Abstract
Post-translational modifications have major importance for the structure and function of a multiplicity of proteins. Phosphorylation is a widespread phenomenon among eukaryotic proteins. Whereas O-phosphorylation on the side chains of serine, threonine, and tyrosine in proteins is well known and has been studied extensively, to our knowledge the endogenous phosphorylation of hydroxyproline has not previously been reported. In the present work, we provide evidence for the first time that O-phosphohydroxyproline (Hyp(P)) is a proteinogenic amino acid. To detect Hyp(P) in proteins we generated a Hyp(P)-specific polyclonal antibody. We could identify Hyp(P) in various proteins by Western blot analysis, and we characterized the sequence position of Hyp(P) in the protein α-crystallin A by electrospray ionization-tandem mass spectrometry. Our experiments clearly demonstrate hydroxylation and subsequent phosphorylation of a proline residue in α-crystallin A in the eye as well as in heart tissue of rat.
Keywords: Hydroxyproline, Mass Spectrometry (MS), Peptide Conformation, Post-translational Modification, Protein Phosphorylation, Hydroxyamino Acids, Hydroxylation, Phosphoamino Acids, Phosphoprotein
Introduction
A wide range of biological processes are controlled by reversible protein phosphorylation, and conservative estimates indicate reversible phosphorylation targets of up to one-third of cellular proteins (1). To understand protein regulation through phosphorylation it is essential to find and characterize all sites and types of phosphorylation on proteins and to identify the kinases involved.
Besides the three genetically encoded hydroxyamino acids serine (Ser), threonine (Thr), and tyrosine (Tyr), there are two other hydroxyamino acids, hydroxyproline (Hyp)2 and hydroxylysine (Hyl). They are not encoded in the genome, although they can be identified in mature proteins. They are generated by enzymatic hydroxylation of proline (Pro) and lysine (Lys) residues (2). All genetically encoded hydroxyamino acids in polypeptide chains, such as serine, threonine, and tyrosine have been shown that they can undergo post-translational phosphorylation (3, 4). Although phosphorylation of Hyl has been described in collagens (5) phosphohydroxyproline (Hyp(P)) has not been identified in native proteins.
In addition to the O-phosphorylation of the hydroxyl group, O-glycosylation is another important modification. There is a growing speculation that phosphorylation and glycosylation share a reciprocal relationship. Both Hyp and Hyl are known to undergo glycosylation in a few proteins. In collagens up to 70% of Hyl residues are modified with galactose and glycosyl-galactose (6). Hyl was also found to be phosphorylated in collagens (5). In extensins (hydroxyproline-rich glycoproteins) of the cell wall in plants, ∼50% of protein mass is glycosylated at Hyp residues by mono-, di-, tri-(1→2)-β-linked arabinoses (7, 8, 9). Furthermore, cytoplasmic F box-binding protein SKP1 in Dictyostelium contains a pentasaccharide linked to Hyp (10). These findings suggest that subsequent modification of Hyp is widespread and that, in analogy to all other hydroxyamino acids, phosphorylation of Hyp should be possible in native proteins.
Hydroxylation of Pro is the precondition for subsequent phosphorylation and has been studied extensively in collagens (2). The hydroxyl groups are introduced by sequence specific prolyl-3- or prolyl-4-hydroxylases. Prolyl-3-hydroxylase modifies Pro residues located C-terminally of glycine (Gly-Pro motif). Although Pro located at the N-terminal side of Gly (Pro-Gly motif) is hydroxylated by prolyl-4-hydroxylase, both hydroxylases introduce the hydroxyl group in trans configuration. In collagens >90% of Hyp is in trans-4 configuration. It seems to be the most common Hyp isomer whereas only up to 2% are trans-3-Hyp (2). The alternative cis configuration of Hyp was not described in native proteins so far. Although Hyp was found in a multiplicity of other proteins, the corresponding hydroxylases are mostly unknown.
Several experiments have shown that enzymatic phosphorylation of Hyp residues in synthetic peptides by a Ser-specific kinase is possible, although the kinetics of Hyp phosphorylation were very low compared with those of Ser and Thr (11–13). The configuration of Hyp played a crucial role in these experiments. Enzymatic phosphorylation of trans-4-Hyp appeared to be possible, but not the phosphorylation of cis-4-Hyp (12). All of these findings implicate that Hyp(P) is a proteinogenic amino acid.
Here, we present the phosphorylation of Hyp in proteins by a specific polyclonal antibody. Furthermore we identified for the first time the sequence position of Hyp(P) by mass spectrometry in α-crystallin A. Because the isomeric configuration of the Hyp residue remained unclear for the majority of Hyp-containing proteins, we outline the mass spectrometrical characterization of peptides containing various Hyp(P) isomers. Thus, it was interesting to know whether MS/MS fragmentation was able to characterize the configuration of Hyp(P) in post-translationally modified proteins and to find parameters that would allow for the identification of Hyp(P) in unknown proteins. This would permit to distinguish between Hyp(P) and other phosphoamino acids.
EXPERIMENTAL PROCEDURES
Materials
Amino acid derivatives were purchased from Sigma-Aldrich and polystyrene AM RAM resin was obtained from Rapp Polymere (Tübingen, Germany). Peptide synthesis was performed with reagents and solvents from Applied Biosystems. All reagents and solvents for phosphorylation were obtained from Sigma-Aldrich or Merck and were of the highest available purity.
Protein Purification
Total protein was prepared from aorta, heart, and eye tissue of a female adult rat in consideration of the conservation of O-phosphoesters. O-Phosphates are acid-stable and more or less unstable in alkali for example, whereas N-phosphates are acid-labile (14). Thus, it is important to prevent the phosphoamino acid residues from dephosphorylation by unsuitable chemical conditions or by phosphatases because in most cases only a small percentage of the potential phosphoprotein is phosphorylated at a specific point in time (15). The rat was maintained in the animal facility of the Heinrich-Heine-University, Düsseldorf, Germany. The tissues were frozen in liquid nitrogen and pulverized. Homogenization was performed with a buffer containing 50 mm Tris-HCl (pH 7.4), 150 mm sodium chloride, 1% Nonidet P-40, 0,25% sodium deoxycholate, 1 mm sodium orthovanadate, 1 mm EDTA, 1 mm benzamidine, 10 μm PMSF, and 1 mm DTT. After centrifugation at 28,000 × g, soluble proteins of the supernatant were precipitated in ice-cold acetone/trifluoroacetic acid (9:1 v/v). Advanced protein purification was performed by SDS-PAGE.
Electrospray Mass Spectrometry
Electrospray experiments were performed with an electrospray ionization-QqTOF hybrid mass spectrometer (QSTAR XL; Applied Biosystems) equipped with a nanoflow electrospray source. In preliminary steps, protein bands were excised from SDS gels and digested in situ with modified trypsin (Sigma-Aldrich). Resulting peptides were desalted and concentrated by reversed phase C18 ZipTips (Millipore). To reduce complexity of the samples, elution was performed gradually in 10% MeOH steps.
Synthesis of cis-4/trans-3-Fmoc-Hyp Isomers and Peptides
Fmoc derivatives of cis-4-hydroxy-l-proline and trans-3-hydroxy-l-proline were synthesized according to the protocol of Paquet (16). Peptides were synthesized on a 433A peptide synthesizer (Applied Biosystems) using standard Fmoc chemistry. When Fmoc-hydroxy-l-proline derivatives with an unprotected hydroxyl group were introduced activation was employed by N,N′-diisopropylcarbodiimide/1-hydroxybenzotriazole. After cleavage and deprotection, the peptides were purified by reversed phase HPLC and lyophilized. Peptide identity was confirmed by mass spectrometry.
Peptide Phosphorylation
The side chain-unprotected peptide resins were phosphitilated by dibenzyl-N,N-diisopropylphosphoramidite/tetrazole yielding phosphopeptides after oxidation by tert-butylhydroperoxide followed by trifluoroacetic acid cleavage/deprotection (17). The resulting phosphopeptides were confirmed by mass spectrometry.
Generation of Polyclonal Antibodies
The generation of mono- or polyclonal antibodies against Ser(P), Thr(P), and Tyr(P) is described by several authors (18, 19). Antibodies against Hyp(P) were not available so far. Therefore, a Hyp(P)-specific polyclonal antibody was generated. We used a short synthetic sequence of collagen, including five Hyp(P) residues instead of Hyp for immunization. The sequence of the peptide was Gly-Pro-Hyp(P)-Gly-Pro-Hyp(P)-Gly-Pro-Hyp(P)-Gly-Pro-Hyp(P)-Gly-Pro-Hyp(P). After coupling the peptide to keyhole limpet hemocyanin it was used for immunization of a rabbit. Polyclonal antiserum was obtained from SEQLAB (Göttingen, Germany). Polyclonal antibodies were separated and enriched from the serum by protein A-Sepharose. The resulting specificity of the antibodies we obtained meets the requirements for Western blot analysis. No further affinity purification using synthetic phosphopeptides was done.
RESULTS
Detection of Hyp(P)-containing Proteins by Western Blot Analysis
To detect Hyp(P)-containing proteins, we generated a Hyp(P)-specific antibody. The specificity of the generated polyclonal antibody was tested by ELISA and dot blot analysis. The polyclonal antibody showed high specificity for Hyp(P) in all tests independently from the surrounding peptide sequence and without cross-reactivity to different phosphoamino acids such as Ser(P), Thr(P) or Tyr(P) (Fig. 1). Although unlikely, there is a small possibility that the polyclonal antibody could recognize sulfated Hyp. Sulfation can be a post-translational modification of proteins, but to our knowledge the only target amino acid is tyrosine. Sulfation of Hyp has not yet been demonstrated.
FIGURE 1.
Specificity of the polyclonal antibody tested by dot blot analysis. Each spot contained 1 μg of synthetic peptide. Sequences of the peptides are denoted to the right of the spots. Phosphoamino acids in the sequences are boldfaced. Hyp and all phosphoamino acids are denoted in three-letter code, all other amino acids are in one-letter code.
Total protein from aorta, heart, and eye tissue of rat was prepared. Each of the three protein samples was separated by SDS-PAGE and assayed immunologically by Western blot analysis. The Hyp(P)-specific antibody recognized Hyp(P) in a few protein bands (Fig. 2). To verify these results and to confirm the specificity of the antibody, two aliquots of the protein preparations were pretreated with various phosphatases prior to SDS-PAGE. Pretreatment with the serine-/threonine phosphatase calcineurin did not alter the recognition pattern of proteins by the Hyp(P)-specific antibody compared with untreated proteins. In contrast, the Hyp(P)-specific antibody did not recognize any protein when the protein samples were pretreated with alkaline phosphatase which unspecifically dephosphorylates any known phosphoester, including Hyp(P) (Fig. 2).
FIGURE 2.
Detection of Hyp(P)-containing proteins by Western blot analysis. SDS-PAGE (8–15% gradient) of total protein of heart, aorta, and eye from rat is shown. The Western blot was performed with a Hyp(P)-specific polyclonal antibody. Aliquots of total protein without pretreatment, pretreated with calcineurin, and pretreated with alkaline phosphatase were compared. Protein bands denoted with roman numerals were identified by MS/MS analysis as (I) β-crystallin B2, (II) β-crystallin A4, (III) α-crystallin B, (IV) α-crystallin A.
The Western blot showed specific recognition of Hyp(P) in different tissues: in three protein bands from heart tissue and four protein bands from eye tissue (Fig. 2). These seven proteins were analyzed by mass spectrometry. They all were identified as members of the protein family of crystallins (Table 1).
TABLE 1.
Proteins recognized by Hyp(P)-specific antibody in Western blot
| Protein band | Heart | Eye |
|---|---|---|
| I | β-Crystallin B2 | β-Crystallin B2 |
| II | β-Crystallin A4 | β-Crystallin A4 |
| III | α-Crystallin B | α-Crystallin B |
| IV | α-Crystallin A |
Identification of Hyp(P) in α-Crystallin A
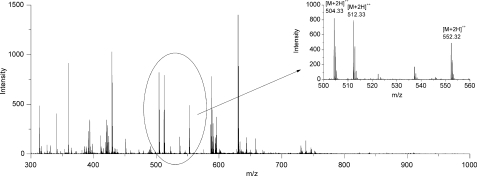
To confirm the results of Western blot analysis and to identify Hyp(P) in the amino acid sequence of the detected proteins, MS analysis was performed. Hydroxylation or phosphorylation resulted in a mass increase of 16 Da and 80 Da, respectively. The mass spectrum of α-crystallin A indicated a signal of a doubly charged ion with [M+2H]2+ 504.33 m/z that was accompanied by signals with [M+2H]2+ 512.33 m/z and [M+2H]2+ 552.32 m/z (Fig. 3). These characteristic mass differences of 8 m/z and 40 m/z for doubly charged ions (16 Da and 80 Da for singly charged ions) gave a strong indication for peptide hydroxylation and phosphorylation.
FIGURE 3.
MS spectrum of eye α-crystallin A after trypsin digestion. Peptide signals within the ellipsoid are shown in the inset. Mass differences of 8 m/z and 40 m/z of doubly charged ions indicate a possible peptide hydroxylation and phosphorylation, respectively.
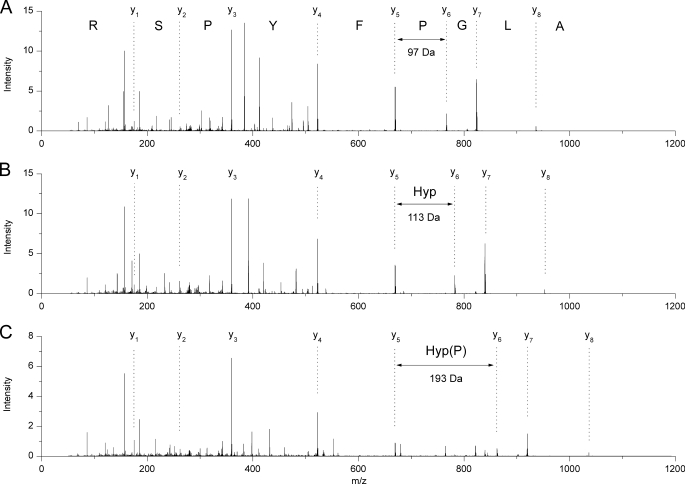
MS/MS analysis of the doubly charged ion with [M+2H]2+ 504.33 m/z identified the peptide sequence ALGPFYPSR corresponding to amino acid residues 13–21 of rat α-crystallin A (Fig. 4A). Despite the different masses of precursor ions with [M+2H]2+ 512.33 m/z and with [M+2H]2+ 552.32 m/z the corresponding peptides had the same sequence but each with another modification. The ion with [M+2H]2+ 512.33 m/z contained Hyp instead of Pro, resulting in the sequence ALG(Hyp)FYPSR (Fig. 4B), whereas the ion with [M+2H]2+ 552.32 m/z contained Hyp(P) at this position, resulting in the sequence ALG(Hyp(P))FYPSR (Fig. 4C). MS/MS spectra of the three peptides showed the entire y series of fragment ions. The completeness of y series facilitated the comparison of peptide sequences. Fragment ions y1 to y5 were identical in all three peptides. Post-translational modification of Pro was indicated by mass differences between fragment ions y5 and y6. In case of Pro, the mass difference between y5 and y6 was 97 Da. Mass differences of 113 Da and 193 Da between y5 and y6 corresponded to the modified forms of Pro, meaning Hyp and Hyp(P), respectively. All of the following fragment ions were transposed to the corresponding higher m/z values.
FIGURE 4.
Comparison of the MS/MS spectra of the precursor ions with [M+H]2+ 504.33 m/z, [M+2H]2+ 512.33 m/z, and [M+2H]2+ 552.32 m/z. All spectra show complete y series. The three peptides have identical amino acid sequences. The mass shift between fragment ions y5 and y6 is characteristic for Pro (A), Hyp (B), and Hyp(P) (C) at the relevant sequence position.
Thus, α-crystallin A from eye tissue contained three subpopulations with different structural states at amino acid position 16 (Table 2). The amino acid Pro appears in nonmodified state as well as the modified forms Hyp and Hyp(P). Comparison of the signal intensities of the three peptides in the overview mass spectra (Fig. 3) shows that the unmodified (m/z 504.33) and the hydroxylated peptide (m/z 512.33) exist in more or less equal amounts. The phosphohydroxylated peptide (m/z 552.32) is about 80% of the signal intensities of proline and hydroxyproline and thus constitutes a significant proportion of the total protein. Both modifications of Pro16 were also observed in α-crystallin A from heart tissue. In all other protein bands recognized by the polyclonal antibody no further Hyp(P) could be verified by MS so far.
TABLE 2.
Sequence of rat α-crystallin A chain (Swiss-Prot accession number P02490)
The underlined residues denote the sequence section where the modifications were found. At position 16, the amino acid Pro (boldfaced) was observed in unmodified, hydroxylated, and phosphorylated state.
| MDVTIQHPWFKRALGPFYPSRLFDQFFGEGLFEYDL |
| LPFLSSTISPYYRQSLFRTVLDSGISEVRSDRDKFV |
| IFLDVKHFSPEDLTVKVLEDFVEIHGKHNERQDDHG |
| YISREFHRRYRLPSNVDQSALSCSLSADGMLTFSGP |
| KVQSGLDAGHSERAIPVSREEKPSSAPSS |
Characterization of Hyp(P) Isomers
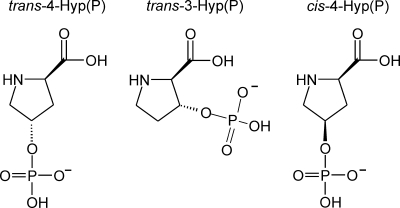
Hydroxylation of Pro is a frequently observed post-translational modification in various proteins (2). Well investigated is hydroxylation of Pro in collagen, the most abundant protein in vertebrates (2). Here, the oxidation is catalyzed by a position-specific prolyl-3- or prolyl-4-hydroxylase. The hydroxyl groups are introduced in trans position by these enzymes (2). Whether hydroxyl groups can also be introduced in cis position by hydroxylases is unknown. To determine whether the detected Hyp(P) in α-crystallin A was in cis or trans conformation and which position at the proline ring was modified (Fig. 5), three sets of three peptides were synthesized for mass spectrometric comparison (Table 3). Three peptides were corresponding to the short α-crystallin A sequence of the Hyp(P)-containing peptide. Three other peptides had the same sequence but were elongated by an additional arginine at the N terminus corresponding to a miscleavage of native α-crystallin A sequence by trypsin. The last three peptides corresponded to a completely different sequence. In each set one peptide contained trans-4-Hyp(P), another one trans-3-Hyp(P), and the last one cis-4-Hyp(P). We characterized the fragmentation behavior of the three Hyp(P) isomers in the different synthetic peptides to find parameters that would allow the identification of Hyp(P) configuration in native proteins. Furthermore, it was of interest to study the fragmentation behavior of Hyp(P) compared to the well known fragmentation behavior of Ser(P), Thr(P), and Tyr(P).
FIGURE 5.
Isomers of Hyp(P).
TABLE 3.
Synthetic peptides for mass spectrometric characterization of Hyp(P) isomers
| [M+H]+ | α-Crystallin A peptides |
| 1103.48 | ALG(trans-4-Hyp(P))FYPSR |
| 1103.48 | ALG(trans-3-Hyp(P))FYPSR |
| 1103.48 | ALG(cis-4-Hyp(P))FYPSR |
| [M+H]+ | α-Crystallin A peptides (elongated) |
| 1259.59 | RALG(trans-4-Hyp(P))FYPSR |
| 1259.59 | RALG(trans-3-Hyp(P))FYPSR |
| 1259.59 | RALG(cis-4-Hyp(P))FYPSR |
| [M+H]+ | Tau peptides |
| 1871.06 | KKVAVVR(trans-4-Hyp(P))LPQVQSVR |
| 1871.06 | KKVAVVR(trans-3-Hyp(P))LPQVQSVR |
| 1871.06 | KKVAVVR(cis-4-Hyp(P))LPQVQSVR |
In general, aliphatic phosphoesters such as Ser(P) and Thr(P) eliminate HPO3 and H3PO4 during MS/MS fragmentation. In contrast, the aromatic phosphoester Tyr(P) shows only the loss of HPO3. These rules are commonly used to distinguish between aliphatic and aromatic phosphoamino acids. Despite the ring structure of Hyp(P), in our measurements of Hyp(P)-containing peptides a fragmentation behavior was observed similar to the aliphatic phosphoesters Ser(P) and Thr(P). All Hyp(P)-containing peptides showed both fragment ions: [M+H-HPO3]+ with a Δ of 80 Da and [M+H-H3PO4]+ with a Δ of 98 Da (Fig. 6).
FIGURE 6.
MS/MS spectra of synthetic peptides. The spectra show the fragmentation patterns of the peptide dephosphorylation. Peptides A–C corresponded to the short α-crystallin A sequence of the Hyp(P)-containing peptide. Peptides D–F had the same sequence but were elongated by an additional arginine at the N terminus. Tau peptides G–I corresponded to a completely different sequence (see Table 3). Peptides in the top row (A, D, and G) contained trans-4-Hyp(P); peptides in the middle row (B, E, and H) contained trans-3-Hyp(P); and the peptides in the bottom row (C, F, and I) contained cis-4-Hyp(P).
MS/MS fragmentation of the single amino acid Hyp(P) showed a fragmentation pattern similar to that of Hyp(P)-containing peptides: both fragment ions [M+H-HPO3]+ and [M+H-H3PO4]+ were observed. In negative mode, the MS/MS spectra of Hyp(P) showed signals of [PO3]− with 79 Da and [H3PO4]− with 97 Da. Thus, the fragmentation behavior of the single amino acid Hyp(P) was again more similar to the fragmentation behavior of the single amino acids Ser(P) and Thr(P) than to that of single Tyr(P). For this reason, a clear distinction between Hyp(P) and other phosphoamino acids based on the fragmentation behavior seems not to be possible.
Synthetic peptides derived from the α-crystallin A sequence displayed an inconsistent fragmentation pattern. The peptides with trans-4-Hyp(P) and cis-4-Hyp(P) showed a loss of HPO3 as a major process. In contrast, the peptide with trans-3-Hyp(P) showed a loss of H3PO4 as a major process (Fig. 6, A–C). Elongation of the peptide sequence by an additional arginine at the N terminus completely changed the fragmentation pattern. All three peptides showed a loss of HPO3 as major process independent of their isomeric status of Hyp(P). Loss of H3PO4 was diminished in all three peptides (Fig. 6, D–F). In the Tau peptides a loss of H3PO4 was the major process in all three peptides regardless of the isomeric status of Hyp(P) (Fig. 6, G–I). This means that neither the position at the proline ring nor the stereoisomerism of the phosphate group showed a clear influence on the fragmentation behavior of peptides. In fact, the sequence of the examined peptides was the dominant factor in all cases. Thus, in case of Hyp(P) in native α-cystallin A we could not determine neither the position of the phosphate group on the proline ring nor the conformation.
In summary, we identified for the first time Hyp(P) in a protein. So far, the identification of Hyp(P) in a protein by MS was hindered by the fact that Hyp, Leu, and isoleucine show the same mass differences in MS/MS analysis. Detection with a Hyp(P)-specific antibody enabled us to focus our search on α-crystallin A. Here, we could identify the sequence position of Hyp(P). Nevertheless, it was not possible to determine the isomeric status of Hyp(P) by comparative MS/MS measurement. For this purpose 31P NMR spectroscopy is still necessary.
DISCUSSION
For the first time the presence and sequence position of hydroxylated and phosphorylated forms of the common amino acid Pro were identified in the native protein α-crystallin A in eye and heart tissue of the rat. In addition, in some other members of the crystallin family preliminary evidence for the presence of Hyp(P) was found. This was achieved initially by generation of a Hyp(P)-specific polyclonal antibody. Antibodies against Ser(P) and Thr(P) are generally not specific enough to detect a phosphorylated Ser or Thr side chain because this epitope is too small. No antibody is known that can recognize Ser(P) or Thr(P) in a sequence-independent manner. In contrast, antibodies against Tyr(P) are specific enough to detect one phosphorylated Tyr side chain independently from the amino acid sequence (18, 19). Similar to Tyr(P), the amino acid side chain of Hyp(P) is sufficiently characteristic to gain amino acid residue-specific antibodies within the polyclonal mixture to detect Hyp(P) epitopes independently of the surrounding amino acid sequence.
With the identification of Hyp and Hyp(P) in α-crystallin A, new questions emerge. The enzyme that oxidizes Pro16 to Hyp is still unknown. A comparison with vertebrate collagen sequences suggests a modification of Pro16 by prolyl-3-hydroxylase because Pro is located C-terminally of Gly. In consequence, the observed Hyp ought to be trans-3-Hyp. Although the isomer trans-4-Hyp was described at the C-terminal side of Gly in annelid collagen (20), it is questionable that Pro16 in α-crystallin A is hydroxylated by prolyl-3-hydroxylase or prolyl-4-hydroxylase because crystallins are intracellular proteins (21). The activity of both mentioned hydroxylases is associated with the endoplasmic reticulum where they are required for collagen biosynthesis (2). Apart from collagen a number of other secreted proteins are hydroxylated on prolyl residues in the endoplasmic reticulum (22). So far, only hypoxia-inducible factor is known as intracellular protein modified on two prolyl residues by hypoxia-inducible factor prolyl hydroxylases (23–25). Here, both hydroxylated prolyl residues are located on the C-terminal side of alanine. In Fasciola hepatica, a trematode, secreted cathepsin L-like proteinases contain trans-3-Hyp residues in different sequences: (i) Val-Hyp-Asp, (ii) Val-Hyp-Glu, and (iii) Tyr-Hyp-Tyr (26). Another yet unknown hydroxylase could be involved in hydroxylation of Pro16 in α-crystallin A.
The mechanism of phosphorylation of Hyp is also completely unknown. A specific kinase was not described so far. There are also no reports describing an unspecific phosphorylation of Hyp by a kinase that is specific for a different hydroxyamino acid. Alternatively, an enzyme-free phosphorylation of Hyp by inositol pyrophosphate is imaginable. Until now, only the phosphate transfer to Ser by inositol pyrophosphate was proven (27). However, this way of phosphorylation could be possible for other hydroxyamino acids, too.
Among the ubiquitous crystallins, there are three groups of proteins: α-crystallins, β-crystallins, and γ-crystallins. α-Crystallins (αA and αB) share 55% sequence similarity and immunoreactivity. The βA polypeptides are 63% homologous, and the basic, βB, are 56% homologous. The γ-crystallins are 70–80% homologous. The β- and γ-crystallins have a common evolutionary origin and share sequence and structural similarities and immunogenicity. Thus, on sequence and structural affinities alone, there are only two groups of proteins, the α-crystallins and the β/γ-crystallins that characterize the mammalian lens (21).
α-Crystallins account for up to 50% of the total soluble protein of the ocular lens. There are two α-crystallins. α-Crystallin A is 173 amino acids long, and α-crystallin B is 175 amino acids long. αA is slightly more acidic than αB. These two proteins exist in a ratio of three (αA) to one (αB) in the lens (21). In rodents, an additional protein, αAins-crystallin is encoded in the αA-crystallin gene through alternative splicing of the “insert” (ins) exon that codes for 23 amino acids. These 23 amino acids are introduced into the α-crystallin A polypeptide between amino acids 63 and 64. The αAins-crystallin is therefore 196 amino acids long (21). The sequence similarity of α-crystallins to small heat shock proteins has led to the establishment of the α-crystallin family of small heat shock proteins (sHsps), which share the α-crystallin domain (28–30). The α-crystallin domain is the primary sequence of the protein in the C-terminal half of the sHsps (31, 32).
The α-crystallin aggregate in lens (containing both αA and αB at a 3:1 ratio) is highly heterogeneous (300–1000 kDa). Each α-crystallin polypeptide has a molecular mass of about 20 kDa; 15–50 subunits thus make the α-crystallin aggregates. Both α-crystallins undergo post-translational modifications (phosphorylation, aspartate racemization, deamidation, glycation, and truncation) (33–36). A number of models have been proposed for the structure of the aggregate that contains αA- and αB-crystallins, but these proteins remain to be crystallized (37, 38). One sHsp member that has been crystallized, the bacterial 16.5-kDa protein, reveals a highly symmetrical aggregate, composed of one homogeneous population of structures (39, 40). This suggests that heterogeneity in the α-crystallin aggregates results predominantly from the N-terminal domain sequences.
The functional role of Hyp(P) in α-crystallin A and other proteins remains elusive. At least the function of α-crystallin A is known, and phosphorylation plays an important role in the process (38, 41, 42). α-Crystallins A and B belong to the sHsps and are involved especially in eye lens tissue in the prevention of misfolding and aggregation of other proteins (43–45). Thereby they act as chaperones and protect the cytoskeleton (46, 47). Possibly, the observed hydroxylation and consecutive phosphorylation of Pro16 in α-crystallin A are involved in these processes, too. The occurrence of Hyp(P) in eye as well as in heart tissue suggests that the phosphorylation of Hyp is not tissue-specific.
It would be desirable to know more Hyp(P)-containing proteins and more sequence positions of Hyp(P). This would facilitate the search for Hyp-specific kinases and the elucidation of Hyp(P) function. Only one hint of Hyp(P) in a native protein was reported before, in natSil-2, a silica-forming phosphoprotein in diatoms (48). However, the discovery of Hyp(P) in mammalian tissue of eye and heart as well as in diatoms points to a widespread occurrence of Hyp(P).
Protein phosphorylation is mostly reported for Ser, Thr, and Tyr. Consecutive phosphorylation of post-translationally generated Hyp and also Hyl is so far not in the spotlight. Maybe Hyp(P) and Hyl(P) are more frequent than generally expected.
Footnotes
- Hyp
- hydroxyproline
- Hyp(P)
- phosphohydroxyproline
- Hyl
- hydroxylysine
- Hyl(P)
- phosphohydroxylysine
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- MS/MS
- tandem mass spectrometry
- sHsp
- small heat shock protein.
REFERENCES
- 1.Hubbard M. J., Cohen P. (1993) Trends Biochem. Sci. 18, 172–177 [DOI] [PubMed] [Google Scholar]
- 2.Kivirikko K. I., Pihlajaniemi T. (1998) Adv. Enzymol. Relat. Areas Mol. Biol. 72, 325–398 [DOI] [PubMed] [Google Scholar]
- 3.Francis S. H., Corbin J. D. (1994) Annu. Rev. Physiol. 56, 237–272 [DOI] [PubMed] [Google Scholar]
- 4.Fantl W. J., Johnson D. E., Williams L. T. (1993) Annu. Rev. Biochem. 62, 453–481 [DOI] [PubMed] [Google Scholar]
- 5.Urushizaki Y., Seifter S. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 3091–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kresina T. F., Miller E. J. (1979) Biochemistry 18, 3089–3097 [DOI] [PubMed] [Google Scholar]
- 7.Knox J. P. (1995) FASEB J. 9, 1004–1012 [DOI] [PubMed] [Google Scholar]
- 8.Kieliszewski M. J., Lamport D. T. (1994) Plant J. 5, 157–172 [DOI] [PubMed] [Google Scholar]
- 9.Roberts K., Grief C., Hills G. J., Shaw P. J. (1985) J. Cell Sci. Suppl. 2, 105–127 [DOI] [PubMed] [Google Scholar]
- 10.Teng-umnuay P., Morris H. R., Dell A., Panico M., Paxton T., West C. M. (1998) J. Biol. Chem. 273, 18242–18249 [DOI] [PubMed] [Google Scholar]
- 11.Feramisco J. R., Kemp B. E., Krebs E. G. (1979) J. Biol. Chem. 254, 6987–6990 [PubMed] [Google Scholar]
- 12.Pearson R. B., Floyd D. M., Hunt J. T., Lee V. G., Kemp B. E. (1988) Arch. Biochem. Biophys. 260, 37–44 [DOI] [PubMed] [Google Scholar]
- 13.Prorok M., Sukumaran D. K., Lawrence D. S. (1989) J. Biol. Chem. 264, 17727–17733 [PubMed] [Google Scholar]
- 14.Martensen T. M. (1984) Methods Enzymol. 107, 3–23 [DOI] [PubMed] [Google Scholar]
- 15.Kalume D. E., Molina H., Pandey A. (2003) Curr. Opin. Chem. Biol. 7, 64–69 [DOI] [PubMed] [Google Scholar]
- 16.Paquet A. (1982) Can. J. Chem. 60, 976–980 [Google Scholar]
- 17.Hoffmann R., Hoffmann T., Tholey A., Schulte A. C., Kalbitzer H. R. (1997) J. Pept. Res. 49, 163–173 [DOI] [PubMed] [Google Scholar]
- 18.Quadroni M., James P. (2000) Exs 88, 199–213 [DOI] [PubMed] [Google Scholar]
- 19.Sickmann A., Meyer H. E. (2001) Proteomics 1, 200–206 [DOI] [PubMed] [Google Scholar]
- 20.Kivirikko K. I., Myllylä R. (1982) Methods Enzymol. 82, 245–304 [DOI] [PubMed] [Google Scholar]
- 21.Bhat S. P. (2003) Prog. Drug Res. 60, 205–262 [DOI] [PubMed] [Google Scholar]
- 22.Adams E., Frank L. (1980) Annu. Rev. Biochem. 49, 1005–1061 [DOI] [PubMed] [Google Scholar]
- 23.Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr. (2001) Science 292, 464–468 [DOI] [PubMed] [Google Scholar]
- 24.Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., Kriegsheim A. V., Hebestreit H. F., Mukherji M., Schofield C. J., Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) Science 292, 468–472 [DOI] [PubMed] [Google Scholar]
- 25.Yu F., White S. B., Zhao Q., Lee F. S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9630–9635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wijffels G. L., Panaccio M., Salvatore L., Wilson L., Walker I. D., Spithill T. W. (1994) Biochem. J. 299, 781–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saiardi A., Bhandari R., Resnick A. C., Snowman A. M., Snyder S. H. (2004) Science 306, 2101–2105 [DOI] [PubMed] [Google Scholar]
- 28.Boelens W. C., Van Boekel M. A., De Jong W. W. (1998) Biochim. Biophys. Acta 1388, 513–516 [DOI] [PubMed] [Google Scholar]
- 29.Krief S., Faivre J. F., Robert P., Le Douarin B., Brument-Larignon N., Lefrere I., Bouzyk M. M., Anderson K. M., Greller L. D., Tobin F. L., Souchet M., Bril A. (1999) J. Biol. Chem. 274, 36592–36600 [DOI] [PubMed] [Google Scholar]
- 30.Kappe G., Verschuure P., Philipsen R. L., Staalduinen A. A., Van de Boogaart P., Boelens W. C., De Jong W. W. (2001) Biochim. Biophys. Acta 1520, 1–6 [DOI] [PubMed] [Google Scholar]
- 31.Ingolia T. D., Craig E. A. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 2360–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narberhaus F. (2002) Microbiol. Mol. Biol. Rev. 66, 64–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spector A. (1984) Invest. Ophthalmol. Vis. Sci. 25, 130–146 [PubMed] [Google Scholar]
- 34.Groenen P. J., Merck K. B., De Jong W. W., Bloemendal H. (1994) Eur. J. Biochem. 225, 1–19 [DOI] [PubMed] [Google Scholar]
- 35.Sax C. M., Piatigorsky J. (1994) Adv. Enzymol. Relat. Areas. Mol. Biol. 69, 155–201 [DOI] [PubMed] [Google Scholar]
- 36.Schaefer H., Marcus K., Sickmann A., Herrmann M., Klose J., Meyer H. E. (2003) Anal. Bioanal. Chem. 376, 966–972 [DOI] [PubMed] [Google Scholar]
- 37.Abgar S., Backmann J., Aerts T., Vanhoudt J., Clauwaert J. (2000) Eur. J. Biochem. 267, 5916–5925 [DOI] [PubMed] [Google Scholar]
- 38.Horwitz J. (2003) Exp. Eye Res. 76, 145–153 [DOI] [PubMed] [Google Scholar]
- 39.Kim K. K., Kim R., Kim S. H. (1998) Nature 394, 595–599 [DOI] [PubMed] [Google Scholar]
- 40.Koteiche H. A., McHaourab H. S. (1999) J. Mol. Biol. 294, 561–577 [DOI] [PubMed] [Google Scholar]
- 41.Augusteyn R. C., Murnane L., Nicola A., Stevens A. (2002) Clin. Exp. Optom. 85, 83–90 [DOI] [PubMed] [Google Scholar]
- 42.Aquilina J. A., Benesch J. L., Ding L. L., Yaron O., Horwitz J., Robinson C. V. (2004) J. Biol. Chem. 279, 28675–28680 [DOI] [PubMed] [Google Scholar]
- 43.De Jong W. W., Leunissen J. A., Voorter C. E. (1993) Mol. Biol. Evol. 10, 103–126 [DOI] [PubMed] [Google Scholar]
- 44.MacRae T. H. (2000) Cell Mol. Life Sci. 57, 899–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ito H., Kamei K., Iwamoto I., Inaguma Y., Nohara D., Kato K. (2001) J. Biol. Chem. 276, 5346–5352 [DOI] [PubMed] [Google Scholar]
- 46.Vicart P., Caron A., Guicheney P., Li Z., Prevost M. C., Faure A., Chateau D., Chapon F., Tome F., Dupret J. M., Paulin D., Fardeau M. (1998) Nat. Genet. 20, 92–95 [DOI] [PubMed] [Google Scholar]
- 47.Quinlan R. A., Sandilands A., Procter J. E., Prescott A. R., Hutcheson A. M., Dahm R., Gribbon C., Wallace P., Carter J. M. (1999) Eye 13, 409–416 [DOI] [PubMed] [Google Scholar]
- 48.Poulsen N., Sumper M., Kroger N. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12075–12080 [DOI] [PMC free article] [PubMed] [Google Scholar]