Abstract
S100A4, a member of the S100 family of proteins, plays an important role in matrix remodeling by up-regulating the expression of matrix metalloproteinases (MMPs). We have previously shown that S100A4 is overexpressed in diseased cartilage and that extracellular S100A4 stimulates MMP-13 production, a major type II collagen-degrading enzyme, via activation of receptor for advanced glycation end product signaling. In the present study, using human articular chondrocytes, we show that intracellular S100A4 translocated into the nucleus upon interleukin-1β (IL-1β) stimulation and translocation required post-translational modification of S100A4 by the sumo-1 protein. Two sumoylation sites were identified on the S100A4 molecule, Lys22 and Lys96. Mutation of these lysine residues abolished the ability of S100A4 to be sumoylated and to translocate into the nucleus. Blocking of sumoylation and nuclear transport of S100A4 inhibited the IL-1β-induced production of MMP-13. Nuclear S100A4 was bound to the promoter region of MMP-13 in IL-1β-treated cells. Thus, we demonstrate a novel mechanism for sumoylated S100A4 as a regulator of expression of the MMP-13 gene.
Keywords: Chromatin Immunoprecipitation (ChiP), Interleukin, Matrix Metalloproteinase, Nuclear Translocation, Sumoylation, Cartilage, S100A4 Protein
Introduction
S100 proteins are low molecular weight, acidic calcium-binding proteins characterized by the presence of two EF-hand calcium binding domains (1). Members of this family have both extracellular and intracellular functions and play an important role in protein phosphorylation, enzyme activation, cell motility, cellular growth and differentiation, and calcium homeostasis (1). S100 proteins are expressed only in vertebrates and are distributed in various tissues in a cell-specific manner (2). Altered expression of S100 proteins has been linked to several human diseases, including cancer and neurodegenerative and inflammatory disorders (3, 4). Recently, we and others (5–8) have shown that S100 proteins are expressed in cartilage and their expression is up-regulated in rheumatoid arthritis and osteoarthritis.
S100A4 was originally isolated as a gene that was differentially expressed in mouse adenocarcinoma cells (9) and subsequently found in other tissues (10, 11). S100A4, like a typical member of the S100 family, exerts its function via protein-protein interactions. S100A4 interacts with F-actin, tropomyosin, and with the heavy chain of nonmuscle myosin II, and plays an active role in cell motility and adhesion in metastatic tumor cells (12). S100A4 also binds to the p53 tumor suppressor and modulates its DNA binding capacity, thereby regulating its function (13). Extracellular S100A4 interacts with cell surface receptors and stimulates neurite outgrowth in astrocytes (14) and angiogenesis in endothelial cells (15).
Importantly, studies have found that S100A4 plays a major role in matrix remodeling by regulating the expression of matrix metalloproteinases (MMPs)2 (16, 17). We have previously shown that extracellular S100A4 stimulates increased production of MMP-13 via its interaction with receptor for advanced glycation end products (RAGE) in chondrocytes (6). MMPs are a large family of structurally related calcium- and zinc-dependent proteolytic enzymes that are involved in the degradation of many different components of the extracellular matrix and play a key role in diverse cellular processes ranging from morphogenesis to tumor invasion to tissue remodeling (18). Given their important role in cellular functions, the expression and activity of MMPs are tightly regulated at multiple levels of gene transcription, synthesis, and extracellular activity.
Interleukin-1β (IL-1β), a major proinflammatory cytokine, promotes expression of MMP-13 in articular chondrocytes (19). However, the underlying mechanism of IL-1β-mediated regulation of MMP-13 is not clearly understood. MMP-13 is one of the major proteases that degrade type II collagen and is overexpressed in chondrocytes isolated from osteoarthritic cartilage tissue (20, 21). Thus, elucidating how IL-1β activates MMP-13 gene expression in chondrocytes would be critical for understanding the pathogenesis of cartilage destruction in arthritis. In the current study, we demonstrate that treatment of chondrocytes with IL-1β promotes nuclear translocation of S100A4, which required the post-translational modification of sumoylation. More importantly, nuclear S100A4 was bound to the chromatin region that corresponds to the promoter region of the MMP-13 gene, suggesting a transcriptional regulatory role for S100A4.
EXPERIMENTAL PROCEDURES
Reagents
Recombinant human IL-1β and an enzyme-linked immunosorbent assay (ELISA) kit for human pro-MMP-13 were purchased from R&D Systems (Minneapolis, MN). Human S100A4 antibody was purchased from Dako (Carpinteria, CA). Sumo-1 antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-lactose dehydrogenase was from Fitzgerald Industries International (Acton, MA). Pronase and anti-lamin B were purchased from Calbiochem and collagenase P from Roche Applied Science. An RNeasy Mini kit was obtained from Qiagen (Valencia, CA). The sumoylation kit was purchased from Enzo Life Sciences International, Inc. (Plymouth Meeting, PA). A chromatin immunoprecipitation (ChIP) kit was purchased from Upstate Biotechnology, Inc. (Lake Placid, NY). Human recombinant S100A4 was purchased from BioVision (Mountain View, CA). S100A4, MMP-13, and TATA box-binding protein primers and SYBR Green polymerase chain reaction (PCR) Master Mix were from SuperArray Bioscience (Frederick, MD). QuikChange XL Site-directed Mutagenesis Kit was from Stratagene (La Jolla, CA). The NE-PER nuclear and cytoplasmic extraction kit was purchased from Pierce Biotechnology. Alexa 488-conjugated secondary antibody, DAPI, and Live/Dead cell survival assay kit were purchased from Molecular Probes (Eugene, OR). Nitrocellulose membranes and enhanced chemiluminescence detection kits were purchased from Amersham Biosciences. Cell culture medium and supplements were purchased from Invitrogen.
Chondrocyte Isolation and Culture Conditions
Human ankle cartilage was obtained from tissue donors within 48 h of death through the National Disease Research Interchange (Philadelphia, PA) in accordance with institutional protocols. Only tissue from donors without a history of known arthritis was used. The tissue was graded on a scale of 0–4 for evidence of morphological changes as previously described (22) and all tissue for this study was either grade 0 or 1. Tissue from a total of 60 donors with ages ranging from 40 to 90 years was used. Cells from at least three independent donors were used in each experiment.
Chondrocytes were isolated under aseptic conditions by sequential enzymatic digestion at 37 °C using 2 mg/ml of Pronase in serum-free DMEM/F-12/antibiotics for 1 h followed by overnight digestion with collagenase-P at 0.25 mg/ml in DMEM/F-12 (5% FBS). Viability of isolated cells was determined using trypan blue and cells were counted using a hemocytometer. Monolayer cultures were established by plating cells in 6-well plates at 2 × 106 cells/ml in DMEM/F-12 medium supplemented with 10% FBS. Cells were maintained for ∼5–7 days with feedings every 2 days until they reached 100% confluency prior to experimental use.
Chondrocyte cultures were made serum-free overnight before treating with 10 ng/ml of IL-1β for 16–18 h to measure MMP-13 production, or for 0–60 min to study the nuclear transport of S100A4. After incubation, conditioned medium was collected and analyzed for MMP-13 by ELISA. Cellular contents were separated into cytoplasmic and nuclear fractions as described below. In some experiments, after IL-1β stimulation total RNA was prepared and mRNA expression levels of S100A4 and MMP-13 were analyzed by real time PCR.
Cytoplasmic and Nuclear Extracts
Cytoplasmic and nuclear fractions were prepared by using the NE-PER nuclear and cytoplasmic extraction kit according to the manufacturer's protocol. In brief, the cell pellet was re-suspended in ice-cold reagent and incubated on ice for 10 min. After incubation, the contents of the tube were mixed and centrifuged at 16,000 × g for 5 min, and the supernatant was collected (cytoplasmic fraction). The insoluble fraction, which contains nuclei, was re-suspended in another reagent, incubated on ice for 40 min, centrifuged at 16,000 × g for 10 min, and the supernatant (nuclear fraction) collected. Each of these fractions was then analyzed for purity by immunoblotting for specific protein markers (for cytoplasmic fraction, lactose dehydrogenase; and for nuclear fraction, lamin). Samples containing 10 μg of total protein were separated by SDS-PAGE, transferred to nitrocellulose, and probed for specific proteins. Immunoreactive bands were detected using the ECL system. All immunoblotting experiments were repeated at least 3 times with similar results.
Immunofluorescence/Confocal Microscopy
Chondrocytes (50,000 cells/coverslip) were seeded on glass coverslips for 48–72 h and serum starved for 18–24 h before treatment with IL-1β for 0–120 min. After incubation, cells were washed and fixed in 4% paraformaldehyde for 15 min at 4 °C. Cells were permeabilized in 0.2% Triton X-100 for 5 min and blocked with 10% goat serum. S100A4 was detected with anti-S100A4 and Alexa 488-conjugated secondary antibodies. Images were captured with a Zeiss confocal laser scanning microscope and analyzed by Zeiss AxioVision software. For nuclear staining, we used DAPI.
Constructs and Mutagenesis
pEGFP-S100A4 containing the S100A4 coding region inserted between HindIII and BamHI sites was a gift from Dr. Anne Bresnick (Albert Einstein College of Medicine, Bronx, New York). Lysine 22, 48, and 96 were mutated to arginine using the QuikChange XL Site-directed Mutagenesis Kit and appropriate mutant primers. A double mutant was generated by subsequent mutagenesis. All mutations were confirmed by sequence analysis. Dominant-negative pCDNA-HA-Ubc9 (C93S and C97S) was a gift from Drs. Marc Piechaczyk and Guillaume Bossis (Institute of Molecular Genetics of Montpellier, France).
In Vitro and in Vivo Sumoylation Assay
In vitro sumoylation assays were performed using the sumoylation kit according to the manufacturer's protocol. In brief, 200 nm of purified recombinant human S100A4 protein was incubated with reaction mixture containing 50 mm Tris-HCl (pH 7.4), 2 mm DTT, 5 mm ATP, 10 mm MgCl2, Aos-1 (150 ng), His-Uba2 (400 ng), GST-Ubc9 (500 ng), and GST-SUMO-1 for 1 h at 30 °C. The control reaction was carried out in the absence of ATP. After the incubation period, S100A4 protein sumoylation was identified by immunoblotting using the anti-sumo antibody provided with the kit.
For in vivo assays, chondrocytes were treated with IL-1β for 0–60 min. After the incubation period, the nuclear extracts were prepared as described above. Extracts (250 μg of protein) were immunoprecipitated with anti-sumo-1 antibody followed by immunoblotting with anti-S100A4.
Chromatin Immunoprecipitation Assay (ChIP)
A ChIP assay was performed to assess in vivo DNA-protein interactions at the MMP-13 promoter, using a ChIP assay kit according to the manufacturer's instructions. Briefly, 6 million cells were harvested and fixed in 1% (v/v) formaldehyde in minimal culture medium for 10 min at room temperature. After washing with cold phosphate-buffered saline, cells were lysed in 1% SDS for 30 min on ice. The lysates were sonicated to shear DNA using a Branson 250 sonicator (two 15-s pulses at 40% power in an ice bath, with 1 min between each pulse). These shearing conditions generate DNA fragments ranging in size from 500 to 1,000 bp. Chromatin solution was precleared with protein G-coated magnetic beads for 2 h at 4 °C. Ten microliters of the chromatin solution was reserved as the “input” sample. The remaining chromatin was immunoprecipitated overnight at 4 °C with 3 μg of antibody specific to S100A4, rabbit immunoglobulin G (IgG) as a negative control (provided with the kit), or dimethylated histone H3 lysine 9 (positive control). The chromatin-Ab complexes captured on the beads were washed several times and then eluted in 50 μl of elution buffer. The immunoprecipitated and input sample cross-links were reversed by incubation for 2.5 h at 65 °C. After treatment with proteinase K at for 1 h at 37 °C, the reaction was stopped and the resulting DNA stored at −20 °C until analyzed by standard and real time PCR as described below.
Semiquantitative PCR
PCR analysis was performed in a 50-μl volume containing 5 μl of ChIP DNA, 1 μm of each primer, 2 mm MgCl2, 0.2 μm dNTP, and 0.04 units/μl of AmpliTaq Gold DNA polymerase. The PCR conditions were as follows: 1 cycle at 94 °C for 5 min, 35 cycles at 94, 58, and 72 °C for 30 s each, and a final cycle at 72 °C for 5 min. PCR products were analyzed by agarose gel electrophoresis. The primers used in the PCR were designed to amplify the promoter region of the human MMP-13 gene (primer sets MMP-13 promoter, 5′-GCCAGATGGGTTTTGAGAC-3′ and 5′-GTGATGCCTGGGGACTGTT-3′) as previously used to identify the MMP-13 promoter (23).
Quantitative Real Time PCR
Total RNA was isolated using an RNeasy Mini kit (Qiagen, Valencia, CA). Two micrograms of total RNA was reverse transcribed for 1 h at 42 °C using an avian myeloblastosis virus reverse transcriptase and oligo(dT) primer. Then 2 μl of reverse transcriptase was combined in a reaction mixture with 1 μl of the specific primer pair, 12.5 μl of 2 μl of SYBR Green PCR Master Mix, and water to a final reaction volume of 25 μl. Predesigned primers (SA Bioscience) for S100A4, MMP-13, and TATA box-binding protein were used for the analysis. PCR were then run in triplicate, with 40 cycles of amplification, on an ABI Prism 7000 real time PCR instrument (Applied Biosystems, Foster City, CA). A negative control containing primers, water, and Master Mix, but no complementary DNA, was included. An amplification plot was generated using ABI software. The expression levels of S100A4 and MMP-13 were normalized to the expression of TATA box-binding protein measured in parallel samples.
For ChIP experiments, immunoprecipitated DNA was analyzed using PCR primers specific to the MMP-13 promoter obtained commercially (SA Bioscience) that spans −0.1 kb from the start codon. The PCR (25 μl) contained 5 μl of ChIP DNA, 12.5 μl of 2× TaqMan Universal Master Mix containing DNA polymerase and dNTP, and 100 nm of primer set. The PCR conditions were as follows: 2 min at 50 °C, 10 min at 95 °C, followed by 40 cycles with 15 s at 95 °C and 1 min at 60 °C (combined annealing and extension), using the ABI Prism 7000 sequence detection system (Applied Biosystems). An isotype-matched IgG-immunoprecipitated DNA sample was also amplified as a negative control (data not shown). Relative enrichment of DNA sequences was calculated by normalizing the averaged cycle threshold values to the input DNA. These values are presented as fold-change relative to DNA from unstimulated cells (control).
Analysis of MMP-13 Production by ELISA
Conditioned medium obtained after 16–18 h of treatment with IL-1β from various experiments was collected and analyzed for MMP-13 by ELISA according to the manufacturer's protocol and using duplicate wells for each sample. Samples were diluted (if needed) with culture medium to obtain a value within the linear range of the assays.
Chondrocyte Transfection
Chondrocytes were transfected by the nucleofection method, using a human chondrocyte nucleofection kit (Amaxa, Gaithersburg, MD) as described previously (6). Briefly, 2 million isolated cells were resuspended in transfection reagent and nucleofected with 2 μg of construct plasmid DNA or 100 nm of siRNA for S100A4. After the recovery period of 48 h, cells were made serum-free overnight, stimulated with IL-1β, and analyses for S100A4 and MMP-13 conducted as described above.
Chondrocyte Survival Assay
Chondrocyte survival was measured using a Live/Dead cell survival assay as described previously (24).
Statistical Analysis
Data sets were analyzed by analysis of variance using StatView 5.0 software (SAS Institute, Cary, NC). p values of <0.05 were considered statistically significant.
RESULTS
Nuclear Localization of S100A4 upon IL-1β Treatment
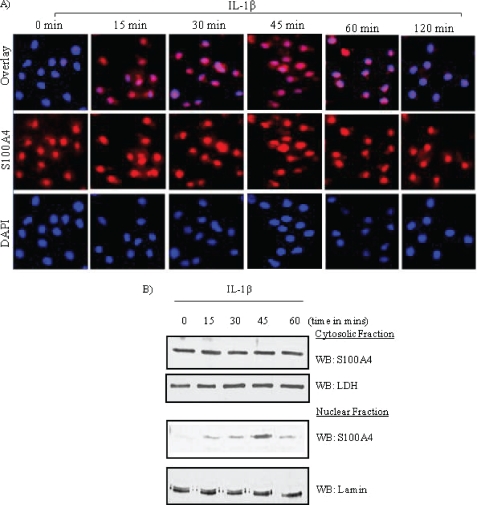
Chondrocytes were treated with IL-1β for 0–120 min and nuclear translocation of S100A4 was analyzed by confocal microscopy and immunoblotting. Immunofluorescence data showed that under basal conditions S100A4 was predominantly localized in the cytoplasm; very little staining was seen in the nucleus. However, stimulation of chondrocytes with IL-1β resulted in translocation of S100A4 into the nucleus as early as 15 min, reaching a maximum at 45 min (Fig. 1A). S100A4 started disappearing from the nucleus by 60 min, and very little or no S100A4 was detected in the nucleus 2 h after stimulation.
FIGURE 1.
S100A4 translocates into the nucleus upon IL-1β stimulation in chondrocytes. Chondrocytes cultured in monolayer were treated with IL-1β (10 ng/ml) for 0–120 min. A, after incubation, cells were fixed and visualized by confocal immunofluorescence microscopy. Images presented are double stained. Blue, nuclear stain with DAPI; red, S100A4 as detected with anti-S100A4 and Alexa 488-conjugated secondary antibodies. B, cytoplasmic and nuclear fractions were prepared as described under “Experimental Procedures” and immunoblotted for S100A4 using an anti-S100A4 antibody. Blots were stripped and reprobed with anti-human lamin B and lactose dehydrogenase (LDH) as a control. WB, Western blot.
Similar to the immunofluorescence, immunoblotting the nuclear fraction with S100A4 antibody demonstrated an immunoreactive band at 15 min after IL-1β treatment, reaching a maximum intensity at 45 min and then decreasing at 60 min (Fig. 1B). However, immunoblots showed no noticeable change in S100A4 protein levels in the cytoplasmic faction. Taken together, these results suggest that a portion of the cytosolic S100A4 translocates into the nucleus upon IL-1β stimulation in chondrocytes. The purity of each fraction obtained was validated by immunoblotting for specific markers: for cytoplasmic fractions, we used lactose dehydrogenase as a marker (Fig. 1B), and for nuclear fractions we used lamin (25, 26).
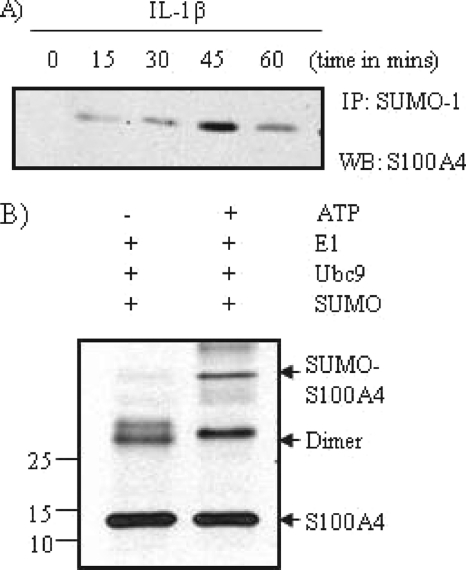
S100A4 Undergoes Sumoylation in Vivo and in Vitro
To better understand the molecular mechanism of nuclear translocation of S100A4, we examined if S100A4 can undergo sumo modification in chondrocytes. Cells were treated with IL-1β for 0–60 min and total cell extracts (250 μg of protein) were immunoprecipitated with anti-sumo-1 antibody followed by immunoblotting with anti-S100A4 (Fig. 2A). An immunoreactive band was seen in the extracts after a 15–60-min stimulation with IL-1β, suggesting that S100A4 undergoes sumo modification. To further demonstrate that S100A4 undergoes sumo modification, we used an in vitro sumoylation kit. A slow moving immunoreactive band of S100A4 was recognized by the anti-sumo antibody (Fig. 2B). When in vitro sumoylation was performed without ATP as a control, no sumoylation was noted (Fig. 2B). This data suggests that sumoylation of S100A4 in vitro is not a random event, but a specific process dependent on ATP. ATP is required in the first step of the sumoylation process for the activation of SUMO proteins by the E1 heterodimer AOS1-UBA2 enzyme complex (27). Taken together, these in vivo and in vitro data suggest that S100A4 undergoes sumoylation after IL-1β stimulation.
FIGURE 2.
S100A4 undergoes sumoylation. A, chondrocytes were treated with IL-1β (10 ng/ml) for 0–60 min, and nuclear extracts were prepared as described under “Experimental Procedures.” Extracts (250 μg of protein) were immunoprecipitated (IP) with anti-sumo-1 antibody followed by immunoblotting with anti-S100A4. B, the in vitro sumoylation reaction was carried out using a sumoylation kit according to the manufacturer's protocol. Sumoylated S100A4 protein was identified by immunoblot using anti-sumo antibody. WB, Western blot.
S100A4 Has a Monopartite Nuclear Localization Sequence and Two Sumoylation Sites
Numerous studies have shown that nuclear proteins, or proteins that crisscross the nuclear membrane, contain a stretch of basic amino acid sequence known as the nuclear localization sequence (NLS). Proteins with a NLS are transported across the nuclear envelope by a family of transport proteins called karyopherins or importins. In addition, nucleocytoplasmic transport is also modulated by post-translational modification of the cargo protein, including phosphorylation, acetylation, and sumoylation, that could mask or expose the NLS of the protein.
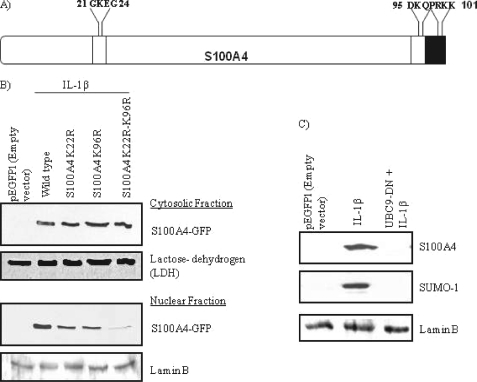
We examined the S100A4 protein for a nuclear localization sequence that could enable it to move across the nuclear membrane. Analysis of the amino acid sequence of S100A4 by PSORT II software (28) showed that the C-terminal end of the molecule contains a stretch of basic amino acids (PRKK) (Fig. 3A) that meet the criteria of being a potential monopartite nuclear localization sequence that could facilitate importin-mediated transport into the nucleus (28). In addition, SUMOplot analysis showed two lysine residues (Lys22 and Lys96), one on each end of the molecule, that could be potentially modified by sumo proteins. Thus, our analysis suggests that S100A4 could undergo sumoylation and translocate into the nucleus.
FIGURE 3.
Sumoylation of S100A4 is an essential step for its nuclear translocation. A, schematic representation of sumoylation and NLS sites on the S100A4 protein. Open box represents two sumoylation sites on either end of the molecule and the filled box at the C terminus represents the monopartite nuclear localization sequence. B, chondrocytes were nucleofected with or without wild type or mutant (K22R, K96R, and K22R/K96R) S100A4 constructs and stimulated with IL-1β (10 ng/ml) for 45 min. After incubation, cytosolic and nuclear extracts were prepared and immunoblotted with anti-S100A4. Blots were stripped and reprobed with anti-human lactose dehydrogenase (LDH) and lamin B as a control. C, cells were transfected with DN-Ubc9 (UBC9-DN) and stimulated with IL-1β for 45 min. Nuclear extracts were then immunoblotted with anti-S100A4 and anti-sumo-1. Blots were stripped and reprobed for lamin B as a control.
Sumoylation of S100A4 Is Required for IL-1β-mediated Nuclear Transport and MMP-13 Production
To further examine if sumoylation of S100A4 plays an important role in its nuclear translocation upon IL-1β stimulation, we mutated acceptor lysine residues (Lys22 and Lys96) to arginine residues. The translocation of S100A4 single mutants (K22R and K96R) into the nucleus was decreased compared with wild type, whereas the IL-1β-stimulated translocation of the S100A4 double mutant (K22R/K96R) was completely abolished (Fig. 3B). These results suggest that sumoylation of both lysine residues is required for translocation of S100A4 into the nucleus upon IL-1β stimulation. In addition, overexpression of the DN-Ubc9 (Ubc9: an E2 sumo-conjugating enzyme) construct, which is incapable of conjugating the sumo moiety to the target protein, inhibited the sumoylation of S100A4 and blocked its nuclear translocation (Fig. 3C).
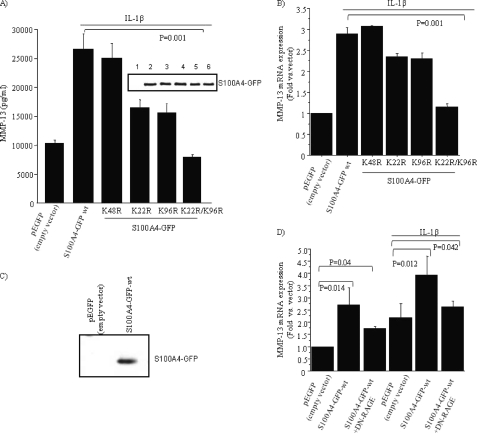
Because previous studies have linked intracellular S100A4 with MMP expression and IL-1β has been shown to stimulate increased production of MMP-13 in chondrocytes (19), we were interested in examining the effect of the sumoylation status of S100A4 on IL-1β-mediated MMP-13 production. Overexpression of S100A4 single mutants (K22R and K96R) reduced production of MMP-13 (both in protein and mRNA levels) compared with wild type S100A4, whereas transfection of the double mutant (K22R/K96R) reduced MMP-13 levels to the basal level (Fig. 4, A and B). As a control we overexpressed an S100A4-K48R mutant, which contained a mutation of a lysine residue present in the middle of the S100A4 molecule away from the two sumoylation sites. This mutant had no effect on IL-1β-induced increased MMP-13 production (Fig. 4, A and B). To further confirm that the observed effect was not due to transfection efficiency of S100A4 wild type and S100A4 sumo-mutant constructs, we analyzed the cell lysates for the S100A4 protein by immunoblotting. Our results showed that all constructs expressed equal amounts of S100A4 protein (Fig. 4A, inset). Taken together, these results suggest that the sumoylation of S100A4 plays an important role in IL-1β-mediated production of MMP-13 in chondrocytes.
FIGURE 4.
Sumoylation of S100A4 is required for IL-1β-mediated induction of MMP-13. A, chondrocytes were transfected with S100A4 WT and S100A4 sumo-mutant constructs by nucleofection and stimulated with or without IL-1β (10 ng/ml) overnight. The conditioned medium was collected and analyzed for MMP-13 production by ELISA and the cell lysates were immunoblotted with anti-S100A4 (inset). The number above each lane corresponds to the order of results shown in the bar graphs. B, expression of mRNA for MMP-13 was examined by real time PCR using MMP-13-specific primers. C, in this experiment, cells were transfected with pEGFP or with S100A4-GFP (wt) construct and after 48 h conditioned medium was collected and immunoblotted with anti-S100A4 antibody. D, cells were transfected with S100A4 WT and dominant-negative RAGE and stimulated with or without IL-1β (10 ng/ml) overnight. MMP-13 mRNA expression was measured by real time PCR. Values are the mean ± S.E. of 3 independent experiments.
Overexpression of S100 proteins could lead to secretion from the cells (29) and extracellular S100A4 has been shown to stimulate MMP-13 expression through activation of RAGE signaling (6). As expected, immunoblotting the conditioned medium of chondrocytes transfected to overexpress S100A4 demonstrated an immunoreactive band not present in control transfected cells (Fig. 4C).
Consistent with our previous studies (6), the increased production of S100A4 was associated with increased expression of MMP-13, which was inhibited by coexpression of dominant-negative RAGE (Fig. 4D). A further increase in MMP-13 expression was noted in cells stimulated with IL-1β after S100A4 overexpression, which was only partially inhibited by dominant-negative RAGE (Fig. 4D). These results suggest that extracellular S100A4-RAGE signaling had some contribution to the observed S100A4-IL-1β-mediated production of MMP-13 in our transfection studies. However, we think this effect was purely due to the forced overexpression of the S100A4 construct in our experimental system. In previous studies, we failed to detect any endogenous S100A4 protein in the conditioned medium of normal chondrocytes after IL-1β stimulation (30). Thus, this data supports the hypothesis that extracellular S100A4 stimulates MMP-13 production via RAGE, whereas IL-1β stimulates intracellular S100A4 to increase MMP-13 expression through sumoylation and nuclear translocation.
S100A4 Expression Is Required for IL-1β-mediated Production of MMP-13 in Human Chondrocytes
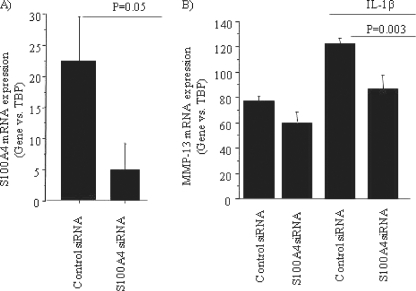
To further confirm the role of S100A4 in IL-1β-mediated expression of MMP-13, we knocked down the expression of S100A4 by using siRNA oligonucleotides and examined the effect on MMP-13 mRNA levels with and without IL-1β stimulation. Total RNA isolated from the cells that received S100A4 siRNA showed decreased expression of S100A4 (Fig. 5A). This was associated with some decrease in basal MMP-13 expression and significant inhibition of IL-1β-stimulated MMP-13 (Fig. 5B).
FIGURE 5.
Knockdown of S100A4 expression inhibits IL-1β-mediated production of MMP-13. Chondrocytes were transfected with siRNA for S100A4 (100 nm) or an equal amount of control siRNA followed by stimulation with or without IL-1β (10 ng/ml) for 16–18 h. After incubation, total RNA was prepared and expression of mRNA for S100A4 (A) and MMP-13 (B) was analyzed by real time PCR using specific primers for S100A4 and MMP-13. Data were normalized with mRNA expression for TATA box-binding protein.
Nuclear S100A4 Binds to the MMP-13 Promoter Region
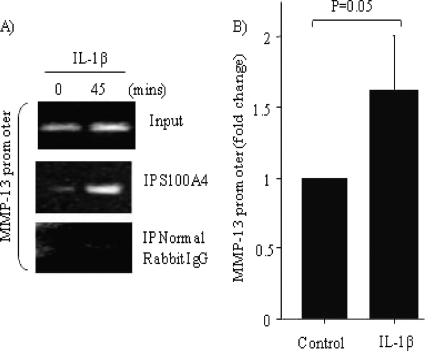
We next examined if S100A4 binds to chromatin that contains the MMP-13 promoter region. Thus, we immunoprecipitated endogenous S100A4 from chondrocytes treated with or without IL-1β and amplified the co-purified DNA using PCR. The ChIP isolates contained the promoter region of MMP-13, as detected by primers used in reverse transcriptase-PCR (Fig. 6A) and quantitative real time PCR (Fig. 6B). Taken together, these data suggest that endogenous S100A4 associates with chromatin containing MMP-13 promoter within 45 min of stimulation with IL-1β, which is within the time frame where we observed S100A4 translocation to the nucleus.
FIGURE 6.
Nuclear S100A4 binds to the promoter region of the MMP-13 gene. Chondrocytes were stimulated with or without IL-1β (10 ng/ml) for 45 min and ChIP was performed as described under “Experimental Procedures.” Endogenous S100A4 was immunoprecipitated (IP) using anti-human S100A4 rabbit polyclonal antibody; as an experimental control, normal rabbit IgG was used. Immunoprecipitated DNA was then analyzed by (A) reverse transcriptase-PCR with specific primers derived from the promoter region of MMP-13 gene and (B) by quantitative real-time PCR using a predesigned set of primers that spans −0.1 kb from site of start codon. Values are the mean ± S.E. of 3 independent experiments.
DISCUSSION
Several studies have suggested a link between intracellular S100A4 protein levels and expression of MMPs (16, 17, 31). However, the molecular mechanism of S100A4-mediated regulation of MMP expression has not been clearly understood. In this study, we show that S100A4 translocates into the nucleus upon IL-1β stimulation in human articular chondrocytes, a process that required the post-translational modification of sumoylation. More importantly, we found that nuclear S100A4 associated with a chromatin region that corresponds to the promoter region of the human MMP-13 gene, suggesting that nuclear S100A4 plays a role in the regulation of MMP-13 expression at the transcriptional level.
S100A4 is predominantly a cytosolic protein; however, a few studies have identified S100A4 in the nucleus of tumor cells (32, 33). Posttranslational modification of proteins has been shown to be important both in regulating the function of the target protein and its cellular localization (34). In our current study, we provide several lines of evidence to show that S100A4 undergoes sumoylation required for its nuclear translocation. Analysis of the S100A4 primary amino acid sequence revealed two possible lysine residues (Lys22 and Lys96) that can accept sumo moieties. Mutating these residues (K22R and K96R) or overexpression of a DN-Ubc9 construct not only inhibited IL-1β-mediated sumoylation of S100A4, but also blocked its nuclear translocation. Ubc9 is an E2-conjugating enzyme that transfers the sumo protein to the acceptor lysine residue on the target protein (35). Expression of DN-Ubc9 (C93S and L97S) has been previously shown to inhibit sumoylation (36). In support of our data, another study showed the presence of modified forms of S100A4 in nuclear extracts of the colorectal cancer cell line SW620, suggesting the presence of post-translational modification (37). However, that study was unable to identify any particular post-translational modification.
Sumoylation is a post-translational protein modification similar to ubiquitination, yet distinct from it. Sumoylation has been implicated in cytoplasmic nuclear transport, in addition to its role in other physiological functions including protein-protein interaction, modulation of enzyme activity and stability, gene expression, chromatin structure, and signal transduction (38). Sumoylation was first described for RanGap1, a nuclear import factor (39, 40). However, the real evidence that sumoylation is required for nuclear transport of a protein came from a study on Sp100, a component of nuclear dots. A single-point mutant in the NLS of Sp100 abolished not only nuclear import but also sumoylation. Importantly, when importation of the Sp100 mutant was re-established by fusing it to the SV40 large t-antigen NLS, sumoylation was restored (41). Taken together, these studies suggest that sumoylation plays an important role in nuclear transport of target proteins.
Previous studies have shown that members of S100 proteins undergo post-translational modification including ubiquitination, transamidation, and oxidative modifications (42). He et al. (43) showed that S100A10 undergoes polyubiquitination in the absence of its binding partner annexin A2, which targeted S100A10 for proteosomal degradation. S100A8 and S100A9 are known to be modified oxidatively to form Nϵ-carboxymethylysine protein adducts that induce inflammatory response by increasing NF-κB-dependent proinflammatory gene expression (44). S100A11 is covalently modified by transamidation in chondrocytes and promotes chondrocyte hypertrophy and matrix catabolism. A mutant S100A11 protein that was incapable of undergoing transamidation could not induce chondrocyte hypertrophy (45). In one report (46), S100A6 was found to be glutathionylated and cysteinylated in cells exposed to ionizing radiation. Interestingly, these modifications correlated with its translocation from the nucleus to the cytoplasm upon radiation. These studies suggest that post-translational modifications of S100 proteins are important in their biological localization and functions.
We also found that disruption of sumoylation of S100A4 or knocking down the expression of S100A4 inhibited IL-1β-mediated production of MMP-13, implicating nuclear S100A4 in the regulation of MMP-13 expression. Our ChIP data show that nuclear S100A4 is associated with the region of the MMP-13 promoter that contains a TATA box, the core transcriptional unit (at approximately −30 bp), and an AP-1 site (at approximately −70 bp) that binds to dimers of the Fos and Jun families of proteins (47). Studies have shown that IL-1β-mediated induction of MMP-13 occurs through activation of the AP-1 transcription factor (48). Interestingly, interaction of other transcription factors with AP-1 proteins or directly binding at the AP-1 site may also regulate tissue-specific expression of MMP-13. The transcription factor Runt domain factor-2 (Runx-2) binds at a unique site on the MMP-13 promoter, and also interacts with AP-1 to mediate MMP-13 transcription (49, 50). Recently, YB-1, a member of the Y-box family of DNA-binding proteins, was shown to directly bind at the AP-1 site on MMP-13 and regulate its transcription (51). Thus, the AP-1 binding site on the promoter region of the MMP-13 gene plays a critical role in MMP-13 gene expression.
Although our studies indicate that S100A4 may interact with the MMP-13 promoter (including the AP-1 binding site), we speculate that this interaction is indirect. S100 proteins do not have a DNA binding motif in their primary structure. However, they play a role in transcriptional regulation of genes by interacting with certain transcription factors, including MyoD and E12, which contain the basic helix-loop-helix domain (2). Based on these studies, we hypothesize that S100A4 may function as a co-transcription factor in the regulation of MMP-13 gene expression. However, further studies are needed to confirm this hypothesis.
In summary, using human articular chondrocytes, we provide evidence that S100A4 translocates into the nucleus upon IL-1β stimulation and that this nuclear translocation requires sumoylation. Nuclear S100A4 interacts with the MMP-13 promoter region and plays an important role in transcriptional regulation of the MMP-13 gene. Because MMP-13 is one of the key factors in the pathogenesis of osteoarthritis, further studies are warranted to delineate this novel regulator mechanism that could potentially identify new targets for therapeutic intervention in osteoarthritis.
Acknowledgments
We are grateful to the Gift of Hope Organ and Tissue Donor Network (Elmhurst, IL) and the National Disease Research Interchange (Philadelphia, PA) for providing tissue. We thank Mary Zhao for technical help and Karen Klein, MA, ELS (Research Support Core, WFUHS) for editorial contributions to this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants P30AG21332 (to Wake Forest University Claude D Pepper Older Americans Independence Center) and R01AG16697 (to R. F. L.) and the Wake Forest University Health Sciences Venture Fund (to R. R. Y.).
- MMP
- metalloproteinase
- NLS
- nuclear localization sequence
- DN
- dominant-negative.
REFERENCES
- 1.Schäfer B. W., Heizmann C. W. (1996) Trends Biochem. Sci. 21, 134–140 [DOI] [PubMed] [Google Scholar]
- 2.Donato R. (2001) Int. J. Biochem. Cell. Biol. 33, 637–668 [DOI] [PubMed] [Google Scholar]
- 3.Marenholz I., Heizmann C. W., Fritz G. (2004) Biochem. Biophys. Res. Commun. 322, 1111–1122 [DOI] [PubMed] [Google Scholar]
- 4.Salama I., Malone P. S., Mihaimeed F., Jones J. L. (2008) Eur. J. Surg. Oncol. 34, 357–364 [DOI] [PubMed] [Google Scholar]
- 5.Loeser R. F., Yammani R. R., Carlson C. S., Chen H., Cole A., Im H. J., Bursch L. S., Yan S. D. (2005) Arthritis Rheum. 52, 2376–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yammani R. R., Carlson C. S., Bresnick A. R., Loeser R. F. (2006) Arthritis Rheum. 54, 2901–2911 [DOI] [PubMed] [Google Scholar]
- 7.Cecil D. L., Johnson K., Rediske J., Lotz M., Schmidt A. M., Terkeltaub R. (2005) J. Immunol. 175, 8296–8302 [DOI] [PubMed] [Google Scholar]
- 8.Foell D., Roth J. (2004) Arthritis Rheum. 50, 3762–3771 [DOI] [PubMed] [Google Scholar]
- 9.Ebralidze A., Tulchinsky E., Grigorian M., Afanasyeva A., Senin V., Revazova E., Lukanidin E. (1989) Genes Dev. 3, 1086–1093 [DOI] [PubMed] [Google Scholar]
- 10.Grigorian M. S., Tulchinsky E. M., Zain S., Ebralidze A. K., Kramerov D. A., Kriajevska M. V., Georgiev G. P., Lukanidin E. M. (1993) Gene 135, 229–238 [DOI] [PubMed] [Google Scholar]
- 11.Takenaga K., Nakamura Y., Sakiyama S. (1994) Cell Struct. Funct. 19, 133–141 [DOI] [PubMed] [Google Scholar]
- 12.Garrett S. C., Varney K. M., Weber D. J., Bresnick A. R. (2006) J. Biol. Chem. 281, 677–680 [DOI] [PubMed] [Google Scholar]
- 13.Grigorian M., Andresen S., Tulchinsky E., Kriajevska M., Carlberg C., Kruse C., Cohn M., Ambartsumian N., Christensen A., Selivanova G., Lukanidin E. (2001) J. Biol. Chem. 276, 22699–22708 [DOI] [PubMed] [Google Scholar]
- 14.Novitskaya V., Grigorian M., Kriajevska M., Tarabykina S., Bronstein I., Berezin V., Bock E., Lukanidin E. (2000) J. Biol. Chem. 275, 41278–41286 [DOI] [PubMed] [Google Scholar]
- 15.Ambartsumian N., Klingelhöfer J., Grigorian M., Christensen C., Kriajevska M., Tulchinsky E., Georgiev G., Berezin V., Bock E., Rygaard J., Cao R., Cao Y., Lukanidin E. (2001) Oncogene 20, 4685–4695 [DOI] [PubMed] [Google Scholar]
- 16.Bjørnland K., Winberg J. O., Odegaard O. T., Hovig E., Loennechen T., Aasen A. O., Fodstad O., Maelandsmo G. M. (1999) Cancer Res. 59, 4702–4708 [PubMed] [Google Scholar]
- 17.Bjørnland K., Bratland A., Rugnes E., Pettersen S., Johansen H. T., Aasen A. O., Fodstad Ø., Ree A. H., Maelandsmo G. M. (2001) J. Pediatr. Surg. 36, 1040–1044 [DOI] [PubMed] [Google Scholar]
- 18.Sternlicht M. D., Werb Z. (2001) Annu. Rev. Cell. Dev. Biol. 17, 463–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mengshol J. A., Vincenti M. P., Coon C. I., Barchowsky A., Brinckerhoff C. E. (2000) Arthritis Rheum. 43, 801–811 [DOI] [PubMed] [Google Scholar]
- 20.Mitchell P. G., Magna H. A., Reeves L. M., Lopresti-Morrow L. L., Yocum S. A., Rosner P. J., Geoghegan K. F., Hambor J. E. (1996) J. Clin. Invest. 97, 761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shlopov B. V., Lie W. R., Mainardi C. L., Cole A. A., Chubinskaya S., Hasty K. A. (1997) Arthritis Rheum. 40, 2065–2074 [DOI] [PubMed] [Google Scholar]
- 22.Muehleman C., Bareither D., Huch K., Cole A. A., Kuettner K. E. (1997) Osteoarthritis Cartilage 5, 23–37 [DOI] [PubMed] [Google Scholar]
- 23.Yan J., Erdem H., Li R., Cai Y., Ayala G., Ittmann M., Yu-Lee L. Y., Tsai S. Y., Tsai M. J. (2008) Cancer Res. 68, 5460–5468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Del Carlo M., Jr., Loeser R. F. (2002) Arthritis Rheum. 46, 394–403 [DOI] [PubMed] [Google Scholar]
- 25.Goldman R. D., Gruenbaum Y., Moir R. D., Shumaker D. K., Spann T. P. (2002) Genes Dev. 16, 533–547 [DOI] [PubMed] [Google Scholar]
- 26.Ramirez V. D., Gautron J. P., Epelbaum J., Pattou E., Zamora A., Kordon C. (1975) Mol. Cell. Endocrinol. 3, 339–350 [DOI] [PubMed] [Google Scholar]
- 27.Geiss-Friedlander R., Melchior F. (2007) Nat. Rev. Mol. Cell. Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 28.Nakai K., Horton P. (1999) Trends Biochem. Sci. 24, 34–36 [DOI] [PubMed] [Google Scholar]
- 29.Arumugam T., Simeone D. M., Schmidt A. M., Logsdon C. D. (2004) J. Biol. Chem. 279, 5059–5065 [DOI] [PubMed] [Google Scholar]
- 30.Yammani R. R., Long D., Loeser R. F. (2009) Arthritis Rheum. 60, 792–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saleem M., Kweon M. H., Johnson J. J., Adhami V. M., Elcheva I., Khan N., Bin Hafeez B., Bhat K. M., Sarfaraz S., Reagan-Shaw S., Spiegelman V. S., Setaluri V., Mukhtar H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 14825–14830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flatmark K., Pedersen K. B., Nesland J. M., Rasmussen H., Aamodt G., Mikalsen S. O., Bjørnland K., Fodstad Ø., Maelandsmo G. M. (2003) J. Pathol. 200, 589–595 [DOI] [PubMed] [Google Scholar]
- 33.Kikuchi N., Horiuchi A., Osada R., Imai T., Wang C., Chen X., Konishi I. (2006) Cancer Sci. 97, 1061–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farley A. R., Link A. J. (2009) Methods Enzymol. 463, 725–763 [DOI] [PubMed] [Google Scholar]
- 35.Hay R. T. (2005) Mol. Cell. 18, 1–12 [DOI] [PubMed] [Google Scholar]
- 36.Garaude J., Farrás R., Bossis G., Charni S., Piechaczyk M., Hipskind R. A., Villalba M. (2008) J. Immunol. 180, 5983–5990 [DOI] [PubMed] [Google Scholar]
- 37.Haugen M. H., Flatmark K., Mikalsen S. O., Malandsmo G. M. (2008) BMC Cancer 8, 172–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gill G. (2004) Genes Dev. 18, 2046–2059 [DOI] [PubMed] [Google Scholar]
- 39.Matunis M. J., Coutavas E., Blobel G. (1996) J. Cell. Biol. 135, 1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahajan R., Gerace L., Melchior F. (1998) J. Cell. Biol. 140, 259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sternsdorf T., Jensen K., Reich B., Will H. (1999) J. Biol. Chem. 274, 12555–12566 [DOI] [PubMed] [Google Scholar]
- 42.Lim S. Y., Raftery M. J., Goyette J., Hsu K., Geczy C. L. (2009) J. Leukocyte Biol. 86, 577–587 [DOI] [PubMed] [Google Scholar]
- 43.He K. L., Deora A. B., Xiong H., Ling Q., Weksler B. B., Niesvizky R., Hajjar K. A. (2008) J. Biol. Chem. 283, 19192–19200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andrassy M., Igwe J., Autschbach F., Volz C., Remppis A., Neurath M. F., Schleicher E., Humpert P. M., Wendt T., Liliensiek B., Morcos M., Schiekofer S., Thiele K., Chen J., Kientsch-Engel R., Schmidt A. M., Stremmel W., Stern D. M., Katus H. A., Nawroth P. P., Bierhaus A. (2006) Am. J. Pathol. 169, 1223–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cecil D. L., Terkeltaub R. (2008) J. Immunol. 180, 8378–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orre L. M., Pernemalm M., Lengqvist J., Lewensohn R., Lehtiö J. (2007) Mol. Cell. Proteomics 6, 2122–2131 [DOI] [PubMed] [Google Scholar]
- 47.Benbow U., Brinckerhoff C. E. (1997) Matrix Biol. 15, 519–526 [DOI] [PubMed] [Google Scholar]
- 48.Liacini A., Sylvester J., Li W. Q., Zafarullah M. (2002) Matrix Biol. 21, 251–262 [DOI] [PubMed] [Google Scholar]
- 49.Hess J., Porte D., Munz C., Angel P. (2001) J. Biol. Chem. 276, 20029–20038 [DOI] [PubMed] [Google Scholar]
- 50.Porte D., Tuckermann J., Becker M., Baumann B., Teurich S., Higgins T., Owen M. J., Schorpp-Kistner M., Angel P. (1999) Oncogene 18, 667–678 [DOI] [PubMed] [Google Scholar]
- 51.Samuel S., Beifuss K. K., Bernstein L. R. (2007) Biochim. Biophys. Acta 1769, 525–531 [DOI] [PMC free article] [PubMed] [Google Scholar]