Abstract
The γ-glutamyl carboxylase utilizes four substrates to catalyze carboxylation of certain glutamic acid residues in vitamin K-dependent proteins. How the enzyme brings the substrates together to promote catalysis is an important question in understanding the structure and function of this enzyme. The propeptide is the primary binding site of the vitamin K-dependent proteins to carboxylase. It is also an effector of carboxylase activity. We tested the hypothesis that binding of substrates causes changes to the carboxylase and in turn to the substrate-enzyme interactions. In addition we investigated how the sequences of the propeptides affected the substrate-enzyme interaction. To study these questions we employed fluorescently labeled propeptides to measure affinity for the carboxylase. We also measured the ability of several propeptides to increase carboxylase catalytic activity. Finally we determined the effect of substrates: vitamin K hydroquinone, the pentapeptide FLEEL, and NaHCO3, on the stability of the propeptide-carboxylase complexes. We found a wide variation in the propeptide affinities for carboxylase. In contrast, the propeptides tested had similar effects on carboxylase catalytic activity. FLEEL and vitamin K hydroquinone both stabilized the propeptide-carboxylase complex. The two together had a greater effect than either alone. We conclude that the effect of propeptide and substrates on carboxylase controls the order of substrate binding in such a way as to ensure efficient, specific carboxylation.
Keywords: Allosteric Regulation, Carboxylation, Enzyme Mechanisms, Protein Carboxylation, Vitamin K
Introduction
The vitamin K-dependent γ-glutamyl carboxylase is an endoplasmic reticulum integral membrane enzyme. It post-translationally modifies certain proteins important in several physiologically important areas, including blood coagulation. The modification involves the addition of a carboxyl group to the γ-carbon of glutamic acid residues in the amino portion of the vitamin K-dependent substrate.
In addition to the glutamic acid and CO2, which provides the added carboxyl group, other substrates required for the reaction are vitamin K hydroquinone (KH2)4 and oxygen. During carboxylation KH2 is converted to the epoxide; thus carboxylase is also an epoxidase.
The primary interaction between carboxylase and most of its protein substrates is through the propeptides of the substrates. Binding is of high affinity, which allows the substrate to bind long enough for multiple carboxylations to occur.
In addition to being the primary binding site for carboxylase, the propeptide also exerts an effect on carboxylase catalysis. Factors IX and X and prothrombin propeptides, when bound to carboxylase, increase the activity of the enzyme toward small glutamate-containing substrates (1–5).
For the most part these results suggest that the propeptides induce a conformational change or stabilize a more active conformation in the carboxylase active site. The efficiency of the change appears to be similar for the propeptides of prothrombin and factors IX and X. The fact that these propeptides have different sequences implies that the conserved residues control both binding and allosterism. However, even deleting the Phe-16 residue, which is important for binding, only decreases affinity but not stimulation of activity (1).
Thermodynamic arguments indicate that if the propeptide effect has to do with substrate binding, then substrate binding should affect propeptide binding. Presnell et al. (6) demonstrated that under catalytic conditions, koff for the propeptide of factor IX was slower than in the absence of catalysis and that the Kd for that peptide was lower under catalytic conditions. Lin et al. (7) showed that Glu-containing peptides stabilize the propeptide-carboxylase complex. There are other reports that seem to imply that other substrate binding events affect the carboxylase conformation and its activity (4, 8–10).
In the present study we have extended the investigation of the effect of propeptide and substrates on carboxylase function, including measuring the Kd of several propeptides in the absence of substrates, determining the effect of the substrates (KH2 and FLEEL) on propeptide dissociation from carboxylase and the effect of various propeptides with widely varying affinities for carboxylase on FLEEL activity and kinetics. Our goal was to determine the following: 1) Is the wide variation of carboxylase affinity of the propeptides similar in the absence of catalysis as it has been shown to be during catalysis? 2) Is stabilization of the propeptide-carboxylase complex by FLEEL a general phenomenon among propeptides, and do substrates other than FLEEL affect the stability of the complexes alone or in combination? 3) Is the substrate effect on the propeptide-carboxylase complex similar for all propeptides? 4) Is the effect on small substrate carboxylation similar for all propeptides as suggested for the propeptides of factors IX and X and prothrombin? 5) Are there amino acid residues necessary for these functions other than those already recognized as important for binding to carboxylase, and finally, how do these effects contribute to enzyme specificity and/or efficiency?
EXPERIMENTAL PROCEDURES
Materials
All of the chemicals used were reagent grade. The pentapeptide substrate, FLEEL, was purchased from Bachem (Philadelphia, PA). Propeptides labeled with 5,6-carboxyfluorescein on the amino terminus, based upon the sequences of the human, mouse, and pufferfish propeptides, were synthesized by Mimotopes (Clayton, Victoria, Australia). All of the peptides were confirmed to be at least 95% pure using high performance liquid chromatography and mass spectrometry analysis. Labeling of the peptides with 5,6-carboxyfluorescein did not have an effect on the propeptide-carboxylase interaction (6). 1,2-Dioleoyl-sn-glycero-3-phosphocholine was purchased from Avanti Polar Lipids (Alabaster, AL). Vitamin K1 was purchased from Abbott Laboratories (Chicago, IL) and was reduced to KH2 as described previously (11). Alkamuls EL-620 was graciously donated by Rhodia (Cranbury, NJ). NaH14CO3 was from ICN pharmaceuticals (Costa Mesa, CA).
Carboxylase was produced using Sf9 insect cells and standard molecular biology methods and purified as described (12). The concentration of the active enzyme was determined as described previously (6). All of the experiments were performed at 4 °C unless described otherwise.
Data Analyses
Kinetic and dissociation constants were estimated using nonlinear least squares analyses of multiple data sets in SigmaPlot version 9.0 (Systat Software, Inc.). The Kd values for the various propeptides were estimated using the numerical integration software Dynafit (BioKin, Ltd.) (13). We also modeled the propeptide dissociation data mechanistically using Dynafit.
Fluorescence Anisotropy Measurements
The studies to determine propeptide Kd and koff values were performed essentially as described previously (6) with excitation at 490 nm and emission measured using 515-nm cut-off filters. The only difference was that we used an SLM 8000 spectrofluorometer modified by the OLIS Corporation (Athens, GA). Anisotropy values were converted to concentration of bound propeptide or fraction bound as described (6). This method of analysis weights the anisotropies accounting for the intensity differences between the free and bound propeptide. In all cases, unless otherwise stated, references to propeptides are to those that are fluorescein-labeled.
Determination of the Kd Values of Propeptides
All of the samples were prepared in the following buffer: 100 mm MOPS, pH 7.5, 180 mm NaCl, 3.3% glycerol, 6.3 mm DTT, 66 μm EDTA, 0.1% 1,2 dioleoyl-sn-glycero-3-phosphocholine, 0.28% CHAPS, and 0.4% BSA. Propeptides (4 nm) were incubated with various concentrations of carboxylase (2 h), and the anisotropy was determined.
We also determined the Kd values for human factors X and IX and prothrombin in the presence of 2.4 mm FLEEL. The FLEEL was added first to the carboxylase and then the propeptide.
Determination of Dissociation Rate Constants of Propeptides from Carboxylase
We measured the dissociation of the propeptides of prothrombin, protein C, and factor IX from carboxylase. Fluorescence experiments were performed in the following buffer: 50 mm MOPS, pH 7.5, 500 mm NaCl, 5% glycerol, 6.3 mm DTT, 66 μm EDTA, 0.16% PC, 0.16% CHAPS, and 0.2% BSA. The propeptide and enzyme (20 nm fluorescein-labeled propeptide and 100 nm carboxylase) were incubated for 1 h. To initiate dissociation of the propeptide, we added 4 μm unlabeled factor IX propeptide. Anisotropy measurements were taken every 2 s.
To measure the effect of FLEEL, NaHCO3, and/or KH2 on koff, we incubated them either together or in various combinations with the carboxylase and propeptide prior to the start of the experiment. Final concentrations were 4.8 mm FLEEL, 160 μm KH2, and 1.4 mm NaHCO3. We measured dissociation as described above.
Because KH2 is hydrophobic, it is dissolved in a viscous emulsifier, Alkamuls EL-620. Alkamuls alone had a small effect on koff, so we included it in all of the dissociation experiments. The final concentrations were 0.50 mg/ml Alkamuls EL-620, 0.27 mg/ml dextrose, and 0.066 mg/ml benzyl alcohol, identical to the concentrations in experiments with KH2.
Measuring the Effect of Various KH2 Concentrations on Propeptide Dissociation from Carboxylase
We measured the dissociation of the propeptide of factor IX from wild-type carboxylase at various KH2 concentrations. The experiments were performed as described above for measuring koff.
Measurement of the Effect of Propeptides on FLEEL Carboxylation
We measured the effect of 10 propeptides on the rate of FLEEL carboxylation. Activity was determined using 14CO2 incorporation into FLEEL as described (14) in the presence of 80 μm propeptide. We also determined the Km and kcat values of carboxylase for FLEEL in the presence of human factor IX, protein C, and prothrombin propeptides (80 μm). At this high concentration of peptides, essentially all of the carboxylase will be in complex with the propeptide.
RESULTS
Kd Values of Various Carboxylase-Propeptide Complexes
The carboxylase Kd values for the 11 human (plus consensus), three mouse, and two pufferfish propeptides are summarized in Figs. 1 and 2. For comparison, our Kd values are 4–7-fold lower than the previously reported Ki values for several of the human propeptides (12). However, the relationship among the Kd and Ki values are similar. In other words, as with the Ki values, the Kd of factor X was less than that of factor IX, which was less than that of prothrombin.
FIGURE 1.
Sequences of human vitamin K-dependent protein propeptides and their relative Kd values for carboxylase. Varying concentrations of carboxylase were incubated with 4 nm of the fluorescein-labeled propeptide for 2 h at 4 °C in 100 mm MOPS, pH 7.5, 180 mm NaCl, 3.3% glycerol, 6.3 mm DTT, 66 μm EDTA, 0.1% PC, 0.28% CHAPS, and 0.4% BSA in a final volume of 300 μl. The resulting fluorescence anisotropies were measured. The fraction of bound propeptide at different carboxylase concentrations and the Kd were determined as described under “Experimental Procedures.” The values shown are from three determinations.
FIGURE 2.
Comparison of the sequences and their Kd values of human factor IX, factor X, and prothrombin propeptides for carboxylase with those of mouse and pufferfish propeptides. 4 nm of the fluorescein-labeled human, mouse, or pufferfish propeptide was incubated with varying concentrations of carboxylase for 2 h at 4 °C in 100 mm MOPS, pH 7.5, 180 mm NaCl, 3.3% glycerol, 6.3 mm DTT, 66 μm EDTA, 0.1% PC, 0.28% CHAPS, and 0.4% BSA in a final volume of 300 μl. The resulting fluorescence anisotropies were measured. The fraction of bound propeptide at different carboxylase concentrations and the Kd were determined as described under “Experimental Procedures.” The values shown are from three determinations.
The difference in the Kd values versus the Ki values may be due to several factors involved with different methods and conditions. For example, the Ki values were determined under catalytic conditions and at 20 °C versus 4 °C, different buffers, and competition versus direct binding.
In Fig. 2 the propeptides of mouse and pufferfish are compared with their human counterparts. The most striking differences are between mouse and pufferfish factor X and mouse and human prothrombin.
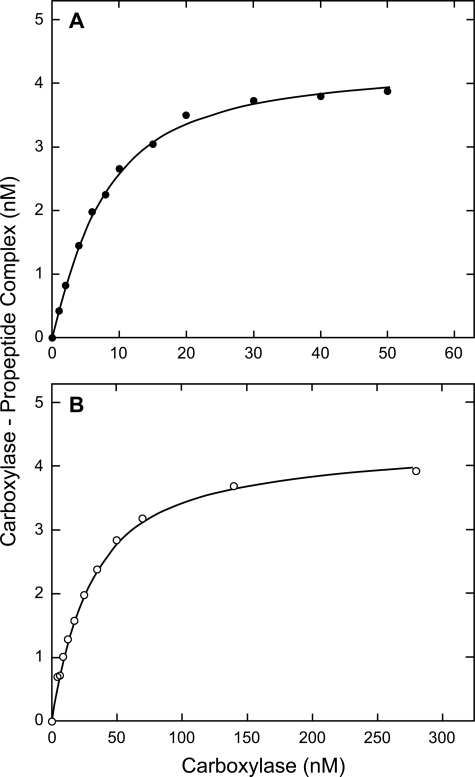
Results of single binding experiments with factor IX and prothrombin propeptides are shown (Fig. 3). The analysis with Dynafit (see “Experimental Procedures”) estimated the Kd and the propeptide concentration; we fixed propeptide-carboxylase stoichiometry at 1. Analysis of the shown factor IX data yielded a Kd of 4.95 nm and a propeptide concentration of 4.37 nm. The intended propeptide concentration was 4 nm. The shown prothrombin data yielded a Kd of 26.3 nm and a propeptide concentration of 4.35 nm.
FIGURE 3.
Data for carboxylase binding to factor IX and prothrombin propeptides. The experiments were performed, and the data were analyzed as described in the legend to Fig. 1 and under “Experimental Procedures.” The data for factor IX and prothrombin are shown in A and B, respectively.
Next, to expand on our previous studies on the propeptide of factor IX, we examined the effect of glutamate substrates on the carboxylase Kd values of the factor X and prothrombin propeptides, in addition to that of factor IX (data not shown). As with the propeptide of factor IX, the other propeptides had lower Kd values in the presence of FLEEL.
The Effect of Substrates on the Dissociation Rate Constants of Propeptides from Carboxylase
In previous studies from this laboratory, the results showed that under catalytic conditions or with FLEEL, the koff for the propeptide of factor IX was slower than when the propeptide was bound to carboxylase in the absence of substrates (6, 7). To determine whether this is a general phenomenon, we determined the koff values for the propeptides of protein C and prothrombin as well as that of factor IX in the presence and absence of FLEEL.
In all cases the data described below were normalized to one for maximum bound propeptide. The actual maximum bound propeptide varied from ∼80% for the propeptide of prothrombin in the absence of substrate to increasing values for the higher affinity situations with substrates and for factor IX and protein C propeptides.
In the absence of substrates, the residuals for a single exponential fit of the fraction propeptide bound versus time indicated a systematic error in that fit (data not shown). The deviation from the fit was similar for all three propeptides. The data fit well to a double exponential equation. This result suggested that there may be at least two conformational forms of carboxylase, each with different affinity for a propeptide.
We previously showed that a slow loss of carboxylase activity occurs at 20 °C (6). In our present study (results not shown), we found that the biphasic curves were not caused by an active and inactive form of carboxylase. On incubation at 20 °C for up to 6 h, the amount of propeptide bound gradually decreased. However, inactivation did not affect the rate of dissociation of the propeptide of prothrombin nor the biphasic nature of the data.
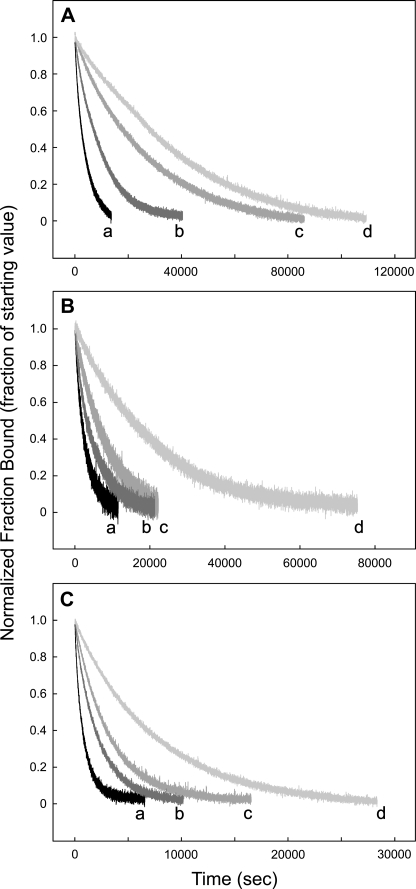
With FLEEL the dissociation data fit a single exponential for all three propeptides. The koff values for the three propeptides were all slower than in the absence of FLEEL (Fig. 4 and Table 1).
FIGURE 4.
Comparison of the off rates for human factor IX, protein C, and prothrombin propeptides from carboxylase. 20 nm fluorescein-labeled propeptide was incubated with 100 nm carboxylase for 1 h at 4 °C in 50 mm MOPS, pH 7.5, 500 mm NaCl, 5% glycerol, 6.3 mm DTT, 66 μm EDTA, 0.16% PC, 0.16% CHAPS, and 0.2% BSA in a final volume of 300 μl. The off rate of the labeled-propeptide was measured by adding 4 μm of the unlabeled propeptide and measuring the time-dependent decrease in fluorescence anisotropy. The anisotropy values were converted to the fraction of bound labeled propeptide, and the resulting data were fitted to a single exponential decay as given under “Experimental Procedures.” The off rates for factor IX, protein C, and prothrombin propeptides are shown in A, B, and C, respectively. For each propeptide, the off rates were measured in the presence of Alkamuls EL-620 (lines a), Alkamuls EL-620 and 4.8 mm FLEEL (lines b), 160 μm vitamin K hydroquinone and 1.4 mm sodium bicarbonate (lines c), or 4.8 mm FLEEL, 160 μm vitamin K hydroquinone and 1.4 mm sodium bicarbonate (lines d), as shown.
TABLE 1.
Comparison of the koff values for the dissociation of factor IX, protein C, and prothrombin propeptides from wild-type carboxylase in the presence of KH2 and/or FLEEL
| Addition(s) |
koff × 104a |
||
|---|---|---|---|
| Factor IX | Protein C | Prothrombin | |
| s−1 | |||
| Noneb | 2.5 (1) | 3.7 (1) | 12 (1) |
| KH2 | 1.0 (0.40) | 2.1 (0.57) | 4.8 (0.40) |
| FLEELb | 0.36 (0.14) | 1.1 (0.30) | 3.2 (0.27) |
| KH2 + FLEEL | 0.24 (0.10) | 0.52 (0.14) | 1.3 (0.11) |
a The values within parentheses denote the ratios of koff in the presence of KH2 and/or FLEEL to koff for propeptide alone.
b Alkamuls EL-620 was added to samples with no KH2.
KH2 with NaHCO3 decreased the koff, and in all cases the data fit a single exponential equation (Fig. 4). To determine whether both KH2 and NaHCO3 were necessary for the change in the koff, we performed additional experiments in the presence of either KH2 or NaHCO3 with the propeptide of protein C. The results (results not shown) indicate that NaHCO3 alone has no effect on koff.
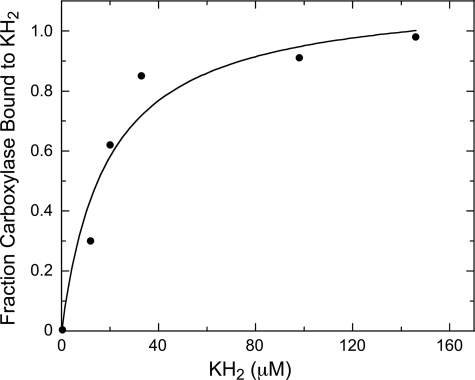
We also used this technique to measure the previously unknown affinity of vitamin K for carboxylase. The change in koff for the carboxylase-propeptide dissociation with increasing KH2 was what one would expect from saturable binding (Fig. 5). The apparent Kd of 19 μm is similar to the reported Km for KH2 (9, 11, 15).
FIGURE 5.
Relative fraction of wild-type carboxylase bound to KH2 at varying KH2 concentrations. 20 nm fluorescein-labeled human factor IX propeptide in 50 mm MOPS, pH 7.5, 500 mm NaCl, 5% glycerol, 6.3 mm DTT, 66 μm EDTA, 0.16% PC, 0.16% CHAPS, and 0.2% BSA was incubated with 100 nm wild-type carboxylase and varying concentrations of KH2 at 4 °C and in a final volume of 329 μl, until binding equilibrium was reached. The off rate of the labeled propeptide from carboxylase at each KH2 concentration was determined by adding unlabeled factor IX propeptide to 3.5 μm and measuring the time-dependent decrease in fluorescence anisotropy. Alkamuls EL-620 was included in the blank with no KH2.
Effect of Propeptides on FLEEL Carboxylation
Next we investigated whether there is a correlation between FLEEL carboxylation rate and the affinity or sequence of the propeptide used to stimulate that carboxylase activity. The activities are similar but do vary (Table 2). However, the rates do not correlate with the propeptide affinity. Although the two with the highest activity are consensus and factor X, which have the lowest Kd values, the next highest stimulation occurs with protein C and prothrombin. The latter two have the highest Kd values.
TABLE 2.
Quantity of carboxylated FLEEL produced during the in vitro carboxylation assay with different propeptides
| Propeptide | Kda | 14CO2 incorporated in FLEEL |
|---|---|---|
| nm | nmol/30 min | |
| Consensus | 0.08 | 0.238 |
| Factor X | 0.59 | 0.236 |
| Matrix Gla protein | 0.98 | 0.195 |
| Protein Z | 1.80 | 0.198 |
| Factor VII | 2.34 | 0.198 |
| PRGP1 | 2.28 | 0.211 |
| Protein S | 2.28 | 0.158 |
| Factor IX | 4.97 | 0.185 |
| Protein C | 18.6 | 0.234 |
| Prothrombin | 25.8 | 0.225 |
a The values are from Fig. 1.
To further test this, we determined the Km and kcat for FLEEL in the presence of high concentrations of factor IX, protein C, and prothrombin propeptides (Table 3). These results are consistent with the previous experiment in that the kinetics are similar despite a range of Kd values from 5 to 25 nm. We measured the rates and kinetic constants at propeptide concentrations (>3000 times Kd) ensuring that almost all carboxylase is bound to each propeptide.
TABLE 3.
Km and kcat values for FLEEL carboxylation by wild-type carboxylase in the presence of factor IX, protein C, or prothrombin propeptides
| Propeptide | Kma | kcata |
|---|---|---|
| mm | min−1 | |
| Factor IX | 0.29 ± 0.01 | 30 ± 1 |
| Protein C | 0.36 ± 0.03 | 26 ± 2 |
| Prothrombin | 0.31 ± 0.03 | 33 ± 2 |
a The values are from two determinations.
DISCUSSION
Our goal in the present study was to further understanding of the structural and functional interactions of carboxylase with its substrates. All indications are that the propeptides of vitamin K-dependent protein precursors provide most of the energy for their binding to the carboxylase. It is also likely that the propeptide of such a protein constitutes the first site to interact with the enzyme. As the first binding event it most likely sets the stage for binding of the other substrates. In addition, the turnover rate during carboxylation of peptide substrates depends to a certain extent on the propeptide affinity for the carboxylase (7). For these reasons we felt that the carboxylase-bound propeptide would be a good model for the enzyme vitamin K-dependent protein precursor complex. Our idea was to characterize the carboxylase-propeptide interaction without substrates and then study the effect of substrates on the stability of the complex.
As we expected, based on results from previous work, we found the propeptides exhibit a broad range of affinities (12). All have what one would consider a high affinity for the carboxylase, that is, a Kd in the nanomolar range. This is an especially high affinity for a reaction occurring in the endoplasmic reticulum where one would expect high local substrate concentrations. The propeptide residues identified as important earlier (16–18) (residues −6, −9, −10, −15, and −16) tend to be conserved among the propeptides or at least have conservative substitutions. Otherwise there is wide variation in the character of most residues.
In contrast to the variation in affinities, the propeptides we tested all stimulated FLEEL carboxylation to a similar extent. Even for the differences in stimulation there is no correlation between stimulatory effect and propeptide affinity. As with affinity, we were unable to identify the patterns of primary structure necessary for the effect on FLEEL. These results agree with observations regarding the prothrombin and factor X (wild-type and truncated) as well as factor IX propeptides (1–5). The results regarding the relationship between propeptide sequence, affinity for carboxylase, and the ability to stimulate carboxylase activity indicate that binding requires relatively few conserved residues, and once any propeptide binds, the active carboxylase conformations are all similar.
As we discussed in our previous work, at least part of the increase in carboxylase catalytic activity is caused by increased FLEEL binding (19). Because propeptide binding affects FLEEL, FLEEL binding must affect propeptide binding. In fact earlier studies had shown that FLEEL or catalytic conditions slowed the factor IX propeptide dissociation from carboxylase (6, 7).
Our present work extends these earlier studies to investigate the effect of individual substrates on the propeptide-carboxylase complex. In the absence of substrates, the dissociation data did not fit a single exponential equation. On the other hand, when we added substrate(s) to the propeptide-carboxylase mixture, a single exponential equation adequately fit the dissociation data. KH2 and FLEEL decreased the koff for the prothrombin and protein C propeptides as well as for that of factor IX. The effect of both together was greater than either substrate alone. These results and those from the earlier study suggest that substrate binding to the enzyme-propeptide complex causes a shift in equilibrium to a conformation that has higher affinity for the propeptide, and as a result, higher catalytic activity (20).
To determine whether a multiple conformation model fits our results, we analyzed our data using the software Dynafit (13). Dynafit allows testing the fit of data to a given mechanism(s). Our dissociation data fit a mechanism that includes two forms of carboxylase and a mechanism in which two propeptides bind to one carboxylase. Although an earlier report suggested that carboxylase might bind two propeptide molecules (21), Presnell et al. (6) showed that the carboxylase binds only one propeptide. Therefore we conclude that a model in which at least two forms of carboxylase exist, which have different affinities for propeptide and presumably propeptide-containing substrates, best describes this interaction. Comparing our results to those of earlier studies puts our results into the context of how substrate binding affects carboxylase activity and specificity and may put the earlier studies into the context of how substrate binding affects carboxylase structure.
Li et al. (4) showed that epoxidation and carboxylation can uncouple. In other words, when all substrates other than CO2 are present, epoxidation of KH2 occurs without carboxylation. This indicates that if all other substrates are present, the CO2 is not needed until the last step in carboxylation: addition to the γ-carbanion of glutamate. It is consistent with the prediction of Uotila (22), who concludes from kinetic results that CO2 was likely the last substrate to react. This also fits logically with our results that indicate bicarbonate does not affect the koff of propeptide from the carboxylase.
Sugiura et al. (10) showed that carboxylase does not catalyze epoxidation of KH2 in the absence of propeptide or FLEEL. The reaction is fastest, ∼10-fold faster than with any single component, with both FLEEL and propeptide. FLEEL alone is more effective than propeptide alone. As the authors suggest, this indicates that binding of both or either causes a conformational change that increases the epoxidase activity and/or KH2 binding.
The experiments of Bouchard et al. (8) showed that N-ethylmaleimide inhibited carboxylase. Glu-containing substrates, such as FLEEL or a peptide containing the propeptide of prothrombin and the first 10 residues of its Gla domain, increased the rate of inactivation, but propeptide alone did not. They also report that KH2 protects from inactivation only when a Glu-containing substrate is present. These results are in some ways hard to interpret because N-ethylmaleimide is expected to modify Cys residues. It now appears that Cys residues are not directly involved in carboxylase catalysis, as was previously thought. However, these studies are consistent with our hypothesis that substrate binding supports multiple conformations of the carboxylase.
A more recent work by Rishavy et al. (23) implicates two lysine residues (Lys-217 and Lys-218), not cysteines, as important in catalysis. Quantum chemical calculations are consistent with this model, but they also support a second possible mechanism for removal of the KH2 proton involving only one lysine (24). Because both models are similar for our purposes, we will focus on the two-lysine model in this discussion.
Rishavy et al. (23) propose that Lys-218, in combination with Lys-217, is involved in deprotonating KH2. According to this model, in the absence of substrates Lys-218(0) is not charged, but Lys-217(+1) is, perhaps because of the proximity of the two lysines to one another. They further suggest that when the Glu substrate approaches the lysine pair, it interacts with Lys-217(+1), thus allowing Lys-218(0) to acquire a proton. Assuming the KH2 is nearby, Lys-218(0) can then remove the hydroxyl proton that starts the pathway to the vitamin K alkoxide, the strong base necessary for carbanion formation, and on to carboxylation of the Glu residue.
We interpret our results presented here and those of others described above to support a mechanism that might occur as follows. First, the vitamin K-dependent protein binds to carboxylase through its propeptide. As mentioned above the two lysines (Lys-217 and Lys-218) of carboxylase involved in catalysis are adjacent to one another, allowing one residue to remain uncharged (Lys-218(0)). Therefore, when substrate binds, the glutamate(s) to be carboxylated must not be near Lys-217 and Lys-218. If the glutamate were in proximity to the lysines at this point, then the glutamate with a negative charge and the lysine with a positive charge (Lys-217(+1)) could interact. If that occurred in the absence of KH2, then the free lysine, Lys-218(0), could acquire a proton from solvent and thus not be available to remove the KH2 proton necessary for strong base production. Therefore, to set the stage for glutamate interaction with the active site, propeptide binding must first cause the conformational change that promotes KH2 binding near the lysines. Next KH2 binding supports a conformation that brings the glutamate substrate in proximity to Lys-217(+1) and Lys-218(0). Now through its negative charge, the glutamate residue binds Lys-217(+1), freeing Lys-218(0), which can then abstract the hydroquinone proton from the nearby KH2. Glutamate binding to Lys-217(+1) is essential for a second reason; the glutamate negative charge must be neutralized to make its γ-proton removal energetically favorable (25). That step is necessary to produce the carbanion precursor of carboxylation. According to Davis et al. (24), after removal of the proton from the hydroquinone group on KH2, the rest of the reactions producing γ-carboxyglutamic acid are relatively energetically favorable.
Acknowledgment
We thank Da-Yun Jin for technical assistance in preparing the recombinant enzyme.
This work was supported, in whole or in part, by National Institutes of Health Grants HL48318 and HL06350 (to D. W. S.).
- KH2
- vitamin K1 hydroquinone
- MOPS
- 3-(N-morpholino)propanesulfonic acid
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- PC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine.
REFERENCES
- 1.Knobloch J. E., Suttie J. W. (1987) J. Biol. Chem. 262, 15334–15337 [PubMed] [Google Scholar]
- 2.Cheung A., Engelke J. A., Sanders C., Suttie J. W. (1989) Arch. Biochem. Biophys. 274, 574–581 [DOI] [PubMed] [Google Scholar]
- 3.Cheung A., Suttie J. W., Bernatowicz M. (1990) Biochim. Biophys. Acta 1039, 90–93 [DOI] [PubMed] [Google Scholar]
- 4.Li S., Furie B. C., Furie B., Walsh C. T. (1997) Biochemistry 36, 6384–6390 [DOI] [PubMed] [Google Scholar]
- 5.Ulrich M. M., Furie B., Jacobs M. R., Vermeer C., Furie B. C. (1988) J. Biol. Chem. 263, 9697–9702 [PubMed] [Google Scholar]
- 6.Presnell S. R., Tripathy A., Lentz B. R., Jin D. Y., Stafford D. W. (2001) Biochemistry 40, 11723–11733 [DOI] [PubMed] [Google Scholar]
- 7.Lin P. J., Straight D. L., Stafford D. W. (2004) J. Biol. Chem. 279, 6560–6566 [DOI] [PubMed] [Google Scholar]
- 8.Bouchard B. A., Furie B., Furie B. C. (1999) Biochemistry 38, 9517–9523 [DOI] [PubMed] [Google Scholar]
- 9.Soute B. A., Ulrich M. M., Watson A. D., Maddison J. E., Ebberink R. H., Vermeer C. (1992) Thromb. Haemost. 68, 521–525 [PubMed] [Google Scholar]
- 10.Sugiura I., Furie B., Walsh C. T., Furie B. C. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 9069–9074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morris D. P., Soute B. A., Vermeer C., Stafford D. W. (1993) J. Biol. Chem. 268, 8735–8742 [PubMed] [Google Scholar]
- 12.Stanley T. B., Jin D. Y., Lin P. J., Stafford D. W. (1999) J. Biol. Chem. 274, 16940–16944 [DOI] [PubMed] [Google Scholar]
- 13.Kuzmic P. (1996) Anal. Biochem. 237, 260–273 [DOI] [PubMed] [Google Scholar]
- 14.Wu S. M., Soute B. A., Vermeer C., Stafford D. W. (1990) J. Biol. Chem. 265, 13124–13129 [PubMed] [Google Scholar]
- 15.Sugiura I., Furie B., Walsh C. T., Furie B. C. (1996) J. Biol. Chem. 271, 17837–17844 [DOI] [PubMed] [Google Scholar]
- 16.Chu K., Wu S. M., Stanley T., Stafford D. W., High K. A. (1996) J. Clin. Investig. 98, 1619–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oldenburg J., Quenzel E. M., Harbrecht U., Fregin A., Kress W., Müller C. R., Hertfelder H. J., Schwaab R., Brackmann H. H., Hanfland P. (1997) Br. J. Haematol. 98, 240–244 [DOI] [PubMed] [Google Scholar]
- 18.Stanley T. B., Humphries J., High K. A., Stafford D. W. (1999) Biochemistry 38, 15681–15687 [DOI] [PubMed] [Google Scholar]
- 19.Mutucumarana V. P., Acher F., Straight D. L., Jin D. Y., Stafford D. W. (2003) J. Biol. Chem. 278, 46488–46493 [DOI] [PubMed] [Google Scholar]
- 20.Rishavy M. A., Berkner K. L. (2008) Biochemistry 47, 9836–9846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hallgren K. W., Hommema E. L., McNally B. A., Berkner K. L. (2002) Biochemistry 41, 15045–15055 [DOI] [PubMed] [Google Scholar]
- 22.Uotila L. (1988) Arch. Biochem. Biophys. 264, 135–143 [DOI] [PubMed] [Google Scholar]
- 23.Rishavy M. A., Hallgren K. W., Yakubenko A. V., Shtofman R. L., Runge K. W., Berkner K. L. (2006) Biochemistry 45, 13239–13248 [DOI] [PubMed] [Google Scholar]
- 24.Davis C. H., Deerfield D., 2nd, Wymore T., Stafford D. W., Pedersen L. G. (2007) J. Mol. Graph. Model. 26, 409–414 [DOI] [PubMed] [Google Scholar]
- 25.Davis C. H., Deerfield D., 2nd, Stafford D. W., Pedersen L. G. (2007) J. Phys. Chem. 111, 7257–7261 [DOI] [PubMed] [Google Scholar]