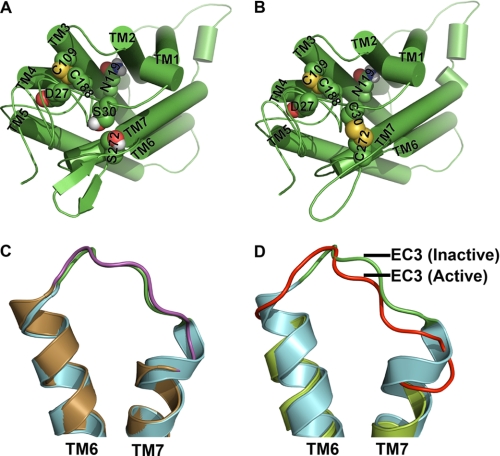
Functional data of C5aR presented in Figs. 3, 4, and 7–9 suggest that Ser272 plays a crucial role for maintaining EC3 structure, which may have a role in ligand binding and activation. It also appears that the S272C of EC3 perhaps favors the formation of a putative disulfide linkage with S30C/D27C by reducing the conformational possibilities of the flexible N terminus. Furthermore, Ser30 or Asp27 could be involved in N terminus-EC3 packing as S30C or D27C mutants rescue the function in S272A and S272T mutations. This argument is pictorially represented via molecular models using one out of the several possible conformations of N terminus for the double mutant S30C-S272C. Fig. 11A represents a conformational snapshot of wild type C5aR, where Ser272 of EC3 is shown within the permissive covalent contact with Ser30 of the N terminus, although Fig. 11B represents the putative disulfide linkage between Cys30 and Cys272 in engineered C5aR. Formation of an EC3-N terminus disulfide linkage may affect the EC3 loop structure and can potentially stabilize the receptor conformation. Introduction of a cysteine at Asp282 in EC3 of bovine rhodopsin and a corresponding cysteine substituted at Asn2 in the N terminus formed a disulfide bridge as evidenced in the crystal structure (51), but it did not significantly alter the conformation of EC3 when compared with the wild type rhodopsin structure (Fig. 11C). However, the engineered mutant receptor displayed higher thermostability (51). In contrast, opsin bound to a G protein-like peptide displayed appreciable conformational changes in EC3 when compared with the “off” structure of dark state rhodopsin (Fig. 11D).
FIGURE 11.
Hypothetical molecular models of C5aR and illustration of conformational changes in EC3 due to disulfide linkage and receptor activation. Molecular models of wild type and engineered C5aR are presented, respectively, in A and B. Asp27, Ser30, Asn119, and Ser272 that have been mutated for engineering the C5aR are highlighted in spheres. B, highlight of both the conserved and the engineered disulfide bond, respectively, between Cys109–Cys188 and Cys30–Cys272 in spheres, in context of other residues. Loop structures have been smoothed for clarity. C, structural alignment of EC3 of wild type (green loop with cyan helices, 1F88) and engineered rhodopsin (magenta loop with sand helices, Protein Data Bank code 2J4Y). Disulfide linkage between Asp282 of EC3 with Asn2 of the N terminus increases the EC3 loop length by straightening the TM7 by one helical loop. D, structural alignment of EC3 of rhodopsin (green loop with cyan helices, Protein Data Bank code 1F88) with EC3 of apparently active opsin (red loop with lime helices, Protein Data Bank code 3DQB). Major conformational changes noted in EC3 are associated with straightening of both TM6 and TM7 helices.
DISCUSSION
This study highlights the importance of EC3-N terminus cysteine pairs in receptor function. In contrast to the highly conserved EC2-TM3 cysteine pair that plays an important structural role in nearly all rhodopsin family receptors, the EC3-N terminus cysteine pairs appear to add functionality to a subset of the rhodopsin family of receptors. The nature of this functionality varies from receptor to receptor. In CXCR4, the EC3-N terminus cysteine pair adds functionality by stabilizing the active state of the receptor. In contrast, when engineered into the C5aR, the cysteine pair exerts the effect of an inverse agonist in C5aR by diminishing the constitutive signaling of the novel C5aR CAM. These studies demonstrate a structural interaction between the EC3 and the N termini in the C5aR and CXCR4, which suggests a novel microswitch region for C5aR and CXCR4. Moreover, the different functional phenotypes obtained for single and double cysteine mutants across the receptors imply a more involved role of EC3-N terminus via cysteine pairs beyond a simple disulfide linkage.
EC3-N terminus cysteine pairs have been studied previously in both chemokine and non-chemokine receptors. Disruption of these cysteine pairs in wild type CXCR4 have been shown to reduce CXCL12 binding affinity (30). In addition, the cysteine pair is not essential for CXCR4 function as a co-receptor for HIV-1 Env-mediated membrane fusion in a strain-specific manner (35). Mutation of the cysteine pair in CCR5 completely abolishes the detectable level of MIP-1β binding, whereas the Cys-mutated receptors supported infection of R5 HIV strains at slightly reduced levels (34). In CCR6, cysteine pair mutation at Cys36 significantly reduced MIP-3α binding and signaling, whereas Cys288-mutated CCR6 showed only blunted signaling (33). Despite expression levels similar to wild type CCR6, neither cysteine-mutated CCR6 supported chemotaxis (33). Similarly, in CXCR2, mutation of Cys286 in the EC3 completely disrupted CXCL8 binding and CXCR2-mediated intracellular signaling (36); however, mutation of the pair residue Cys39 in the N terminus increased both the binding affinity slightly for CXCL8 and intracellular signaling through calcium flux. Interestingly, HEK cells transfected with C39S-mutated CXCR2 demonstrated severely blunted chemotactic function relative to the wild type CXCR2 (36).
Earlier studies suggested cysteine pair mutations do not significantly impair the cell surface expression levels in chemokine receptors, although antibody staining suggested cysteine pair mutation could affect the overall receptor conformation (35). Few studies have directly examined if a disulfide bond links the EC3-N terminus cysteine pair. In CCR6, alkylation of free thiols demonstrated decreased mobility in SDS-PAGE for the wild type CCR6 indicative of cysteine thiol modification, but no shift was observed in the C36S/C288S-mutated CCR6 (33). These results do not rule out the possibility that the cysteines might exist in equilibrium between free thiols and labile disulfide bridges, which could be trapped during prolonged incubation with alkylated agents.
In non-chemokine receptors like angiotensin II AT1 receptor, disruption of the EC3-N terminus cysteine pair conferred constitutive signaling without significantly affecting the expression levels or EC50 value for angiotensin II (62). Similarly, in AT2R mutational disruption or reduction by DTT of the EC3-N terminus cysteine pair resulted in an increased binding affinity for both angiotensin II and receptor-specific antagonist PD123319 (63). The purinergic receptors such as P2Y1 and P2Y12 are another class of non-chemokine receptors, where the EC3-N terminus cysteine pairs appear to play different roles within members of the purinergic family. Previous studies demonstrate an essential role for EC3-N terminus cysteine pairs in P2Y1 ligand binding (28). In contrast, disruption of the EC3-N terminus cysteine pair in P2Y12 did not significantly affect the EC50 value for agonist-mediated inhibition of adenylyl cyclase activity (64). Although the cysteine pair was not essential for P2Y12 activity, the cysteines could be modified by either thiol agents or the active metabolite of the widely prescribed anti-platelet drug, clopidogrel (64).
Given the different roles of cysteine pairs in both chemokine and non-chemokine-type receptor function, we elected to engineer an EC3-N terminus cysteine pair into C5aR. In doing so, we uncovered an essential role for the serine located at the position in EC3 at 7.25, the position at which cysteines most likely occur in receptors containing the cysteine pair. In our studies in yeast, mutations S272A, S272T, and S272C abrogated the C5a-stimulated signaling in these single EC3 point mutants. In mammalian cells the S272C mutant displayed appreciable signaling but at a reduced level in reference to wild type C5aR. This is a novel finding as our previous studies using random saturation mutagenesis suggested that mutations could be tolerated at this position in EC3 (65) in the presence of other point mutations introduced into EC3. The loss in C5a-stimulated signaling in the S272C mutant could be rescued by introducing another cysteine on the N terminus of C5aR (Figs. 3 and 10). In addition, the EC3-N terminus cysteine pair significantly reduced constitutive signaling of the C5aR CAM (Figs. 8 and 10). The ability of the N-terminal cysteine to act as second-site suppressors implies a direct interaction with EC3, which may favor a possible disulfide linkage between the EC3-N terminus cysteine pair in C5aR. The most direct interpretation is that the rescue effect occurs via an intramolecular interaction between the N terminus and EC3. However, the possibility of intermolecular interactions cannot be excluded, because many GPCRs are known to oligomerize, including the C5aR and CXCR4. In addition, our studies in yeast demonstrate that a disulfide bond is not required for the rescue effect because the S30C mutation on N terminus rescued the C5a-stimulated signaling in S272A and S272T mutant in wild type C5aR (Fig. 5). This result was particularly interesting as it implicates the possibility of a noncovalent, conformationally dynamic interaction between EC3 and N terminus in C5aR, a feature that was previously unappreciated. The importance of noncovalent interactions between EC3-N terminus is further evidenced in CXCR4 (Figs. 2 and 10), as the loss of constitutive or CXCL12-mediated signaling in C28S/C274S mutant could be restored either by introducing a salt bridge (C28R/C274E) or π-stacking interactions (C28F/C274F). Previous studies in bovine rhodopsin suggests that the double mutant N2C/D282C can replace the noncovalent interactions between Asp282 (EC3) and Asn2 (N terminus) through a disulfide bond without altering the overall fold of the protein (51).
Our data suggest that EC3 loop structure is susceptible to mutations, and its conformational states can modulate both ligand-dependent and -independent G protein signaling. EC3 forms a helical hairpin with kinked TM6 and TM7 and is positioned such that any conformational transition in EC3 could modulate the sea-saw motion of TM6 with respect to TM7. In fact, as noted in Fig. 11D, EC3 in apparently active opsin demonstrates a major conformational displacement, compared with the inactive rhodopsin, suggesting a possible unappreciated role of EC3 in receptor activation.
The biologic significance of the EC3-N terminus cysteine pair remains an open and important question. The high degree of conservation of the cysteine pair in chemokine receptors suggests a unique role for this structural motif in chemokine biology. Our studies in CXCR4 and C5aR demonstrate that the interface between the N terminus and EC3 might serve as a “microswitch” in many rhodopsin family members. The presence of thiol groups at this microswitch region may confer on chemokine receptors the ability to regulate or fine-tune the signaling properties of the receptors. This raises the interesting possibility that these cysteine pairs might be acting as redox sensors responding to peroxides, reactive oxygen species, reducing agents, or enzymes that modify the disulfide bridges such as protein-disulfide isomerases. Further studies will be required to understand better the effects of the redox environment on cellular signaling.
This work was supported, in whole or in part, by National Institutes of Health Grants GM071634 and GM071634-03S109 (to T. J. B.).
- GPCR
- G protein-coupled receptor
- TM
- transmembrane
- CAM
- constitutive active mutant
- IP3
- inositol triphosphate
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Pierce K. L., Premont R. T., Lefkowitz R. J. (2002) Nat. Rev. Mol. Cell Biol. 3, 639–650 [DOI] [PubMed] [Google Scholar]
- 2.Fredriksson R., Lagerström M. C., Lundin L. G., Schiöth H. B. (2003) Mol. Pharmacol. 63, 1256–1272 [DOI] [PubMed] [Google Scholar]
- 3.Fredriksson R., Schiöth H. B. (2005) Mol. Pharmacol. 67, 1414–1425 [DOI] [PubMed] [Google Scholar]
- 4.Defea K. (2008) Br. J. Pharmacol. 153, S298–S309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neves S. R., Ram P. T., Iyengar R. (2002) Science 296, 1636–1639 [DOI] [PubMed] [Google Scholar]
- 6.Hopkins A. L., Groom C. R. (2002) Nat. Rev. Drug Discov. 1, 727–730 [DOI] [PubMed] [Google Scholar]
- 7.Drews J. (2000) Science 287, 1960–1964 [DOI] [PubMed] [Google Scholar]
- 8.Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., Stevens R. C. (2007) Science 318, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaakola V. P., Griffith M. T., Hanson M. A., Cherezov V., Chien E. Y., Lane J. R., Ijzerman A. P., Stevens R. C. (2008) Science 322, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Science 289, 739–745 [DOI] [PubMed] [Google Scholar]
- 11.Scheerer P., Park J. H., Hildebrand P. W., Kim Y. J., Krauss N., Choe H. W., Hofmann K. P., Ernst O. P. (2008) Nature 455, 497–502 [DOI] [PubMed] [Google Scholar]
- 12.Warne T., Serrano-Vega M. J., Baker J. G., Moukhametzianov R., Edwards P. C., Henderson R., Leslie A. G., Tate C. G., Schertler G. F. (2008) Nature 454, 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Visiers I., Ballesteros J. A., Weinstein H. (2002) Methods Enzymol. 343, 329–371 [DOI] [PubMed] [Google Scholar]
- 14.Ahuja S., Smith S. O. (2009) Trends Pharmacol. Sci. 30, 494–502 [DOI] [PubMed] [Google Scholar]
- 15.Nygaard R., Frimurer T. M., Holst B., Rosenkilde M. M., Schwartz T. W. (2009) Trends Pharmacol. Sci. 30, 249–259 [DOI] [PubMed] [Google Scholar]
- 16.Ahuja S., Crocker E., Eilers M., Hornak V., Hirshfeld A., Ziliox M., Syrett N., Reeves P. J., Khorana H. G., Sheves M., Smith S. O. (2009) J. Biol. Chem. 284, 10190–10201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi L., Liapakis G., Xu R., Guarnieri F., Ballesteros J. A., Javitch J. A. (2002) J. Biol. Chem. 277, 40989–40996 [DOI] [PubMed] [Google Scholar]
- 18.Ballesteros J. A., Jensen A. D., Liapakis G., Rasmussen S. G., Shi L., Gether U., Javitch J. A. (2001) J. Biol. Chem. 276, 29171–29177 [DOI] [PubMed] [Google Scholar]
- 19.Mahalingam M., Martínez-Mayorga K., Brown M. F., Vogel R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17795–17800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sum C. S., Tikhonova I. G., Costanzi S., Gershengorn M. C. (2009) J. Biol. Chem. 284, 3529–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaidehi N., Floriano W. B., Trabanino R., Hall S. E., Freddolino P., Choi E. J., Zamanakos G., Goddard W. A., 3rd (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12622–12627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilchrist A. (2007) Trends Pharmacol. Sci. 28, 431–437 [DOI] [PubMed] [Google Scholar]
- 23.Leach K., Sexton P. M., Christopoulos A. (2007) Trends Pharmacol. Sci. 28, 382–389 [DOI] [PubMed] [Google Scholar]
- 24.Torrice M. M., Bower K. S., Lester H. A., Dougherty D. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 11919–11924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kristiansen K., Dahl S. G., Edvardsen O. (1996) Proteins 26, 81–94 [DOI] [PubMed] [Google Scholar]
- 26.Cain S. A., Coughlan T., Monk P. N. (2001) Biochemistry 40, 14047–14052 [DOI] [PubMed] [Google Scholar]
- 27.Feng Y. H., Noda K., Saad Y., Liu X. P., Husain A., Karnik S. S. (1995) J. Biol. Chem. 270, 12846–12850 [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann C., Moro S., Nicholas R. A., Harden T. K., Jacobson K. A. (1999) J. Biol. Chem. 274, 14639–14647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawson Z., Wheatley M. (2004) Biochem. Soc. Trans. 32, 1048–1050 [DOI] [PubMed] [Google Scholar]
- 30.Zhou H., Tai H. H. (2000) Arch. Biochem. Biophys. 373, 211–217 [DOI] [PubMed] [Google Scholar]
- 31.Zhou N., Luo Z., Luo J., Liu D., Hall J. W., Pomerantz R. J., Huang Z. (2001) J. Biol. Chem. 276, 42826–42833 [DOI] [PubMed] [Google Scholar]
- 32.Kolakowski L. F., Jr., Lu B., Gerard C., Gerard N. P. (1995) J. Biol. Chem. 270, 18077–18082 [DOI] [PubMed] [Google Scholar]
- 33.Ai L. S., Liao F. (2002) Biochemistry 41, 8332–8341 [DOI] [PubMed] [Google Scholar]
- 34.Blanpain C., Lee B., Vakili J., Doranz B. J., Govaerts C., Migeotte I., Sharron M., Dupriez V., Vassart G., Doms R. W., Parmentier M. (1999) J. Biol. Chem. 274, 18902–18908 [DOI] [PubMed] [Google Scholar]
- 35.Chabot D. J., Zhang P. F., Quinnan G. V., Broder C. C. (1999) J. Virol. 73, 6598–6609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Limatola C., Di Bartolomeo S., Catalano M., Trettel F., Fucile S., Castellani L., Eusebi F. (2005) Exp. Cell Res. 307, 65–75 [DOI] [PubMed] [Google Scholar]
- 37.Tran P. B., Miller R. J. (2003) Nat. Rev. Neurosci. 4, 444–455 [DOI] [PubMed] [Google Scholar]
- 38.Guo R. F., Ward P. A. (2005) Annu. Rev. Immunol. 23, 821–852 [DOI] [PubMed] [Google Scholar]
- 39.Brown A. J., Dyos S. L., Whiteway M. S., White J. H., Watson M. A., Marzioch M., Clare J. J., Cousens D. J., Paddon C., Plumpton C., Romanos M. A., Dowell S. J. (2000) Yeast 16, 11–22 [DOI] [PubMed] [Google Scholar]
- 40.Klco J. M., Wiegand C. B., Narzinski K., Baranski T. J. (2005) Nat. Struct. Mol. Biol. 12, 320–326 [DOI] [PubMed] [Google Scholar]
- 41.Gerber B. O., Meng E. C., Dotsch V., Baranski T. J., Bourne H. R. (2001) J. Biol. Chem. 276, 3394–3400 [DOI] [PubMed] [Google Scholar]
- 42.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 43.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J. (2005) J. Comput. Chem. 26, 1701–1718 [DOI] [PubMed] [Google Scholar]
- 44.Angel T. E., Chance M. R., Palczewski K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8555–8560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evans B. J., Wang Z., Broach J. R., Oishi S., Fujii N., Peiper S. C. (2009) Methods Enzymol. 460, 399–412 [DOI] [PubMed] [Google Scholar]
- 46.Berchiche Y. A., Chow K. Y., Lagane B., Leduc M., Percherancier Y., Fujii N., Tamamura H., Bachelerie F., Heveker N. (2007) J. Biol. Chem. 282, 5111–5115 [DOI] [PubMed] [Google Scholar]
- 47.Zhang W. B., Navenot J. M., Haribabu B., Tamamura H., Hiramatu K., Omagari A., Pei G., Manfredi J. P., Fujii N., Broach J. R., Peiper S. C. (2002) J. Biol. Chem. 277, 24515–24521 [DOI] [PubMed] [Google Scholar]
- 48.Fleury Y., Dayem M. A., Montagne J. J., Chaboisseau E., Le Caer J. P., Nicolas P., Delfour A. (1996) J. Biol. Chem. 271, 14421–14429 [DOI] [PubMed] [Google Scholar]
- 49.Derksen D. J., Boudreau M. A., Vederas J. C. (2008) ChemBioChem 9, 1898–1901 [DOI] [PubMed] [Google Scholar]
- 50.Derksen D. J., Stymiest J. L., Vederas J. C. (2006) J. Am. Chem. Soc. 128, 14252–14253 [DOI] [PubMed] [Google Scholar]
- 51.Standfuss J., Xie G., Edwards P. C., Burghammer M., Oprian D. D., Schertler G. F. (2007) J. Mol. Biol. 372, 1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mery L., Boulay F. (1994) J. Biol. Chem. 269, 3457–3463 [PubMed] [Google Scholar]
- 53.Hagemann I. S., Miller D. L., Klco J. M., Nikiforovich G. V., Baranski T. J. (2008) J. Biol. Chem. 283, 7763–7775 [DOI] [PubMed] [Google Scholar]
- 54.Higginbottom A., Cain S. A., Woodruff T. M., Proctor L. M., Madala P. K., Tyndall J. D., Taylor S. M., Fairlie D. P., Monk P. N. (2005) J. Biol. Chem. 280, 17831–17840 [DOI] [PubMed] [Google Scholar]
- 55.Cain S. A., Higginbottom A., Monk P. N. (2003) Biochem. Pharmacol. 66, 1833–1840 [DOI] [PubMed] [Google Scholar]
- 56.Short A. J., Paczkowski N. J., Vogen S. M., Sanderson S. D., Taylor S. M. (1999) Br. J. Pharmacol. 128, 511–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kobilka B. K., Deupi X. (2007) Trends Pharmacol. Sci. 28, 397–406 [DOI] [PubMed] [Google Scholar]
- 58.Bokoch M. P., Zou Y., Rasmussen S. G., Liu C. W., Nygaard R., Rosenbaum D. M., Fung J. J., Choi H. J., Thian F. S., Kobilka T. S., Puglisi J. D., Weis W. I., Pardo L., Prosser R. S., Mueller L., Kobilka B. K. (2010) Nature 463, 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Noda K., Feng Y. H., Liu X. P., Saad Y., Husain A., Karnik S. S. (1996) Biochemistry 35, 16435–16442 [DOI] [PubMed] [Google Scholar]
- 60.Thomas W. G., Qian H., Chang C. S., Karnik S. (2000) J. Biol. Chem. 275, 2893–2900 [DOI] [PubMed] [Google Scholar]
- 61.Buck E., Bourne H., Wells J. A. (2005) J. Biol. Chem. 280, 4009–4012 [DOI] [PubMed] [Google Scholar]
- 62.Correa S. A., Pignatari G. C., Ferro E. S., Pacheco N. A., Costa-Neto C. M., Pesquero J. B., Oliveira L., Paiva A. C., Shimuta S. I. (2006) Regul. Pept. 134, 132–140 [DOI] [PubMed] [Google Scholar]
- 63.Feng Y. H., Saad Y., Karnik S. S. (2000) FEBS Lett. 484, 133–138 [DOI] [PubMed] [Google Scholar]
- 64.Ding Z., Kim S., Dorsam R. T., Jin J., Kunapuli S. P. (2003) Blood 101, 3908–3914 [DOI] [PubMed] [Google Scholar]
- 65.Klco J. M., Nikiforovich G. V., Baranski T. J. (2006) J. Biol. Chem. 281, 12010–12019 [DOI] [PubMed] [Google Scholar]