Abstract
Vitamin D3 up-regulated protein 1 (VDUP1) plays multifunctional roles in diverse cellular responses, particularly in its relation to proliferation, apoptosis, differentiation, and diseases such as cancer and stress-related diseases. In this study, we demonstrated that VDUP1 was up-regulated during the senescence process. Our results showed that overexpression of VDUP1 in young cells caused typical signs of senescence. We also found that VDUP1 knockdown delayed the onset of Ras-induced cellular senescence. Subsequently, we found that FOXO3A, whose activity increased in senescent cells, transcriptionally up-regulates VDUP1 expression and miR-17-5p, whose expression decreased in senescent cells, directly interacted with the 3′-untranslated region of VDUP1 transcripts, and destabilized VDUP1 mRNA in young cells. These results indicated that VDUP1 expression was regulated by FOXO3A at the transcriptional level and by miR-17-5p at the post-transcriptional levels during the senescence process.
Keywords: Fibroblast, Gene Regulation, MicroRNA, Transcription Factors, Transcription Regulation, FOXO3A, Senescence, VDUP1, Fibroblasts, miR-17-5p
Introduction
Cellular senescence is the limited ability of primary human cells to divide when cultured in vitro, which is accompanied by a specific set of changes in cell morphology, gene expression, and function (1). The cellular hypothesis of aging was established by Hayflick in 1975 (41). This hypothesis is supported by evidence that the replicative potential of primary cultured human cells is dependent on donor age and that the growth potential of cultured cells correlates well with the mean maximum life span of the species from which the cells are derived (2), although some conflicting data have been reported (3). The difference in proliferative potential between senescent and cancer cells has led some investigators to suggest that senescence is a cancer protection mechanism. Furthermore, it has been proposed that proteins that control cell cycle progression and that are inactivated in tumor cells, such as the restriction point proteins Rb and p53 and various cyclin-dependent kinase inhibitors, play an important role in the establishment of senescence (4–7).
A diversity of stresses, such as dysfunctional telomeres, severe or irreparable DNA damage, and expression of certain oncogenes can induce cellular senescence (8). Despite the fact that different mechanisms are involved in stress-induced senescence, senescent cells share numerous common features, including an arrest of cell proliferation, an enlarged flattened morphology, expression of a neutral senescence-associated β-galactosidase, and an altered pattern of gene expression.
Vitamin D3 up-regulated protein 1 (VDUP1) was originally identified in HL60 cells stimulated with 1,25-(OH)2D3. It was reported to bind to reduced thioredoxin (TRX)3 by yeast two-hybrid assay; hence, it is considered to be a negative regulator of Trx (9–11). VDUP1 blocks the reducing activity of TRX and inhibits the interaction between TRX and other factors, such as ASK-1 and PAG (9, 11). VDUP1 expression is regulated by environmental conditions, and its expression affects cell growth. VDUP1 is up-regulated by various stresses, including H2O2, irradiation, heat shock, serum starvation, and transforming growth factor-β (11, 12). In addition, anticancer and anti-proliferative reagents, such as 5-fluorouracil, anisomycin, dexamethasone, and SAHA (a potent inhibitor of histone deacetylases), increased VDUP1 expression in cancer cells (13, 14). However, its expression was reduced in chemically induced rat mammary tumors and ferric nitrilotriacetate-induced renal cancer model of rats (15, 16).
In this study, we demonstrated that VDUP1 expression dramatically increased in senescent cells, and functional analysis revealed that the increase of VDUP1 expression contributed to cellular senescence establishment. Subsequent analysis of the mechanism responsible for the up-regulation of its expression showed that VDUP1 expression in senescent cells was regulated by FOXO3A at the transcriptional and miR-17-5p at the post-transcriptional level.
EXPERIMENTAL PROCEDURES
Cell Culture, Viral Transduction, and Cell Proliferation Assay
Human embryonic lung diploid fibroblast 2BS cells (obtained from the National Institute of Biological Products, Beijing, China) were previously isolated from female fetal lung fibroblast tissue and have been fully characterized (17). The cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (Invitrogen), glutamine (2 mm; Invitrogen), and penicillin/streptomycin (100 units/ml/100 μg/ml; Invitrogen) at 37 °C in 5% CO2. The normal human embryo lung fibroblasts (WI-38) were from ATCC and were cultured in the same manner as described for 2BS cells.
For the cell proliferation assay, cells were typically transfected with siRNA and subsequently infected with pBABE-RasV12-puro. The 4 × 105 cells were then seeded into 6-well plates, fixed, and stained 9 days postinfection with Coomassie Blue.
Phoenix cells and pBABE-RasV12-puro were kindly provided by Xiaowei Zhang (Health Science Center, Peking University, China), and the cells were cultured in the same manner as described above. Viral packaging was carried out according to Manuel et al. (18).
Plasmid Constructs
To obtain various deletion constructs of human VDUP1 promoter plasmids, a series of forward primers and a common reverse primer were used to amplify the VDUP1 promoter fragments by PCR from human genomic DNA. The various forward primers used were as follows: −1326, 5′-AA CTC GAG CCT AAA GCA TCT CAC AGC CAG CA-3′; −1028, 5′-AA CTC GAG TTA GTG AGT TGG AAG AGG GGA TGG-3′; −620, 5′-AA CTC GAG CTG AAA GTT CTC CTT TCC CTC AGA GA-3′; −148, 5′-AA CTC GAG AGC ACA CCG TGT CCA CGC-3′; and −89, 5′-AA CTC GAG AGG ACC GGG CAG CCA AT-3′ (the underlined sequence is the XhoI (New England Biolabs, Beverly, MA) recognition site). The common reverse primer is as follows: +265, 5′-AA AAG CTT CTC CAA ATC GAG GAA ACC CCT T-3′ (the underlined sequence is the HindIII (New England Biolabs) recognition site). Each pair of primers was used to amplify various sizes of promoter region. The resulting fragments were subjected to restriction digestion and subcloned into pGL3-basic plasmid (Promega, Madison, WI). These clones were designated as −1326 (full-length), −1028, −620, −148, and −89 relative to the transcriptional start site. To obtain deletion constructs of human VDUP1 3′-UTR reporter plasmids, the various forward primers were used: +19, 5′-AA ACTAGT GCA GCT TTA CCT ACT TGT TT-3′; +381, 5′-AA ACTAGT GAA CTT TGG ACC TTG AAC TT-3′; +692, 5′AA ACTAGT TGG AGC CTA TTG CAC TGT G-3′; +1204, 5′AA ACTAGT ACT GTC CTG TGT CAG AGC A-3′ (the underlined sequence is the SpeI New England Biolabs) recognition site); numbering is relative to the position of the VDUP1 termination codon). The common reverse primer was 5′-AA AAG CTT GGT ACA TAA TAC CCC CAC AT-3′ (the underlined sequence is the HindIII (New England Biolabs)recognition site). The PCR products were ligated into pMIR-REPORT miRNA Expression Reporter Vector (Ambion, Austin, TX), which was designated as pVDUP1–3′UTR. The miR-17-5p luciferase sensor construct (pmiR-17-5p reporter) was engineered by inserting the full 22-bp sequence complementary to the mature miR-17-5p into the SpeI (New England Biolabs)and HindIII (New England Biolabs) sites of pMIR-REPORT vector (Ambion).
VDUP1-pBABE was prepared as follows. The cDNA for wild-type VDUP1 was derived by PCR and subcloned into the EcoRI and SalI sites of pBABE-puro vector (provided by Dr. Xiaowei Zhang). The oligonucleotide sequences for the upstream and downstream PCR primers were as follows: 5′-AA GAA TTC CCA ACT CAG TTC CAT CAT GGT G-3′ and 5′-AA GTC GAC ACT GTT GAG TCT CTG AAA AAG TGA GTG-3′, respectively (the underlined sequences are the EcoRI (New England Biolabs) and SalI (New England Biolabs) recognition sites). pECE vector, HA-FOXO3A WT and two FOXO3A mutants (HA-FOXO3A-TM (T32A, S253A, S315A) and HA-FOXO3A-TM Δ DB(T32A, S253A, S315A, Δ DNA binding domain) were bought from Addgene.
Site-directed Mutagenesis
Bases were mutated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). A promoter mutagenic primer (5′-GGTCGGGCTCCTGGCGCGCAAGGACCGGGCAGC-3′, the underlined bases are the mutated bases) and two 3′UTR mutagenic primers were synthesized (site 1, 5′-CAGTCACTCTCAGCCATAGGAGTGTGTTCACTGTCCTGTGTC-3′; site 2, 5′-GATTCTAAGGTGATGTTCTTAGGAGTGTAATTCCTGTCAAATTTTTTGTTC-3′, the underlined bases are the mutated bases). Pfu Turbo DNA polymerase was used to synthesize the mutagenic promoter and 3′UTR, followed by digestion of the parental plasmids by DpnI according to the manufacturer's instructions. The mutated plasmids were transformed into XL1-Blue competent cells, and the resulting plasmids were isolated and sequenced to confirm the mutations.
RNA Interference
RNA interference was used to knock down FOXO3A in young or senescent cells. The human FOXO3A siRNA target sequences were 5′-CAA CCT GTC ACT GCA TAG T-3′ (FOXO3A siRNA I) and 5′-GAG CTC TTG GTG GAT CAT C-3′ (FOXO3A siRNA II), which corresponded to 636–654 and 690–708 bp relative to the first nucleotide of the start codon of the human FOXO3A coding sequence, respectively. The siRNA sequence targeted against VDUP1 corresponded to bases 864–886 of the mRNA. The sequence was 5′-CTG AGT GAG CAC TTT GGT CTG GC-3′. A nonrelated, scrambled siRNA without any other match in the human genomic sequence was used as a control 5′-TTC TCC GAA CG T GGC ACG A-3′. The miR-17-5p duplex sequence was 5′-CAA AGU G CU UAC AGU GCA GGU AGU dTdT-3′ and the scramble duplex sequence was 5′-AAC ACG CTC GGT CAA AAG GTT TTC dTdT-3′. The siRNA was synthesized by Shanghai Genechem Co., Ltd., China. FOXO3A knockdown was performed by transfection of siRNA into cells using Lipofectamine 2000 (Invitrogen) transfection reagent according to the manufacturer's instructions.
Transient Transfection and Luciferase Assay
All transfections were carried out with the LipofectamineTM 2000 (Invitrogen) according to the manufacturer's instructions. For VDUP1 promoter activity assays, 2BS cells were seeded in a 24-well plate (1 × 105 cells/well), incubated overnight in complete growth medium, and transfected with 0.45 μg of reporter plasmid, 0.05 μg of control reporter (pRL-CMV). After 24 h of incubation, cells were lysed in passive lysis buffer (Promega), and luciferase activity was measured using the luciferase assay system (Promega) with a luminometer (Lumat LB 9501; Berthold). All the luciferase assays were conducted at least three times in triplicate. Firefly luciferase activity was normalized to Renilla luciferase activity for each transfected well.
VDUP1–3′UTR luciferase assays were performed by cotransfection of 2BS cell lines with vectors and synthetic 2′-O-methyl oligoribonucleotides by using Lipofectamine 2000 (Invitrogen). 2′-O-Methyl oligoribonucleotides were synthesized by Shanghai GeneChem Co., Ltd., China (ASO scramble, 5′-GAA AAC CUU UUG ACC GAG CGU GUU-3′; ASO miR-17-5p, 5′-ACU ACC UGC ACU GUA AGC ACU UUG-3′; ASO miR-106, 5′-CUA CCU GCA CUG UAA GCA CUU UU-3′; and ASO miR-372, 5′ACG CUC AAA UGU CGC AGC ACU UU-3′).
Real Time PCR
Total RNA was extracted by using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. The first strand cDNA was synthesized using SuperScriptTM III first strand kits (Invitrogen) for RT-PCR, and VDUP1 mRNA was analyzed by real time PCR with use of primers (forward, 5′-CTC GTG TCA AAG CCG TTA GGA-3′; reverse, 5′-GTC AAG AAA AGC CTT CAC CCA-3′); β-actin as internal control (forward, 5′-AGC GAG CAT CCC CCA AAG TT-3′; reverse 5′-GGG CAC GAA GGC TCA TCA TT-3′). The mirVanaTM qRT-PCR miRNA detection kit and primer sets were purchased from Ambion. cDNA was synthesized from total RNA using gene-specific primers according to the TaqMan microRNA assay protocol (Ambion). qRT-PCR was performed on cDNA generated from total RNA by using the protocol of the mirVana qRT-PCR miRNA detection kit (Ambion). Amplification and detection of specific products were performed with the ABI Prism 7300 sequence detection system with the cycle profile according to the mirVana qRT-PCR miRNA detection kit. As an internal control, U6 primers were used for RNA template normalization.
Western Blotting
Cells were rinsed with PBS (pH 7.2) and lysed in a RIPA buffer containing 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 10 mm EGTA, 1% Nonidet P-40, 0.1% SDS, 1 mm Na3VO4, 50 mm NaF, 2 μg/ml aprotinin, 2 μg/ml leupeptin, and 1 mm phenylmethylsulfonyl fluoride. Lysates were centrifuged at 16,000 × g in an Eppendorf 5417 R centrifuge at 4 °C for 15 min, and supernatants were subjected to protein determination using a bicinchoninic acid protein assay (Pierce). Equal protein amounts of each cell lysate were subjected to 12% SDS-PAGE under reducing conditions (with 2.5% β2-mercaptoethanol) and transferred to PVDF membranes. Separate membranes were probed with the indicated primary antibodies. Immune complexes were detected using HRP-labeled donkey anti-rabbit or anti-goat IgG (Amersham Biosciences) followed by chemiluminescence (Pierce).
Young and senescent cell nuclear and cytoplasmic extracts were prepared using NE-PERTM nuclear and cytoplasmic extraction reagents (Pierce) according to the manufacturer's instructions. Twenty μg of total protein from each preparation were separated by SDS-PAGE, followed by Western blotting.
Such antibodies were used for Western blot as follows: β-actin (Cell Signaling catalog no. 4967), VDUP1 (Santa Cruz Biotechnology, catalog no. sc-33099), FOXO3A (Cell Signaling, catalog no. 9467), FOXO1 (Cell Signaling, catalog no. 2880), AKT (Cell Signaling catalog no. 9272), phospho-AKT (Ser-473) (Cell Signaling, catalog no. 9271), and LMNB2 (Abcam, catalog no. ab16048).
Chromatin Immunoprecipitation
Chromatin immunoprecipitations were performed using the chromatin immunoprecipitation assay kit (Upstate). Antibodies used for ChIP were anti-FOXO3A (Cell Signaling, catalog no. 9467) and FOXO1 (Cell Signaling, catalog no. 2880). Primer pairs used for PCR were as follows: FOXO3A-binding site (−180 to +29), forward primer, 5′-TCC AGA GCG CAA CAA CCA TT-3′; reverse primer, 5′-TCA AGC AGG AGG CGG AAA C-3′; non-FOXO3A-binding site (−1175 to −950), forward primer, 5′-TTT CTA AAC CGT TTA GGG AA-3′; reverse primer, 5′-AAA GGA TGC TTC ACT CTT TT-3′.
SA-β-gal and SAHF Staining
For detection of SA-β-gal activity, cells were washed twice with PBS (pH 7.2), fixed with 4% formaldehyde for 5 min at 22 °C, and washed with PBS (pH 6.0) again. Cells were then incubated overnight at 37 °C without CO2 in freshly prepared staining buffer (1 mg/ml X-gal, 40 mm citric acid/sodium phosphate (pH 6.0), 5 mm potassium ferrocyanide, 5 mm potassium ferricyanide, 150 mm NaCl, 2 mm MgCl2).
For SAHF staining, cells were washed twice with PBS (pH 7.2), fixed with 4% formaldehyde for 5 min at 22 °C, and washed with PBS again. Cells were incubated in 0.1% Triton X-100/PBS for 5 min at 22 °C and stained by DAPI for 5 min according to Ref. 19.
Cell Cycle Analysis
For analysis of the cell cycle, cells were washed with PBS, detached with 0.25% trypsin, and fixed with 70% ethanol overnight. Fixed cells were subsequently washed, treated with 5 μg/ml RNase A (Sigma), and stained with 50 μg/ml propidium iodide. The DNA content was measured by fluorescence-activated cell sorting (FACS) on a FACScan flow cytometry system (BD Biosciences). The data were analyzed using CellFIT software.
Northern Blot Analysis
Cells were treated with 5 μg/ml actinomycin D (Sigma). Total RNA was prepared at the times indicated. Twenty-microgram RNA samples were denatured, size-fractionated by electrophoresis in 1.2% agarose/formaldehyde gels, and transferred onto Magna nylon transfer membrane (Osomonics®). For the detection of endogenous VDUP1 mRNA, 3′UTR of VDUP1 was excised from pVDUP1–3′UTR plasmid and labeled with [α-32P]dCTP with Prime-a-Gene labeling system (Promega). For the detection of 18 S rRNA and pRL-CMV, PCR products was extracted from the gel and labeled as above (primers used were as follows: 18 S rRNA forward, 5′ CGA GCC GCC TGG ATA CC 3′; reverse, 5′ ACA TCT AAG GGC ATC ACA GAC CTG 3′; pRL-CMV forward, 5′ GAT TCA GAA AAA CAT GCA GA 3′; reverse, 5′ CGA TTC AAT AAA CAT TTT TGG T 3′). Hybridization and washes were performed according to ExpressHyb Hybridization Solution manual (Clontech). Signals were visualized and quantitated with a PhosphorImager (GE Healthcare).
RESULTS
VDUP1 Expression Is Up-regulated in Senescent Cells
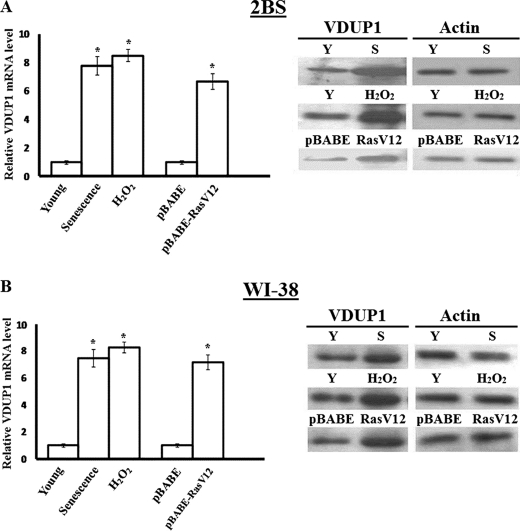
As cell proliferation inhibition is an important characteristic of senescent fibroblasts, it was observed whether VDUP1 expression increases in senescent cells. Results from real time PCR and Western blots analysis showed that VDUP1 was elevated 5-fold in replicative senescent cells (Fig. 1A). Overexpression of certain oncogenes or DNA damage can induce a phenotype that closely resembles replicative senescence (20), so the young 2BS cells were treated by expressing oncogenic RAS[RAS-H(V12)] or with a sublethal dose(200 μm of H2O2), allowing them to develop a senescent phenotype (18, 21). The VDUP1 expression in pre-senescent cells increased about 6–10-fold compared with control cells. We also obtained similar results in the WI-38 cell line (Fig. 1B).
FIGURE 1.
Increased expression of VDUP1 in senescent cells. A, real time PCR and Western blot analysis of VDUP1 expression in 2BS cells that are young (Y), replicative-senescent (S), and premature-senescent. Actin was used as a loading control. Left panel, relative VDUP1 mRNA amount in 2BS cells; right panel, Western blot analysis of VDUP1 protein expression in 2BS cells. B, real time PCR and Western blot analysis repeated in WI-38 cells that are young, replicative-senescent, and premature-senescent. Actin was used as a loading control. Left panel, relative VDUP1 mRNA amount in WI-38 cells; right panel, Western blot analysis of VDUP1 protein expression in WI-38 cells. *, p < 0.05.
VDUP1 Induces Senescent Cell Morphology
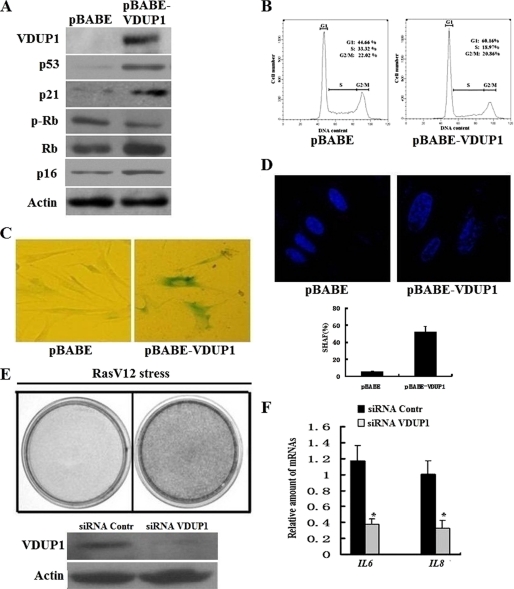
Because VDUP1 level correlated with senescence, we want to understand if this gene could play a causal role during senescence process. To address this question, we produced VDUP1 adventitiously in early passage human fibroblasts. 2BS cells were infected with pBABE-puro or retrovirus vector expressing the VDUP1 gene, pBABE-VDUP1 (Fig. 2A). The results showed that cells expressing VDUP1 arrested cell cycle progression at the G0/G1 phase (Fig. 2B), displayed the morphology of flat cells, stained positive for SA-β-gal (Fig. 2C), and formed SAHF (Fig. 2D) 9 days after retroviral infection (5 days post-selection); moreover, molecular markers of senescence, such as increased p53, p21, p16, and decreased p-Rb levels, were also detected after VDUP1 overexpression (Fig. 2A). These results suggested that overexpression of VDUP1 induced premature senescence in early passage 2BS cells.
FIGURE 2.
VDUP1 regulates cellular senescence. A, Western blot analysis of senescence marker in 2BS cells infected with pBABE-VDUP1. B, cell cycle analysis of 2BS cells infected with pBABE-VDUP1. Young cells were infected with retrovirus containing VDUP1. At 5 days post-selection, cell cycle was analyzed by flow cytometry. C, SA-β-gal activity; D, senescence-associated heterochromatin foci in VDUP1-overexpressing cells. E, cell proliferation assay of oncogenically stressed (RasV12-expressing) 2BS cells with VDUP1 knockdown. Cells were fixed and stained 9 days after infection. Upper panel, cell staining indicated the proliferation assay of oncogenically stressed (RasV12-expressing) 2BS cells with VDUP1 knockdown. Lower panel, Western blot analyzed VDUP1 protein expression after knockdown of VDUP1 protein by siRNA. Contr, control. F, samples from E were analyzed for IL6 and IL8 transcripts by qRT-PCR. Levels are normalized to actin and represented as means ± S.D. *, p < 0.05.
In light of the above results, we further detected whether a decrease in the amount of intracellular VDUP1 delayed senescence. RNA interference was used to deplete the VDUP1 protein. Fig. 2E showed that the amount of VDUP1 protein in the cells was reduced by at least 90% by the specific siRNA duplex as compared with the control siRNA. As seen in Fig. 2E, VDUP1 knockdown delayed the onset of Ras-induced cellular senescence. It has been shown that cellular senescence is accompanied by increased secretion of several key cytokines that play a prominent role in oncogene-induced cellular senescence, such as IL6 and IL8 (22); thus, we tested whether VDUP1 knockdown affected the cytokine production. Our results indicated that VDUP1 knockdown delayed IL6 and IL8 production (Fig. 2F). These results highlight the functional relevance of VDUP1 in senescence establishment.
FOXO3A Regulates VDUP1 Expression at the Transcriptional Level in Senescent Cells
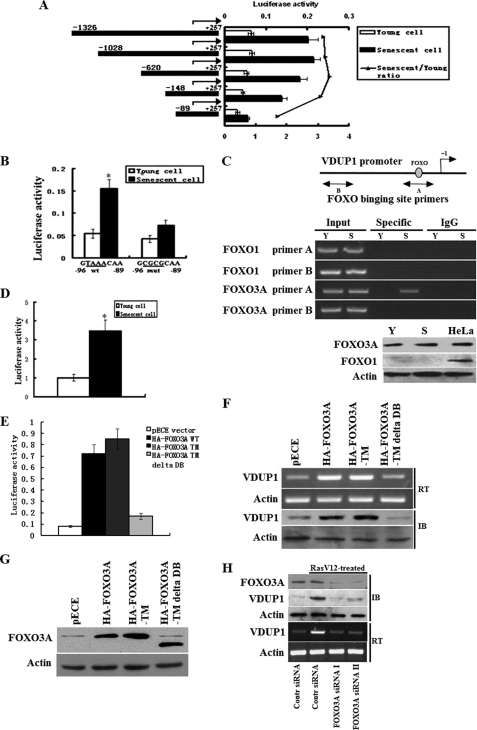
To determine the basal activity of the VDUP1 promoter in young and senescent cells, we fused the 1.5-kb VDUP1 promoter to the luciferase reporter gene and transfected it into young and senescent cells. After 24 h of incubation, cells were harvested and analyzed for luciferase activity. As shown in Fig. 3A, luciferase activity caused by 1.5-kb VDUP1 promoter was ∼3.2-fold greater in senescent than in young cells. To find out the region that plays the role in the up-regulation of VDUP1 in senescent cells, various VDUP1 promoter 5′-deletion constructs were generated (Fig. 3A) and transiently transfected into young and senescent cells. Deletion of segments from the 5′ end, from −1326 to −148, slightly reduced the difference of luciferase activity between young and senescent cells. However, further deletion to −89 bp upstream of the transcription start site decreased the difference of luciferase activity between young and senescent cells dramatically (Fig. 3A). These data suggest that increased expression of VDUP1 in senescent cells requires the promoter region from −148 to −89.
FIGURE 3.
FOXO3A transcriptionally up-regulates VDUP1 expression in senescent cells. A, deletion analysis of a luciferase-based reporter of the VDUP1 promoter in young and senescent cells. In all reporter assays, firefly luciferase-based reporter signal is normalized to the expression of a cotransfected Renilla luciferase control plasmid, pRL-CMV. B, effect of FOXO3A site mutations on the activity of VDUP1 promoter (the FOXO3A site from −96 to −89 GTAAACAA were mutated to GCGCGCAA) in young and senescent cells. C, upper panel, ChIP analysis of FOXO3A binding to VDUP1 promoter in young (Y) and senescent cells (S). Lower panel, Western blot assay showed FOXO1 and FOXO3A expression in young and senescent cells, using HeLa cell as positive control. D, FOXO3A activity increases in senescent fibroblasts, a synthetic promoter containing three FOXO3A-response elements (FHRE) were transfected into young or senescent cells. Luciferase activity was measured 24 h after transfection. E, young 2BS cells were cotransfected with VDUP1 promoter luciferase plasmids and WT or various mutated FOXO3A plasmids, and results indicated FOXO3A-WT and FOXO3A-TM significantly increased VDUP1 promoter plasmid luciferase activity. F, RT-PCR and Western blot analysis of VDUP1 expression after transfected young cells with WT or various mutated FOXO3A plasmids. IB, immunoblot. G, expression of WT and mutated FOXO3A protein. Proteins from experiments shown in E and F were detected by immunoblot (IB) analysis using an anti-FOXO3A antibody; a representative immunoblot is shown. H, effects of FOXO3A knockdown on the VDUP1 mRNA and protein level. Young 2BS cells were transfected with control siRNA or two specific FOXO3A siRNAs; subsequently, the cells were treated by expressing oncogenic RAS[RAS-H(V12)]. RasV12 treatment did not induce VDUP1 mRNA and protein levels in FOXO3A knockdown cells. *, p < 0.05.
To find the specific transcription factor binding in this region, we performed a detailed computer analysis using the MatInspector program and found a FOXO transcription factor binding element. As it has been shown that FOXO1 can regulate VDUP1 expression (23, 24), four base mutations within this element were introduced by site-directed mutagenesis in the −148 construct (Fig. 3B). We transfected the −148 construct and the mutated construct into young and senescent cells and measured the luciferase activity resulting from each construct after normalizing for pRL-CMV control. As shown in Fig. 3B, mutation of the FOXO-binding element decreased about 2.2-fold luciferase activity in senescent cells as compared with the wild-type construct. In contrast, the luciferase activity from the mutant construct slightly decreased as compared with the wild-type construct in young cells. These data suggest that the FOXO-binding site is essential for the up-regulation of VDUP1 in senescent cells.
Next, it was detected whether FOXO bound to the endogenous VDUP1 promoter. Binding of FOXO to the endogenous VDUP1 promoter was determined by ChIP assay. Interaction between FOXO and the VDUP1 promoter region that contains the FOXO-binding sites was detected by a specific PCR assay of immunoprecipitated FOXO-DNA complexes (see “Experimental Procedures”). FOXO1 antibody was used to immunoprecipitate DNA as it has been shown that FOXO1 can regulate VDUP1 expression (23, 24). After amplification of DNA immunoprecipitated by FOXO1 antibody, no PCR amplification product was observed in either young or senescent cells (Fig. 3C). However, PCR amplification product was observed in senescent cells but not in young cells when FOXO3A antibody was used to immunoprecipitate DNA (Fig. 3C). Further analysis showed that no FOXO1 expression was detected in 2BS fibroblasts with HeLa cell as a positive control (Fig. 3C, lower panel), which could explain the observation that no FOXO1 bound to the endogenous VDUP1 promoter. As a negative control, PCR amplification of the promoter region −1175 to −950, which lacks FOXO-binding site, did not yield signal (Fig. 3C), suggesting that FOXO3A can specifically bind to the endogenous VDUP1 promoter in senescent cells.
We next asked whether activity of FOXO3A increased in senescent cells. The synthetic forkhead promoter (FHRE) that contains three insulin-response element sequences driving the expression of the firefly luciferase cDNA (25) was used to examine the FOXO3A activity in young and senescent 2BS cells. This promoter is activated by introduction of the FOXO3A protein (25). We transfected the FHRE luciferase plasmid into young and senescent cells, consistent with an increase in forkhead-mediated transcription in the senescent cells, and the FHRE promoter had much higher activity in the senescent cells (Fig. 3D).
To understand the molecular mechanism that underlies the increase in the FOXO3A activity observed in senescent cells, we observed AKT activity in young and senescent cells because AKT is a major kinase that is known to phosphorylate and inactivate FOXO3A in response to growth factor. In mitogenic conditions, AKT is a primary regulator of FOXO3A. Upon activation following mitogenic stimulation, AKT translocates to the nucleus and phosphorylates FOXO3A, decreasing FOXO3A activity. Thus, defects in either activation or nuclear translocation of Akt could explain the decrease in FOXO3A phosphorylation observed in senescent fibroblasts. We first examined activation of cytoplasmic pAKT by measuring site-specific phosphorylation of the protein. As shown in supplemental Fig. S1A, in both young and senescent cells, serum stimulation efficiently increased cytoplasmic AKT phosphorylation at Ser-473. These data indicate that AKT activation was similar in young and senescent cells. Subsequently, the nuclear abundance of active AKT (pAKT) was examined using a phosphospecific antibody to detect the active form of the enzyme. However, examination of nuclear extracts revealed that only young cells exhibited an increase in the amount of active AKT in the nucleus. In the senescent cells, there was very little active AKT in nuclear extracts following serum stimulation (supplemental Fig. S1B), which is in agreement with reports based on senescent WI-38 cells (26). The above results suggested the failure to translocate active AKT to the nucleus accounted for the increase of FOXO3A activity in senescent cells.
To further confirm that FOXO3A directly up-regulates VDUP1 expression at the transcriptional level, the VDUP1 promoter reporter plasmids were transfected into young 2BS cells together with a pECE vector encoding nothing (Mock) or HA-FOXO3A-WT or FOXO3A mutants (HA-FOXO3A-TM; HA-FOXO3A-TMΔDB). HA-FOXO3A-TM is a triple mutant form of FOXO3A in which all three Akt phosphorylation sites are mutated to alanine (FOXO3A-TM, T32A/S253A/S315A), and the TM construct is expressed as a constitutively active form of FOXO3A that is trapped in the nucleus. FOXO3A-TMΔDB is a form of TM protein that lacks the FOXO3A DNA binding domain (25, 27). As shown in Fig. 3E, WT- and FOXO3A-TM significantly increased luciferase activities of the VDUP1 promoter. However, FOXO3A TMΔDB did not affect luciferase activities of the VDUP1 promoter. To observe if FOXO3A affects endogenous VDUP1 expression, we monitored VDUP1 gene mRNA and protein expression in WT or mutated FOXO3A-expressing young cells. As seen in Fig. 3F, overexpression of WT- or FOXO3A-TM in young 2BS cells resulted in the increase of VDUP1 mRNA and protein levels, whereas FOXO3A TMΔDB had no effect on VDUP1 mRNA and protein levels. These data show that FOXO3A induces VDUP1 expression by binding to DNA. Representative FOXO3A immunoblots are shown in Fig. 3G, showing that the wild-type FOXO3A and mutants used in Fig. 3, E and F, were expressed at similar levels.
To further evaluate the association between FOXO3A and VDUP1 gene expression, young 2BS cells were transfected with control siRNA or two specific FOXO3A siRNAs; subsequently, the cells were treated by expressing oncogenic RAS[RAS-H(V12)]. The amount of FOXO3A protein in the cells was reduced by at least 80% by the two specific siRNAs as compared with the control siRNA (Fig. 3H). As shown in Fig. 3H, RasV12 treatment did not induce VDUP1 mRNA and protein levels in FOXO3A knockdown cells. Collectively, these results demonstrate that FOXO3A is a transcriptional activator of VDUP1 gene in 2BS cells.
Down-regulation of miR-17-5p Expression Is Associated with the Increase in VDUP1 Expression in Senescent Cells
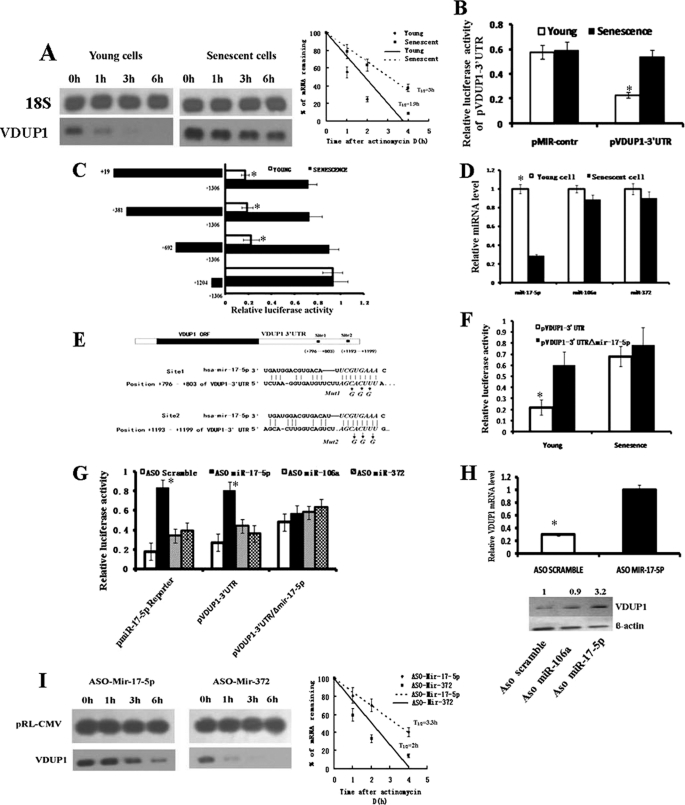
Analysis of promoter activity showed that FOXO3A-induced VDUP1 expression could not explain the difference at the mRNA level between young and senescent cells, and it was suspected that VDUP1 expression was also regulated at the post-transcriptional level. To explore this, we performed mRNA stability assay with the presence of actinomycin D, a transcription inhibitor. Northern blot results showed that VDUP1 mRNA in young cells degraded rapidly, whereas in senescent cells, the bands disappeared at a relatively lower speed (Fig. 4A). These results suggest that VDUP1 expression is post-transcriptionally regulated in young cells. 3′-UTR is a major target site, through which mRNA turnover is regulated. Thus, VDUP1 3′-UTR was cloned downstream of the luciferase reporter gene of pMIR-report plasmid, and the resultant plasmid (pVDUP1–3′UTR) was transfected into young and senescent cells. The results showed that senescent cells had higher luciferase activity than young ones (Fig. 4B), suggesting that 3′-UTR mediated VDUP1 expression in young cells. To identify the functional region in the 3′-UTR of VDUP1 that contributes to VDUP1 up-regulation, various 5′-deleted pVDUP1–3′UTR constructs were generated and transfected into young and senescent cells. As shown in Fig. 4C, deletion of segments from the 5′ end, from +692 to +1204, increased the luciferase activity by 3-fold in young cells, whereas only slight elevation of luciferase activity was observed in senescent cells. These data suggest that regulation elements within this region of 3′UTR regulate VDUP1 expression, and this regulation may have been lost in senescent cells. It has been shown that AU-rich elements (28, 29) and microRNAs (30, 31) play important roles in destabilizing mRNA; however, no AU-rich elements were found in this region by computer analysis. Thus, we focused our attention on microRNA elements. An on-line search of the prediction web sites from the computational biology center of the Memorial Sloan-Kettering Cancer Center and TargetScan demonstrated that 24 putative miRNA target sites were harbored in this region of VDUP1 mRNA 3′-UTR. We selected miRNA based on their known oncogenic potential or miRNAs whose expression increases in tumor cells. We chose three miRNAs to study, miR-17-5p, -106a, and -372, which have been shown to have oncogenic potential. Next, we observed whether the expression of these miRNA decreases in senescent cells by Taqman RT-PCR analysis. Our results showed that miR-17-5p expression decreased ∼3-fold in senescent cells compared with young cells (Fig. 4D). Additionally, miR-106a and miR-372 expressions remained relatively unchanged (Fig. 4D). Thus, the scope of research was narrowed to miR-17-5p. To demonstrate that miR-17-5p interacts with specific target sequence localized in this region of VDUP1 3′-UTR, an additional reporter construct was generated in which the 7-bp “seed” sequence (CGUGAAA) of two putative miR-17-5p target sites were mutated using PCR (Fig. 4E). The resulting construct, pVDUP1–3UTR/ΔmiR-17-5p, was transfected into young and senescent cells; this mutation dramatically increased luciferase activity in young cells, whereas only a slight elevation was observed in senescent cells (Fig. 4F). These results suggest that the loss of miR-17-5p expression likely associates with the up-regulation of VDUP1 expression in senescent cells.
FIGURE 4.
miR-17-5p post-transcriptionally regulates VDUP1 expression. A, VDUP1 mRNA metabolism in young and senescent cells. Representative Northern result is shown. Average half-life and S.E. values were calculated for VDUP1 (young cell = 110 ± 8 min; senescent cell = 180 ± 25 min from three independent half-life experiments). B, effects of the 3′-UTR of VDUP1 on the luciferase activity of reporter construct pVDUP1–3′-UTR in young and senescent cells. pMir-reporter plasmid was used as the empty vector. C, deletion analysis of the 3′UTR of VDUP1 gene in young and senescent cells. D, real time PCR analysis of the expression of miR-17-5p, miR-106a, and miR-372 in young and senescent cells. E, complementarity between miR-17-5p and the two putative VDUP1 3′UTR sites targeted. Site 1 and site 2 represent the nucleotides that are predicted to base pair with nucleotides 2–7 of the miRNA (the miRNA “seed sequence”). Also shown are the mutated bases in the seed sequence of the putative miR-17-5p target sequence present in the VDUP1 3′UTR. F, effects of the mutation of putative miR-17-5p target sequence on the luciferase activity of reporter construct pVDUP1–3′UTR in young and senescent cells. G, effects of inhibition of miR-17-5p by ASO miR-17-5p on the luciferase activity of pVDUP1–3′UTR and pVDUP1–3′UTR/Δmir-17-5p in young cells. The pmiR-17-5p reporter with perfect complementarity to miR-17-5p was used as a positive control. H, effects of miR-17-5p inhibition by ASO miR-17-5p on the mRNA and protein levels of VDUP1 in young cells. The relative gene expression levels of VDUP1 mRNA and protein were normalized to actin. I, effects of ASO mir-17-5p transfection on the stability of endogenous VDUP1 mRNA level. Young cells were cotransfected with plasmid pRL-CMV (as internal control) together with ASO miR-17-5p or ASO miR-372 (as negative control), 12 h later, actinomycin D was added into culture medium, and RNA was extracted at indicated time. Average half-life and S.E. values were calculated for VDUP1 (ASO miR-372 = 120 ± 18 min; ASO miR-17-5p = 204 ± 35 min from three independent half-life experiments).
To further confirm that miR-17-5p inhibits VDUP1 expression, young cells were cotransfected with ASO miR-17-5p together with the wild-type reporter construct pVDUP1–3′UTR or the mutated construct pVDUP1–3′UTR/ΔmiR-17-5p. As expected, miR-17-5p inhibitor increases luciferase activity of pVDUP1–3′UTR but not the mutated construct pVDUP1–3′UTR/ΔmiR-17-5p (Fig. 4G). There was no change in luciferase reporter activity when ASO miR-106a or ASO miR-372 was cotransfected with either of the reporter constructs (Fig. 4G), indicating that the observed elevation was specific to ASO miR-17-5p. Consistent with the luciferase assays, ASO miR-17-5p treatment of young cells also significantly increased VDUP1 mRNA levels compared with the control (Fig. 4H). Correspondingly, VDUP1 expression at the protein level also increased (Fig. 4H). Next, we observed if miR-17-5p down-regulates VDUP1 expression via affecting its mRNA half-life. Young cells were cotransfected with plasmid pRL-CMV (as internal control) together with ASO miR-17-5p or ASO miR-372 (as negative control), and 12 h later, actinomycin D was added into the culture medium, and RNA was extracted at the indicated time. Northern blot showed that ASO miR-17-5p treatment resulted in the increase of VDUP1 mRNA stability, whereas ASO miR-372 did not change VDUP1 mRNA half-life (Fig. 4I). These results suggest that miR-17-5p down-regulates VDUP1 expression via destabilizing its mRNA. To rule out the possibility that miR-17-5p transcriptionally regulates VDUP1 mRNA levels; young cells were cotransfected with the wild-type VDUP1 promoter reporter vector pVDUP1 and either the ASO miR-17-5p or the control. No difference in luciferase activity was observed between samples and control (data not shown). Collectively, these data indicate that miR-17-5p post-transcriptionally down-regulates VDUP1 expression, and that down-regulation of miR-17-5p is associated with up-regulation of VDUP1 in senescent cells.
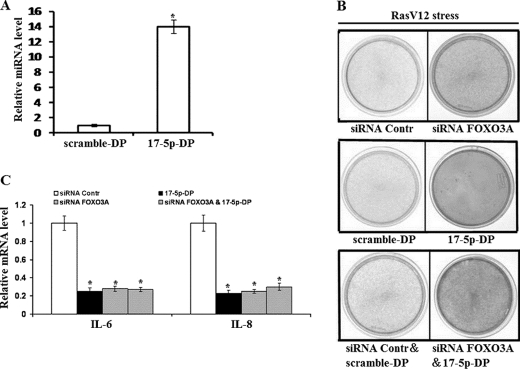
Knockdown of FOXO3A and/or Overexpression of miR-17-5p Delays Cellular Senescence
Because FOXO3A positively and miR-17-5p negatively regulate VDUP1 expression, and VDUP1 up-regulation induces cellular senescence, we next examined whether knockdown of FOXO3A and/or overexpression of miR-17-5p can delay cellular senescence. FOXO3A was depleted by siRNA as described above; ectopic expression of miR-17-5p was done by transfecting miR-17-5p duplex (17-5p-DP) into 2BS cells, and the transfection efficiency and the effect of this oligonucleotide on miR-17-5p level were verified by quantitative RT-PCR (Fig. 5A). Results showed that knockdown of FOXO3A or overexpression of miR-17-5p alone or the combination of knockdown of FOXO3A and overexpression of miR-17-5p delayed RasV12-induced senescence and decreased cytokine IL6 and IL8 production (Fig. 5, B and C). These results are consistent with the FOXO3A and miR-17-5p regulation of VDUP1 expression.
FIGURE 5.
Knockdown of FOXO3A and/or overexpression of miR-17-5p delay cellular senescence. A, quantitative RT-PCR detects miR-17-5p level after transfecting miR-17-5p duplex (17-5p-DP) into 2BS cells. B, cell proliferation assay of oncogenically stressed (RasV12-expressing) 2BS cells with knockdown of FOXO3A and/or overexpression of miR-17-5p, FOXO3A knockdown (upper panel), miR-17-5p overexpression (middle panel), FOXO3A knockdown, and miR-17-5p overexpression (lower panel). Cells were fixed and stained 9 days after infection. C, samples from B were analyzed for IL6 and IL8 transcript levels by qRT-PCR. Levels are normalized to actin expression and represented as means ± S.D.
DISCUSSION
The expression of VDUP1 is frequently associated with conditions of decreased cellular proliferation. Based on the observations that VDUP1 may bind and inhibit TRX activity, and that TRX transgenic mice can achieve significantly extended life span, it has been assumed that VDUP1 is likely a regulator of cellular senescence. In this study, it was found that VDUP1 expression increases during the senescence process (Fig. 1), and adventitious expression of VDUP1 arrests the cell cycle of young cells and increases senescence-associated β-galactosidase activity and senescence-associated heterochromatin foci, which are two important markers of cellular senescence. These data suggest that the increase of VDUP1 expression induces premature senescence. Consistently, inhibition of VDUP1 expression by siRNA delayed Ras-induced senescence and cytokine IL6 and IL8 production. Collectively, we conclude that the increase of VDUP1 expression contributes to cellular senescent phenotypes.
Expression and regulation analysis demonstrates that FOXO3A transcriptionally up-regulates VDUP1 in senescent cells, which is coincident with an increase in FOXO3A activity in senescent cells. It has been reported that transcription factor FOXO1 may regulate VDUP1 expression through a different mechanism. In rat neurons, synaptic NMDA receptor blockade up-regulates VDUP1 expression via activating FOXO1 activity, which promotes vulnerability to oxidative damage (24). In contrast, another report shows that FOXO1 down-regulates VDUP1 expression in a human liver cell line (23). In this study, it was found that VDUP1 was up-regulated by FOXO3A rather than FOXO1 in senescent fibroblasts. Western blot analysis shows that 2BS fibroblasts lack FOXO1 expression. These differences indicate that FOXO regulation of VDUP1 expression is likely to be cell type-specific.
Analysis of promoter activity found that up-regulation of FOXO3A activity alone cannot explain the difference in VDUP1 expression at the mRNA level between young and senescent cells. Thus, we examined whether VDUP1 expression is also regulated at the post-transcriptional level. Results showed that miR-17-5p post-transcriptionally down-regulated VDUP1 expression via mRNA destabilization. Because miR-17-5p expression decreases in senescent cells, its suppressive role is attenuated, which results in an increase of VDUP1 mRNA stability in senescent cells. Down-regulation of miR-17-5p expression appears to augment the effect of FOXO3A on VDUP1 expression in senescent cells. It has been shown that miR-17-5p is able to act as both an oncogene and a tumor suppressor in different cellular contexts (32, 33). This gene is amplified in childhood lymphoma (34). Furthermore, transcription of the miRNA 17-5p-92 cluster is regulated by the oncogene c-myc (35), and its overexpression contributes to tumorigenesis (36, 37). However, miR-17-5p appears to act as a tumor suppressor in some breast and ovarian cancer cell lines (38). Here, our results showed that miR-17-5p acted as an oncogene in normal fibroblasts. Identification of target gene is the key to understanding microRNA function. It has been reported that E2F1–3, NCOA3, AIB1, and RBL2 are targets of miR-17-5p (35, 38–40). In this study, VDUP1 has been identified as a novel target gene of miR-17-5p, allowing insight into the mechanism by which miR-17-5p regulates cell proliferation.
Taken together, our results showed that VDUP1 expression evidently increased in senescent cells. Functional analysis revealed that the increase of VDUP1 expression contributed to cellular senescence establishment. Expression and regulation analysis demonstrated that VDUP1 was jointly regulated by FoxO3a and miR-17-5p at the transcriptional and post-transcriptional levels, respectively. These results provide novel clues for investigating the complex network of underlying pathways resulting in cellular senescence.
Supplementary Material
Acknowledgment
We thank Dr. Xiaowei Zhang for the gift of Phoenix cells and pBABE-RasV12- puro plasmid.
This work was supported by the National Basic Research Programs of China Grant 2007CB507400 and the National Science Foundation of China Grants 30672201 and 306210021.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- TRX
- thioredoxin
- qRT
- quantitative reverse transcription
- miRNA
- microRNA
- ASO
- antisense oligonucleotide.
REFERENCES
- 1.Faragher R. G., Kipling D. (1998) BioEssays 20, 985–991 [DOI] [PubMed] [Google Scholar]
- 2.Röhme D. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 5009–5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cristofalo V. J., Allen R. G., Pignolo R. J., Martin B. G., Beck J. C. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10614–10619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas D. M., Yang H. S., Alexander K., Hinds P. W. (2003) Cancer Biol. Ther. 2, 124–130 [PubMed] [Google Scholar]
- 5.Campisi J. (2005) Cell 120, 513–522 [DOI] [PubMed] [Google Scholar]
- 6.Dimri G. P. (2005) Cancer Cell 7, 505–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Porath I., Weinberg R. A. (2005) Int. J. Biochem. Cell Biol. 37, 961–976 [DOI] [PubMed] [Google Scholar]
- 8.Nishiyama A., Matsui M., Iwata S., Hirota K., Masutani H., Nakamura H., Takagi Y., Sono H., Gon Y., Yodoi J. (1999) J. Biol. Chem. 274, 21645–21650 [DOI] [PubMed] [Google Scholar]
- 9.Yamanaka H., Maehira F., Oshiro M., Asato T., Yanagawa Y., Takei H., Nakashima Y. (2000) Biochem. Biophys. Res. Commun. 271, 796–800 [DOI] [PubMed] [Google Scholar]
- 10.Junn E., Han S. H., Im J. Y., Yang Y., Cho E. W., Um H. D., Kim D. K., Lee K. W., Han P. L., Rhee S. G., Choi I. (2000) J. Immunol. 164, 6287–6295 [DOI] [PubMed] [Google Scholar]
- 11.Han S. H., Jeon J. H., Ju H. R., Jung U., Kim K. Y., Yoo H. S., Lee Y. H., Song K. S., Hwang H. M., Na Y. S., Yang Y., Lee K. N., Choi I. (2003) Oncogene 22, 4035–4046 [DOI] [PubMed] [Google Scholar]
- 12.Butler L. M., Zhou X., Xu W. S., Scher H. I., Rifkind R. A., Marks P. A., Richon V. M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11700–11705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi Y., Nagata T., Ishii Y., Ikarashi M., Ishikawa K., Asai S. (2002) Oncol. Rep. 9, 75–79 [PubMed] [Google Scholar]
- 14.Dutta K. K., Nishinaka Y., Masutani H., Akatsuka S., Aung T. T., Shirase T., Lee W. H., Yamada Y., Hiai H., Yodoi J., Toyokuni S. (2005) Lab. Invest. 85, 798–807 [DOI] [PubMed] [Google Scholar]
- 15.Yang X., Young L. H., Voigt J. M. (1998) Breast Cancer Res. Treat. 48, 33–44 [DOI] [PubMed] [Google Scholar]
- 16.Nishinaka Y., Nishiyama A., Masutani H., Oka S., Ahsan K. M., Nakayama Y., Ishii Y., Nakamura H., Maeda M., Yodoi J. (2004) Cancer Res. 64, 1287–1292 [DOI] [PubMed] [Google Scholar]
- 17.Tang Z., Zhang Z., Zheng Y., Corbley M. J., Tong T. (1994) Mech. Ageing Dev. 73, 57–67 [DOI] [PubMed] [Google Scholar]
- 18.Serrano M., Lin A. W., McCurrach M. E., Beach D., Lowe S. W. (1997) Cell 88, 593–602 [DOI] [PubMed] [Google Scholar]
- 19.Zhang R., Poustovoitov M. V., Ye X., Santos H. A., Chen W., Daganzo S. M., Erzberger J. P., Serebriiskii I. G., Canutescu A. A., Dunbrack R. L., Pehrson J. R., Berger J. M., Kaufman P. D., Adams P. D. (2005) Dev Cell 8, 19–30 [DOI] [PubMed] [Google Scholar]
- 20.Campisi J. (2000) In Vivo 14, 183–188 [PubMed] [Google Scholar]
- 21.Chen Q. M., Bartholomew J. C., Campisi J., Acosta M., Reagan J. D., Ames B. N. (1998) Biochem. J. 332, 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuilman T., Michaloglou C., Vredeveld L. C., Douma S., van Doorn R., Desmet C. J., Aarden L. A., Mooi W. J., Peeper D. S. (2008) Cell 133, 1019–1031 [DOI] [PubMed] [Google Scholar]
- 23.de Candia P., Blekhman R., Chabot A. E., Oshlack A., Gilad Y. (2008) PLoS ONE 3, e1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papadia S., Soriano F. X., Léveillé F., Martel M. A., Dakin K. A., Hansen H. H., Kaindl A., Sifringer M., Fowler J., Stefovska V., McKenzie G., Craigon M., Corriveau R., Ghazal P., Horsburgh K., Yankner B. A., Wyllie D. J., Ikonomidou C., Hardingham G. E. (2008) Nat. Neurosci. 11, 476–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. (1999) Cell 96, 857–868 [DOI] [PubMed] [Google Scholar]
- 26.Mawal-Dewan M., Lorenzini A., Frisoni L., Zhang H., Cristofalo V. J., Sell C. (2002) J. Biol. Chem. 277, 7857–7864 [DOI] [PubMed] [Google Scholar]
- 27.Tran H., Brunet A., Grenier J. M., Datta S. R., Fornace A. J., Jr., DiStefano P. S., Chiang L. W., Greenberg M. E. (2002) Science 296, 530–534 [DOI] [PubMed] [Google Scholar]
- 28.Wilusz C. J., Wilusz J. (2004) Trends Genet. 20, 491–497 [DOI] [PubMed] [Google Scholar]
- 29.Chen C. Y., Shyu A. B. (1995) Trends Biochem. Sci. 20, 465–470 [DOI] [PubMed] [Google Scholar]
- 30.Zhang B., Pan X., Cobb G. P., Anderson T. A. (2007) Dev. Biol. 302, 1–12 [DOI] [PubMed] [Google Scholar]
- 31.Lim L. P., Lau N. C., Garrett-Engele P., Grimson A., Schelter J. M., Castle J., Bartel D. P., Linsley P. S., Johnson J. M. (2005) Nature 433, 769–773 [DOI] [PubMed] [Google Scholar]
- 32.Calin G. A., Sevignani C., Dumitru C. D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M., Croce C. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2999–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu J., Getz G., Miska E. A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B. L., Mak R. H., Ferrando A. A., Downing J. R., Jacks T., Horvitz H. R., Golub T. R. (2005) Nature 435, 834–838 [DOI] [PubMed] [Google Scholar]
- 34.Ota A., Tagawa H., Karnan S., Tsuzuki S., Karpas A., Kira S., Yoshida Y., Seto M. (2004) Cancer Res. 64, 3087–3095 [DOI] [PubMed] [Google Scholar]
- 35.O'Donnell K. A., Wentzel E. A., Zeller K. I., Dang C. V., Mendell J. T. (2005) Nature 435, 839–843 [DOI] [PubMed] [Google Scholar]
- 36.He L., Thomson J. M., Hemann M. T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S. W., Hannon G. J., Hammond S. M. (2005) Nature 435, 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dews M., Homayouni A., Yu D., Murphy D., Sevignani C., Wentzel E., Furth E. E., Lee W. M., Enders G. H., Mendell J. T., Thomas-Tikhonenko A. (2006) Nat. Genet. 38, 1060–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hossain A., Kuo M. T., Saunders G. F. (2006) Mol. Cell. Biol. 26, 8191–8201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu Y., Thomson J. M., Wong H. Y., Hammond S. M., Hogan B. L. (2007) Dev. Biol. 310, 442–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sylvestre Y., De Guire V., Querido E., Mukhopadhyay U. K., Bourdeau V., Major F., Ferbeyre G., Chartrand P. (2007) J. Biol. Chem. 282, 2135–2143 [DOI] [PubMed] [Google Scholar]
- 41.Hayflick L. (1975) Fed. Proc. 34, 9–13 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.