Abstract
Fanconi anemia (FA) is a cancer susceptibility syndrome characterized by sensitivity to DNA-damaging agents. The FA proteins (FANCs) are implicated in DNA repair, although the precise mechanisms by which FANCs process DNA lesions are not fully understood. An epistatic relationship between the FA pathway and translesion synthesis (TLS, a post-replication DNA repair mechanism) has been suggested, but the basis for cross-talk between the FA and TLS pathways is poorly understood. We show here that ectopic overexpression of the E3 ubiquitin ligase Rad18 (a central regulator of TLS) induces DNA damage-independent mono-ubiquitination of proliferating cell nuclear antigen (PCNA) (a known Rad18 substrate) and FANCD2. Conversely, DNA damage-induced mono-ubiquitination of both PCNA and FANCD2 is attenuated in Rad18-deficient cells, demonstrating that Rad18 contributes to activation of the FA pathway. WT Rad18 but not an E3 ubiquitin ligase-deficient Rad18 C28F mutant fully complements both PCNA ubiquitination and FANCD2 activation in Rad18-depleted cells. Rad18-induced mono-ubiquitination of FANCD2 is not observed in FA core complex-deficient cells, demonstrating that Rad18 E3 ligase activity alone is insufficient for FANCD2 ubiquitylation. Instead, Rad18 promotes FA core complex-dependent FANCD2 ubiquitination in a manner that is secondary to PCNA mono-ubiquitination. Taken together, these results demonstrate a novel Rad18-dependent mechanism that couples activation of the FA pathway with TLS.
Keywords: Cell Cycle, Checkpoint Control, DNA Repair, DNA Replication, Mutagenesis Mechanisms
Introduction
Genomes are continuously exposed to endogenous and exogenous DNA-damaging agents, and appropriate coordination of DNA replication and repair mechanisms is necessary to prevent genetic instability and cancer (1, 2). Cells have evolved various systems to sense and repair different types of damaged DNA throughout the cell cycle.
During S-phase, DNA lesions that escape repair may persist and cause stalling and collapse of DNA replication forks, a potential cause of DNA double strand breaks. Translesion synthesis (TLS)3 is a DNA damage tolerance mechanism that allows cells to avoid detrimental consequences of persistent replication stalling (3). TLS involves a “polymerase switch” that replaces replicative DNA polymerases with specialized TLS DNA polymerases at stalled replication forks (3–5). In mammalian cells, the four main Y-family TLS polymerases are DNA pol η, pol κ, pol ι, and REV1 (6). Collectively, the Y-family TLS polymerases facilitate replicative bypass of various helix-distorting lesions such as bulky adducts, allowing continued fork progression in cells with DNA damage. However, because of the relatively low fidelity and processivity of TLS DNA polymerases, the ensuing lesion bypass is often error-prone and preserves replication forks at the cost of mutagenesis (3).
The polymerase switching process is dependent on an E3 ubiquitin ligase termed “Rad18.” In response to replication fork stalling, Rad18 mono-ubiquitinates PCNA at Lys-164 (4, 5). All Y-family TLS polymerases have specialized ubiquitin-binding motifs (designated “UBM” and “UBZ” domains) that mediate their associations with mono-ubiquitinated PCNA (7). Rad18 also contributes to TLS via its direct associations with pol η which help guide the TLS polymerase to stalled replication forks (8). Of the four Y-family TLS polymerases, pol η is the first to be recruited to most DNA lesions, in part because of Rad18-mediated chaperone activity. We and others have previously shown that Rad18 and TLS polymerases are important for recovery from replication arrest and timely attenuation of S-phase checkpoint signaling in genotoxin-treated cells (9–11).
Similar to TLS, the Fanconi anemia (FA) DNA repair pathway is implicated in processing and repairing DNA damage acquired during S-phase (12, 13). FA is a rare chromosome instability syndrome associated with increased cancer susceptibility. Gross chromosomal rearrangements following exposure to DNA cross-linking agents, such as mitomycin C (MMC), diepoxybutane, and cisplatin, is a well known characteristic of cells derived from FA patients. At least 13 FA complementation groups (A–C, D1, D2, E–G, I, J, and L–N) have been identified. The FA proteins (FANCs) encoded by the various complementation groups are thought to function in a common biochemical pathway of DNA repair. The FA core complex includes protein products of genes belonging to at least eight FA complementation groups (A–C, E–G, L, and M) and acts as an E3 ligase to mono-ubiquitinate FANCD2 and FANCI in response to DNA damage and replication stress during S-phase. The mono-ubiquitinated FANCD2-I complex, the putative effector of the FA pathway, is then recruited to chromatin, and it is thought to direct DNA repair (13).
The FA pathway has been implicated in nuclear excision repair, homologous recombination, non-homologous end joining, and TLS. A potential link between the FA and TLS pathways was suggested based on the hypomutability of FA patient cell lines after acquisition of psoralen photo-induced lesions (14, 15), indicating that the FA pathway facilitates an error-prone repair process. Moreover, Niedzwiedz et al. (16) demonstrated that DT40 cells deficient in REV1 and REV3 display cross-linker sensitivity phenotypes that are epistatic to FANCC. Also, pol ζ is necessary for FANCD2-FANCI-mediated ICL repair in cell-free Xenopus extracts (17). The finding that a common de-ubiquitinating enzyme (USP1) regulates the mono-ubiquitination of both PCNA and FANCD2 (18, 19) and that Rad6 (an E2 ligase for Rad18 in TLS) status affects FANCD2 mono-ubiquitination (20) also suggest that the coordination of the FA pathway and TLS might be important for the DNA damage response. REV1 focus formation is partially dependent on the FA core complex in human cells, and FA patient-derived core complex mutants show reduced mutagenesis (14, 15, 21). Interestingly, however, FANCD2-deficient cells are hypermutable compared with wild type cells (in the absence or presence of UVC damage) (21), suggesting that the FA pathway might be forked, with the FANCD2-I ubiquitination branch responsible for homologous recombination control and the core complex-dependent branch controlling TLS via REV1 (22).
Thus, although there is considerable evidence that the FA and TLS pathways are linked, the molecular mechanisms that coordinate the FA and TLS pathways are incompletely understood. Furthermore, it is not known whether the FANCs are necessarily regulated distally or proximally to the TLS pathway or whether bidirectional communication takes place between FANCs and TLS proteins. Because a common de-ubiquitinating enzyme (USP1) negatively regulates both the TLS and FA pathways (via de-ubiquitination of PCNA and FANCD2, respectively), we considered the possibility that Rad18 might be responsible for coordinate activation of the two repair pathways. Our results described here indicate that FANCD2 mono-ubiquitination is Rad18-dependent. However, we do not find evidence that FANCD2 is a direct substrate of Rad18. Instead, our results indicate that the FA pathway is activated in a manner that is coupled to PCNA ubiquitination and TLS polymerase recruitment.
EXPERIMENTAL PROCEDURES
Cell Culture
H1299 cells, HCT116 WT RAD18+/+, and RAD18−/− cells were obtained from Dr. Tadahiro Shiomi, Kumamoto University, Japan (23). CRL1162 cells derived from pol η-deficient XPV patients and CRL1162 + XPV cells stably expressing reconstituted WT POLH cDNA were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, streptomycin sulfate (100 μg/ml), and penicillin (100 units/ml). FANCD2-deficient PD20 cells, FANCA-deficient GM6914, and isogenic cell lines stably expressing reconstituted wild type FANCs (obtained from Dr. Alan D'Andrea, Dana Farber Cancer Institute) were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 15% fetal bovine serum, streptomycin sulfate (100 μg/ml), and penicillin (100 units/ml). Sf9 insect cells (derived from Spodoptera frugiperda ovarian cells) were cultured in Sf-900TM III SFM (Invitrogen).
RNA Interference (RNAi)
The following siRNA oligonucleotides (Dharmacon, Lafayette, CO) were used for the experiments in this study: pol η, 5′-GCA GAA AGG CAG AAA GUU A-3′; Rad18, On-Target plus SMART POOL (L-004591, Dharmacon); UBE2T, 5′-GAA GAG AGA GCT GCA CAT GTT-3′; and nontargeting control siRNA oligonucleotides (D-001210-01, Dharmacon). Lipofectamine 2000 (Invitrogen) was used for transfection of all the siRNA oligonucleotides, following the manufacturer's protocol. All tubes, tips, and solutions used for RNA interference experiments were certified RNase-free.
SDS-PAGE and Immunoblotting
Total cell lysates were prepared in lysis buffer containing 50 mm Hepes, pH 7.4, 0.1% Triton X-100, 150 mm NaCl, 1 mm EDTA, 50 mm NaF, 80 mm β-glycerophosphate, supplemented with protease inhibitor and phos-stop (Roche Applied Science). In some experiments, cells were fractionated to generate soluble fractions and whole nuclei, exactly as described by Izumi et al. (24). In brief, monolayers of cells were washed two times with ice-cold phosphate-buffered saline (PBS) and scraped into 500 μl of ice-cold cytoskeleton buffer (10 mm PIPES, pH 6.8), 100 mm NaCl, 300 mm sucrose, 3 mm MgCl2, 1 mm EGTA, 1 mm dithiothreitol, 0.1 mm ATP, 1 mm Na3VO4, 10 mm NaF, and 0.1% Triton X-100) freshly supplemented with protease inhibitor and phos-stop (Roche Applied Science). Cytoskeleton lysates were transferred to microcentrifuge tubes and incubated on ice for 5 min. The cell lysates were centrifuged at 1000 × g for 2 min. The supernatants were removed and further clarified by centrifugation at 10,000 × g for 10 min to obtain Triton X-100-soluble fractions. The Triton X-100-extracted insoluble nuclear fractions were washed once with 1 ml of cytoskeleton buffer and then resuspended in a minimal volume of cytoskeleton buffer. Soluble or nuclear protein samples were separated by SDS-PAGE, transferred to nitrocellulose, and analyzed by immunoblotting with appropriate antibodies. Bound antibodies were visualized using ECL. Films were scanned and analyzed by densitometry using ImageJ software to compare relative expression levels of various proteins.
Fluorescence Microscopy
H1299 or HCT116 cells exponentially growing in two-well chamber slides were infected with Ad-Rad18 for 48 h. The resulting slides were further treated with BPDE or UV genotoxins or left untreated. Cells were washed with PBS and incubated with cold CSK buffer on ice for 10 min. Cell nuclei were fixed with 4% paraformaldehyde in CSK for 15 min at room temperature and washed with PBS. After washing with PBS, cells were permeabilized with 0.5% Triton X-100 for 5 min. Slides were washed again with PBS, and cells were incubated overnight at 4 °C with anti-FANCD2 antibodies (sc-20022, mouse) in 3% bovine serum albumin (BSA) in PBS containing 0.1% Triton X-100 (PBST). The slides were washed three times with 1% BSA/PBST and incubated with anti-mouse IgG-FITC antibodies or anti-mouse IgG-Cy3 antibodies (C2181, Sigma) for 2 h at room temperature. After washing with PBST, the slides were DAPI-stained, mounted with VECTASHIELD® Mounting Medium (Vector Laboratories), imaged, and analyzed with a DeltaVision image restoration microscopy system (dv1301421; Applied Precision).
Genotoxin Treatments
BPDE (NCI Carcinogen Repository, National Institutes of Health) was dissolved in anhydrous Me2SO and added directly to the growth medium as a 1000× stock to give final concentrations of 100, 200, 300, or 500 nm. MMC was dissolved in 50% ethanol added directly to the growth medium as a 1000× stock to give a final concentration of 300 nm. For UVC treatment, the growth medium was removed from the cells and replaced with PBS. The plates were transferred to a UV cross-linker (Stratagene) and then irradiated. The UVC dose delivered to the cells was confirmed with a UV radiometer (UVP, Inc.). The cells were then re-fed with complete growth medium and returned to the incubator.
Adenovirus Construction and Infection
All adenovirus vectors were constructed and purified as described previously (9, 10). Briefly, various cDNAs were subcloned into pAC-CMV. The resulting shuttle vectors were co-transfected into 293T cells with the pJM17 plasmid to generate recombinant adenovirus, and adenovirus particles were purified from 293T cell lysates by polyethylene glycol precipitation, CsCl gradient centrifugation, and gel filtration column chromatography. Adenovirus infections were performed by direct addition of purified virus to the tissue culture medium. Expression of adenovirus-encoded proteins was confirmed by immunoblotting. Cells were routinely infected with 5 × 109 plaque-forming units/ml of adenovirus.
In Vitro Ubiquitination Assays
Recombinant full-length wild type FANCD2 was purified using recombinant baculovirus-infected Sf9 insect cells as described by Park et al. (25). Briefly, Sf9 cells were infected with baculovirus encoding zz-tagged FANCD2 (obtained from Dr. Jeffery Parvin, Ohio State University) for 4 days. The infected cells were harvested and washed in chilled phosphate-buffered saline. Cells were lysed in 15 ml of buffer B (20 mm Hepes, pH 7.9, 20% glycerol, 200 mm KCl, 0.5 mm EDTA, 0.5% Nonidet P-40, 0.5 mm dithiothreitol, supplemented with Complete-mini (Roche Applied Science)). The pre-equilibrated 250 μl of IgG-Sepharose (Amersham Biosciences) was added into the protein lysate and incubated with mixing at 4 °C for 16 h. The resulting protein-IgG-Sepharose complexes were washed twice with buffer B and twice with buffer C (20 mm Hepes, pH 7.9, 20% glycerol, 200 mm KCl, 0.5 mm EDTA, 0.1% Nonidet P-40, 0.5 mm dithiothreitol), four times with TEV cleavage buffer (10 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.1% Nonidet P-40, 0.5 mm EDTA, 0.5 mm dithiothreitol), and then resuspended in 300 μl of TEV cleavage buffer plus 18 μg of TEV enzyme (Invitrogen). Beads containing the bound fusion protein were incubated with the TEV protease at 4 °C for 16 h. The supernatant containing FANCD2 protein was resolved by SDS-PAGE, stained with Coomassie Blue to estimate protein concentration, and used for in vitro ubiquitination assay. As an alternative source of FANCD2 protein for in vitro ubiquitination assay, FANCD2 was immunoprecipitated from PD20 + FLAG-FANCD2 reconstituted cells. Cells were lysed and fractionated using CSK buffer as described above, and the soluble fraction of the cell lysates was used for immunoprecipitation to enrich nonubiquitinated FANCD2 protein. Anti-FLAG M2 Affinity gel (Sigma) was incubated with the soluble cell lysate at 4 °C for 16 h, washed five times with wash buffer (20 mm Tris, pH 7.5, 0.1 m NaCl, 0.1 mm EDTA, 0.05% Tween 20), and eluted with 5 volumes of 0.2 mg/ml FLAG peptide (Sigma). In vitro ubiquitination assays were performed as described by Tateishi and co-workers (8) using recombinant PCNA recombinant protein as a positive control for the Rad6-Rad18-mediated ubiquitination. Briefly, purified recombinant FANCD2 or PCNA proteins were incubated for 1 h at 25 °C with RAD6B-RAD18 complexes (26) in a buffer containing 50 mm Hepes, pH 7.6, 0.05 mm DTT, 1 mm MgCl2, 1 mm ATP, and an ATP-regenerating system together with either 5 mm ubiquitin (Boston Biochem) or GST-ubiquitin. The reaction mixtures were analyzed by SDS-PAGE and immunoblotting with anti-FANCD2 or anti-PCNA antibodies.
Reproducibility
All data shown are representative of experiments that were performed at least three times on separate occasions with similar results.
RESULTS
FA Pathway Activation in Response to BPDE-induced Bulky DNA Adducts
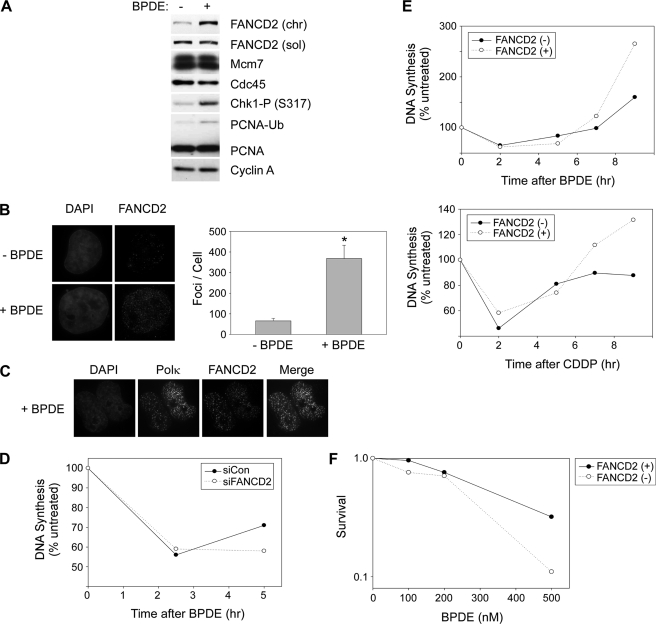
We have previously shown a key role for Rad18 and TLS polymerases in mediating attenuation of S-phase checkpoint signaling elicited by BPDE lesions (9, 10). Because the FA pathway is implicated in TLS (14–16, 21, 22), it was of interest to test the possible role of FANCD2 in the DNA damage response to BPDE (9, 10). H1299 cells were treated with 100 nm BPDE for 2 h to induce DNA damage, and the cell lysates were analyzed for FANCD2 and other DNA replication and S-phase checkpoint proteins. As expected, BPDE induced Chk1 phosphorylation at Ser-317 and decreased chromatin loading of Cdc45 (Fig. 1A), demonstrating activation of S-phase checkpoint signaling. BPDE also induced mono-ubiquitination of PCNA and FANCD2 under these experimental conditions (Fig. 1A, left panels). In response to genotoxins or during S-phase, FANCD2 redistributes to distinct nuclear foci likely corresponding to DNA replication and repair factories (28, 29). We tested the effect of BPDE on FANCD2 subcellular distribution by immunofluorescence microscopy. As shown in Fig. 1B, BPDE treatment increased the number of FANCD2 nuclear foci by 5.5-fold (p = 0.00015). FANCD2 foci were partially co-localized with pol κ (Fig. 1C), the TLS polymerase involved in bypass of BPDE-induced DNA lesions (9, 10). Therefore, BPDE induces concomitant activation of checkpoint signaling, Rad18-dependent TLS, and FANCD2 ubiquitination.
FIGURE 1.
Role of FANCD2 in the cellular response to BPDE. A, H1299 cells were treated with 100 nm BPDE for 2 h or left untreated for controls. Soluble (sol) and chromatin (chr) extracts from the resulting cells were analyzed by SDS-PAGE and immunoblotting with anti-FANCD2, MCM7, Cdc45, phospho-Chk1 (Ser-317), PCNA, and cyclin A antibodies (left panel). The chromatin-bound (mono-ubiquitinated) FANCD2 is designated “FANCD2-Ub.” B, in parallel with the experiment described in A above, H1299 cells growing in chamber slides were treated with 100 nm BPDE for 4 h or left untreated. Cells were washed with CSK buffer to remove soluble proteins, fixed with 4% paraformaldehyde, and stained with anti-FANCD2 antibodies. FANCD2 nuclear foci were visualized by Deltavision immunofluorescence microscopy. FANCD2 foci in 10 representative nuclei from control (−BPDE) and BPDE-treated (BPDE) cells were enumerated. Data points represent the mean number of foci for 10 representative cells, with error bars representing the range. *, p = 0.00015 compared with control cells that did not receive BPDE. C, GFP-pol κ-expressing H1299 cells growing in chamber slides were treated with 100 nm BPDE for 4 h or left untreated. Cells were fixed and stained with anti-FANCD2 antibodies as described under “Experimental Procedures.” FANCD2 and GFP nuclear foci were visualized by Deltavision immunofluorescence microscopy. D, effect of FANCD2 deficiency on DNA synthesis in genotoxin-treated cells. H1299 cells were transfected with siRNA against FANCD2 (or with nontargeting control siRNA oligonucleotides). The resulting cells were treated with 60 nm BPDE, and at various times, the rates of DNA synthesis were determined by measuring the rates of thymidine incorporation. E, DNA synthesis assays similar to those performed in D above were conducted in FANCD2(+) and FANCD2(−) cells at the indicated times after treatment with 50 nm BPDE and 250 μm cisplatin. F, exponentially growing FANCD2(+) and FANCD2(−) cells were treated with different doses of BPDE as indicated. 24 h later, cells were trypsinized and re-plated in replicate on 10-cm plates at a density of 1000 cells/plate. The growth medium on the re-plated cells was replenished every 3 days. After 10 days, colonies of >50 cells were stained with crystal violet and then counted.
TLS deficiency delays the recovery from S-phase checkpoints, most likely due to impaired processing and resolution of stalled fork structures when post-replication repair is compromised (9, 10). Given the suspected relationship between the FA pathway and TLS, it was of interest to determine the potential requirement for FANCD2 in mediating recovery from BPDE-induced S-phase arrest. Therefore, we measured rates of DNA synthesis at different times after BPDE treatment in control and FANCD2-depleted H1299 cells. As shown in Fig. 1D, resumption of DNA synthesis following BPDE treatment was delayed in FANCD2-depleted H1299 cells relative to controls. We also compared rates of DNA synthesis after BPDE treatment in FANCD2(−) PD20 FA patient cells and an isogenic FLAG-FANCD2-complemented cell line (designated FANCD2(+)). Qualitatively similar to the results of experiments with FANCD2-depleted H1299 cells, resumption of DNA synthesis following BPDE-induced S-phase arrest was delayed in the FANCD2(−) cell line relative to FANCD2(+) cells (Fig. 1E). As expected, recovery from S-phase arrest following treatment with the intra-strand cross-linking agent (cis-platinum, cisplatin) was also compromised in FANCD2(−) cells relative to FANCD2(+) controls (Fig. 1E). Furthermore, consistent with a role for FANCD2 in tolerance of BPDE-induced DNA damage, FANCD2(−) cells showed modest sensitivity to BPDE in cell survival assays (Fig. 1F). Other workers have also reported mild UV sensitivity of FA cell lines (30), further indicating a role for FANCs in tolerance of bulky DNA lesions. Taken together, our results show that the FA pathway participates in cellular responses to bulky DNA adducts such as BPDE-induced lesions.
Rad18 Promotes Efficient FANCD2 Mono-ubiquitination
Because FANCD2 associated with chromatin in response to BPDE (Fig. 1), and because FANCD2 deficiency recapitulated the S-phase recovery and survival defects of TLS-deficient cells (9, 10), we hypothesized that TLS and FANC proteins function in a common pathway that confers tolerance of bulky adducts. Therefore, we first tested the possibility that FANCD2 ubiquitination is downstream of Rad18.
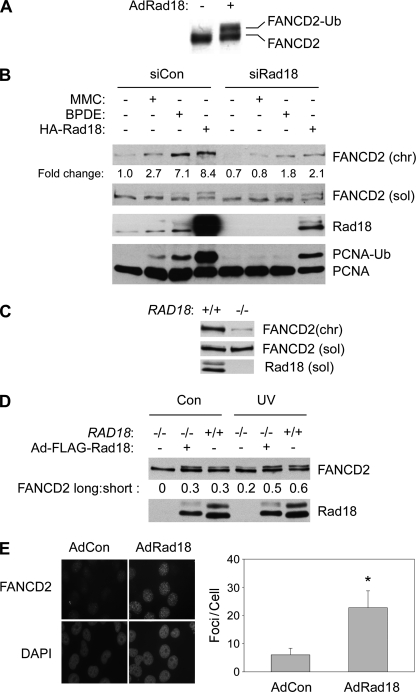
We have previously shown that BPDE-induced PCNA mono-ubiquitination is Rad18-dependent and that overexpressed Rad18 induces DNA damage-independent PCNA ubiquitination (10). As shown in Fig. 2A, Rad18 overexpression in H1299 cells also induced a distinct FANCD2 electrophoretic mobility shift (the hallmark of FANCD2 mono-ubiquitination), even in the absence of DNA damage. Therefore, Rad18 is a potential upstream regulator of the FA pathway.
FIGURE 2.
Rad18 promotes efficient FANCD2 ubiquitylation. A, exponentially growing H1299 cells were infected with adenovirus encoding HA-Rad18 (or with empty vector control “AdCon” virus). 48 h post-infection, total cell lysates were analyzed by SDS-PAGE and immunoblotting for FANCD2. B, control (siCon) and Rad18-depleted (siRad18) cells were treated with 300 nm MMC or 300 nm BPDE for 6 h. Some cultures were additionally infected with AdHA-Rad18 adenovirus for 48 h prior to genotoxin treatment, as indicated. Chromatin (chr) and soluble (sol) fractions from the resulting cells were analyzed by SDS-PAGE and immunoblotting with antibodies against FANCD2, Rad18, and PCNA. C, soluble and chromatin fractions from exponentially growing HCT116 RAD18+/+ and RAD18−/− cells were analyzed by SDS-PAGE and immunoblotting with antibodies against FANCD2 and Rad18. D, HCT116 RAD18−/− cells were infected with adenovirus encoding FLAG-Rad18 for 48 h. The resulting cells were treated with 10 J/m2 UVC (or were left untreated for controls (Con)). 2 h after UV treatment, cells were lysed, and whole cell extracts were analyzed by SDS-PAGE and immunoblotting with antibodies against FANCD2 and Rad18. E, RAD18−/− HCT116 cells growing in chamber slides were infected with adenovirus encoding FLAG-Rad18 or with an “empty” vector control adenovirus (AdCon) for 48 h. The resulting slides were washed with CSK, and FANCD2 nuclear foci were visualized as described for Fig. 1B. FANCD2 foci in 50 representative nuclei from AdCon- and AdRad18-infected cells were enumerated. Data points represent the mean number of foci for 50 representative cells, with error bars representing the range. *, p < 0.0001 compared with control cells that were infected with control adenovirus. Note that we typically observe ∼10-fold fewer basal and genotoxin-inducible FANCD2 nuclear foci in HCT116 cells when compared with H1299 cells (for example compare Figs. 1B, 2E, and 4C).
Endogenous RAD18 Facilitates Mono-ubiquitination of FANCD2
To test whether endogenous Rad18 contributes to FA pathway activation, we determined the effect of depleting endogenous Rad18 on genotoxin-inducible association of mono-ubiquitinated FANCD2 with chromatin.
As expected, genotoxin treatments (BPDE or MMC) induced robust mono-ubiquitination of chromatin-associated PCNA and increased the chromatin association of FANCD2 in control (siCon) transfected cells (Fig. 2B, 2nd and 3rd lanes). Interestingly, both PCNA mono-ubiquitination and FANCD2 chromatin binding were reduced in Rad18-depleted H1299 cells (Fig. 2B, 6th and 7th lanes). The protein levels of soluble (nonubiquitinated) FANCD2 were unaffected by Rad18 status.
To complement our Rad18 siRNA experiments, we examined basal and genotoxin-induced levels of FANCD2 ubiquitination in RAD18−/− and RAD18+/+ HCT 116 cells (23). Basal levels of chromatin-associated (mono-ubiquitinated) FANCD2 were 5-fold lower in RAD18−/− cells relative to RAD18+/+ HCT116 cells (Fig. 2C). Levels of soluble FANCD2 protein were similar between RAD18+/+ and RAD18−/− cell lines. It was formally possible that the impaired chromatin association of FANCD2 in RAD18−/− cells was due to RAD18-independent differences between the two cell lines. Potentially, genetic changes resulting from long term cell culture of the RAD18−/− cells might have resulted in disruption of the FA pathway. Therefore, we reconstituted Rad18 expression in RAD18−/− cells using transient adenovirus-mediated gene delivery. The expression levels of Rad18 expression in the reconstituted cells were similar to Rad18 levels in parental RAD18+/+ HCT116 cells (Fig. 2D). As expected, levels of mono-ubiquitinated FANCD2 (upper bands of FANCD2 doublet designated “FANCD2-long”) were decreased in RAD18−/− cells relative to wild type cells (Fig. 2D, 1st and 3rd lanes). However, reconstitution of RAD18 in RAD18-null cells restored mono-ubiquitination of FANCD2, resulting in a distinct band shift (Fig. 2D, 2nd lane). UV treatment did not induce FANCD2 mono-ubiquitination as efficiently in RAD18−/− cells when compared with RAD18+/+ cells (Fig. 2D, 4th and 6th lanes). However, in Rad18-complemented RAD18−/− cells, UV radiation induced a level of FANCD2 mono-ubiquitination that was comparable with that observed in wild type cells (Fig. 2D, 5th lane).
FANCD2 mono-ubiquitination is necessary for its redistribution to nuclear foci (28, 29). We performed immunofluorescence microscopy to monitor the effect of Rad18 complementation on the subcellular localization of FANCD2 in RAD18−/− cells. As shown in Fig. 2E, ectopically expressed FLAG-Rad18 induced a 4-fold increase in the number of FANCD2 nuclear foci (p < 0.0001), further consistent with a role for Rad18 in facilitating efficient FANCD2 mono-ubiquitination. Taken together, our results demonstrate that Rad18 promotes FANCD2 mono-ubiquitination and formation of FANCD2 nuclear foci, consistent with a role for Rad18 in regulating the FA pathway.
Purified RAD6-RAD18 Complex Is Insufficient to Mono-ubiquitinate FANCD2 in Vitro
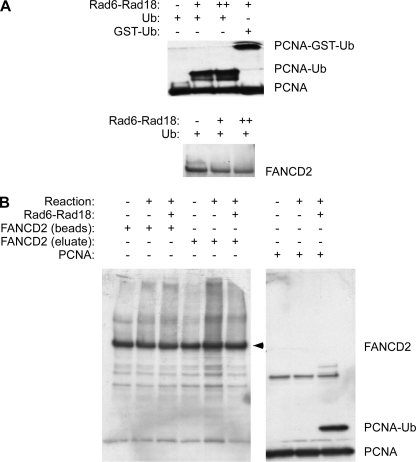
The results of Fig. 2 indicated that Rad18 contributes to basal and genotoxin-induced FANCD2 ubiquitination in human cells. In co-immunoprecipitation experiments, we noticed both basal and genotoxin-inducible association of RAD18 and FANCD2 (data not shown). Therefore, we performed in vitro ubiquitination assays to test the possibility that FANCD2 is a direct target of RAD18 E3 ligase activity. Recombinant human FANCD2 (expressed and purified from insect cells) and immunoprecipitated FLAG-FANCD2 from human PD20 cells were both tested as possible substrates of Rad18-Rad6. Recombinant PCNA was used as a positive control substrate for ubiquitination by Rad18 (8, 26).
As expected, the recombinant Rad6-Rad18 complex mono-ubiquitinated PCNA effectively, as shown by the appearance of an electrophoretically shifted form of PCNA (Fig. 3A). Using GST-tagged ubiquitin, we also verified that the band shift truly resulted from mono-ubiquitination of PCNA by the Rad6-Rad18 complexes (Fig. 3A, 4th lane). However, in parallel in vitro ubiquitination assays containing recombinant FANCD2 as a candidate substrate, we did not detect the mono-ubiquitinated species of FANCD2 (Fig. 3A, 2nd and 3rd lanes). Neither insect cell-derived FANCD2 (Fig. 3A) nor immunopurified FLAG-FANCD2 prepared from human PD20 fibroblasts were in vitro substrates for Rad18-Rad6 (Fig. 3B, 3rd and 6th lanes). Although these results do not exclude the possibility that FANCD2 is a physiological substrate of Rad18 within intact cells, they indicate that active purified Rad6-Rad18 is insufficient to ubiquitinate FANCD2. We infer that either additional proteins are necessary to facilitate direct ubiquitination of FANCD2 by Rad18 or that Rad18 activates the FA pathway indirectly. Experiments described below suggest that Rad18-mediated FA pathway activation occurs indirectly and is secondary to other TLS events.
FIGURE 3.
Recombinant Rad18-Rad6 complex ubiquitinates PCNA but not FANCD2 in vitro. A, Rad18-Rad6 complexes were incubated with recombinant PCNA or recombinant baculovirus-encoded FANCD2 purified from Sf9 cells as described under “Experimental Procedures.” Reaction products were analyzed by SDS-PAGE and immunoblotting with antibodies against PCNA or FANCD2. B, Rad18-Rad6 complexes were incubated with immunoprecipitated FANCD2 complexes from FLAG-FANCD2-complemented PD20 cells. Immobilized FLAG-FANCD2 bound to the anti-FLAG affinity matrix (“beads”) and FLAG-FANCD2 eluted using FLAG peptides (“eluate”) were both tested as potential Rad18 substrates as indicated.
Role of RAD18-mediated PCNA Mono-ubiquitination in FA Pathway Activation
We hypothesized that Rad18-induced mono-ubiquitination of FANCD2 might occur indirectly, secondarily to PCNA mono-ubiquitination and recruitment of TLS polymerases. The SAP (SAF-A/B, Acinus, and PIAS) domain of RAD18 (residues 248–282) is necessary for the efficient mono-ubiquitination of PCNA and pol η recruitment (26) but is dispensable for other Rad18 functions (31). Therefore, to investigate a correlation between PCNA ubiquitination and FA pathway activation, we tested the effect of a SAP domain mutation (ΔSAP) on Rad18-induced FANCD2 ubiquitination.
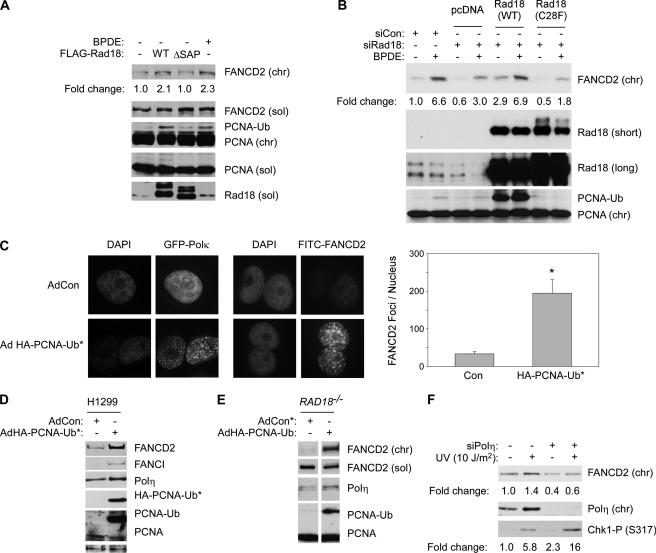
As shown in Fig. 4A, ectopic expression of the WT form of RAD18 induced DNA damage-independent mono-ubiquitination of PCNA and FANCD2. As expected, overexpression of Rad18 ΔSAP did not promote PCNA mono-ubiquitination and also failed to induce FANCD2 mono-ubiquitination. These data suggested that mono-ubiquitination of PCNA might be a prerequisite for RAD18-induced mono-ubiquitination of FANCD2.
FIGURE 4.
Mono-ubiquitinated PCNA and pol η promote efficient FANCD2 mono-ubiquitination. A, H1299 cells were transiently transfected with vectors encoding FLAG-Rad18 WT or FLAG-Rad18Δ SAP or with an empty vector (pcDNA) for control. 48 h later, the resulting cells were analyzed by SDS-PAGE and immunoblotting with antibodies against FANCD2, PCNA, and FLAG (to detect RAD18). B, H1299 cells were transfected with plasmids encoding Myc-Rad18 (WT) or Myc-Rad18 (C28F) or with an empty vector (pcDNA) for control. 24 h later the cells were transfected with siRNA directed against the 3′UTR of Rad18 or with a nontargeting control siRNA (siCon). After an additional 24 h, the cells were treated with 600 nm BPDE (or DMSO for controls). 6 h later, cell extracts were prepared and analyzed by SDS-PAGE and immunoblotting with antibodies against FANCD2, PCNA, and RAD18. C, H1299 cells were infected with adenovirus vectors encoding GFP-pol κ and HA-PCNA-Ub* fusion protein. The subcellular distribution of GFP-pol κ or FANCD2 in the resulting cultures was determined by DeltaVision fluorescence microscopy. Images of representative cells are shown. FANCD2 foci in 10 representative nuclei from AdCon- and AdPCNA-UB*-infected cells were enumerated. Data points represent the mean number of foci for 10 representative cells, with error bars representing the range. *, p < 0.0001 compared with control cells that were infected with control adenovirus (AdCon). D, H1299 cells were infected with AdHA-PCNA-Ub* or a control empty adenovirus vector (AdCon). After 48 h, chromatin and soluble extracts from the resulting cells were analyzed by SDS-PAGE and immunoblotting with antibodies against FANCD2, FANCI, pol η, and PCNA. E, HCT116 RAD18−/− cells were infected with AdHA-PCNA-Ub*. After 48 h, extracts from the resulting cells were analyzed by SDS-PAGE and immunoblotting using antibodies against FANCD2, pol η and PCNA. F, H1299 cells expressing FLAG-Rad18 were transfected with siPolη or control nontargeting siRNA (siCon). The resulting cells were treated with UVC (10 J/m2). Cell extracts were prepared 2 h after UV treatment and analyzed by SDS-PAGE and immunoblotting using antibodies against FANCD2, pol η, and phospho-Chk1 (Ser-317).
To further test the role of Rad18 E3 ubiquitin ligase activity in promoting FANCD2 mono-ubiquitination, we used a RING domain point mutant of Rad18 (designated “Rad18 C28F”). The Rad18 C28F mutant lacks ubiquitin ligase activity, yet supports DNA repair via homologous recombination repair (31). In the experiment shown in Fig. 4B, partial depletion of Rad18 using siRNA reduced chromatin association of FANCD2 by ∼55%. The ectopic expression of siRNA-resistant WT Rad18 fully corrected the reduced basal and genotoxin-inducible chromatin association of FANCD2. However, when expressed at similar levels to WT Rad18, the Rad18 C28F mutant failed to induce PCNA mono-ubiquitination and did not correct the defective FANCD2 chromatin association of Rad18-depleted cells (Fig. 4B). We conclude that Rad18 E3 ubiquitin ligase activity is necessary to promote efficient FANCD2 ubiquitination. Moreover, these data show that the Rad18-mediated FA pathway activation is separable from the E3 ligase-independent role of Rad18 in DNA repair via homologous recombination (31).
We reasoned that if RAD18-induced FANCD2 mono-ubiquitination occurs secondarily to PCNA mono-ubiquitination, expression of a PCNA-ubiquitin fusion protein that mimics native mono-ubiquitinated PCNA (7) may confer FA pathway activation independently of Rad18. We generated an adenovirus encoding HA-tagged PCNA-ubiquitin fusion protein (designated “PCNA-Ub*”). To test whether the PCNA-Ub* protein recruits TLS polymerases independently of genotoxin treatment, we co-expressed HA-PCNA-Ub* and GFP-pol κ in H1299 cells and monitored the effect of the PCNA-Ub* fusion protein on GFP-pol κ distribution.
As shown in Fig. 4C, PCNA-Ub* expression redistributed GFP-pol κ to nuclear foci in the absence of DNA damage, verifying that the PCNA-ubiquitin fusion protein mimics native mono-ubiquitinated PCNA. In parallel fluorescence microscopy experiments, PCNA-Ub* induced a 5.8-fold increase in the number of basal FANCD2 nuclear foci (p < 0.0001). Images of representative cells containing PCNA-Ub*-induced nuclear foci are shown in Fig. 4C.
The effect of PCNA-Ub* on the subcellular distribution of FANCD2 further suggests that PCNA ubiquitination may contribute to activation of the FA pathway. As shown in Fig. 4D, increased chromatin association of FANCD2 and FANCI in response to PCNA-Ub* expression was also evident by immunoblotting. As expected, chromatin loading of endogenous pol η was increased in response to PCNA-Ub* (Fig. 4C).
The H1299 cells used for experiments shown in Fig. 4, A–D, express functional wild type Rad18 protein. To test whether the expression of PCNA-Ub* can induce mono-ubiquitination of FANCD2 in the absence of Rad18, we infected RAD18−/− cells with AdHA-PCNA-Ub*. As shown in Fig. 4E, chromatin loading of the TLS polymerase pol η was increased upon PCNA-Ub* expression (albeit to a lesser degree than in H1299 cells, probably because of the important role of Rad18 in guiding pol η to stalled replication forks). Interestingly, chromatin association of FANCD2 was increased in response to PCNA-Ub* expression, even in a RAD18−/− background (Fig. 4E). Taken together, our results suggest that Rad18-dependent mono-ubiquitination of FANCD2 occurs secondarily to Rad18-mediated PCNA modification.
pol η Promotes Efficient FANCD2 Mono-ubiquitination
It was of interest to determine whether FANCD2 ubiquitination is coupled directly to PCNA ubiquitination or to more distal event(s) such as TLS polymerase recruitment. The Y-family TLS polymerases pol κ, pol η, pol ι, as well as REV1 are all thought to associate with ubiquitinated PCNA via specialized ubiquitin-binding motifs. Potentially, recruitment of any one (or all) Y-family polymerase may be linked to subsequent activation of the FA pathway. We chose to focus on pol η as a candidate mediator of FANCD2 activation for several reasons as follows. Of the Y-family members, pol η has the highest affinity for PCNA and is likely the first TLS polymerase recruited to all stalled replication forks (32). Moreover, pol η is a versatile TLS polymerase, which can cope with many types of lesions. Indeed, for some lesions (including UV-induced cyclobutane pyrimidine dimers), pol η performs insertion and extension steps without the requirement for additional TLS enzymes (3).
To test whether pol η mediates Rad18/PCNA-Ub-induced FANCD2 mono-ubiquitination, we depleted pol η by siRNA-mediated knockdown in H1299 cells. Interestingly, depletion of pol η attenuated the genotoxin-inducible association of FANCD2 with chromatin (Fig. 4F). As expected from our previous work (9), UV-inducible Chk1 phosphorylation was increased (by 2.7-fold) as a result of pol η depletion (Fig. 4F). Therefore, the reduced activation of FANCD2 in pol η-depleted cells was not due to a general decrease in DNA damage signaling. Taken together, these data support our hypothesis that recruitment of TLS polymerases to DNA replication forks stalled by bulky lesions is coupled to the activation of the Fanconi anemia pathway.
Effect of Rad18 Status on the FA Core Complex Proteins
We considered possible mechanisms by which TLS polymerase recruitment might be linked to FANCD2 ubiquitination. The intact FA core complex is necessary for mono-ubiquitination of FANCD2 in vivo, although USP1 de-ubiquitinates FANCD2 both in vivo and in vitro (18, 19, 33–36). Potentially, Rad18-dependent TLS could promote FANCD2 ubiquitination via increased FA core complex activity, reduced USP1-dependent de-ubiquitylation, or via both mechanisms. Therefore, we determined whether FA core complex proteins are differently regulated in RAD18+/+ cells compared with RAD18−/− cells.
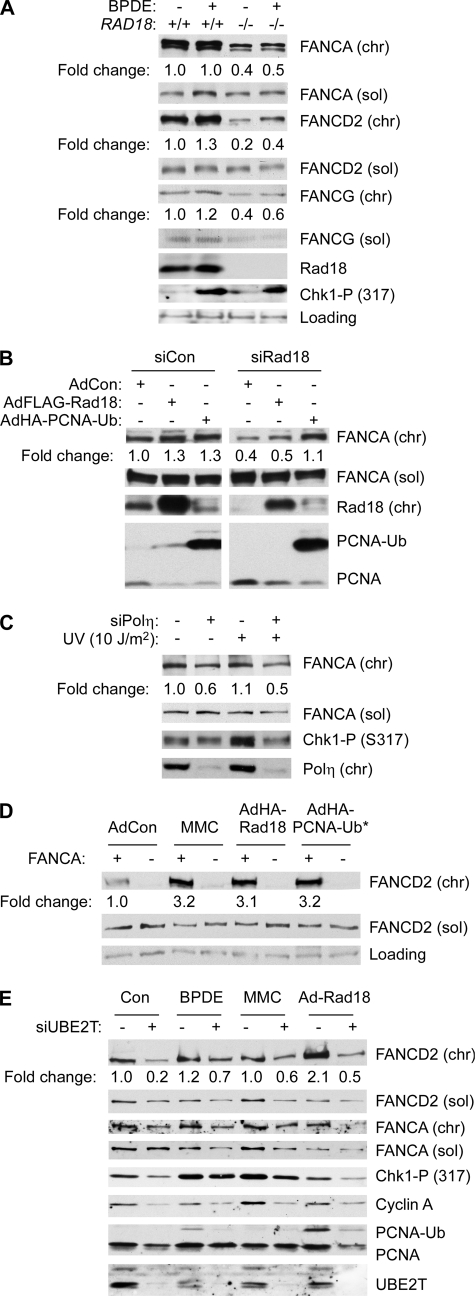
As shown in Fig. 5A, levels of chromatin-bound FANCA were ∼2-fold higher in RAD18+/+ cells relative to RAD18−/− cells (although levels of soluble FANCA were similar between the two cell lines). We have also observed reduced recruitment of FANCA to chromatin in Rad18-depleted H1299 cells (data not shown). For the purpose of comparison, we also determined protein levels of FANCG in the same experiment. As shown in Fig. 5A, chromatin-associated FANCG was 1.5-fold more abundant in RAD18+/+ cells relative to the RAD18-null line. However, for reasons that are unclear, expression of soluble FANCG was also reduced in RAD18−/− cells relative to the parental RAD18+/+ cell line. Nevertheless, these results are consistent with a role for Rad18 in regulating recruitment of the FA core complex to chromatin.
FIGURE 5.
Relationship between TLS and FA core complex components. A, exponentially growing HCT116 RAD18+/+ and RAD18−/− cells were treated with 100 nm BPDE for 2 h. Soluble (sol) and chromatin (chr) extracts from the resulting cells were analyzed by SDS-PAGE and immunoblotting with antibodies against FANCA, FANCD2, FANCG, RAD18, and phospho-Chk1 (Ser-317). B, control (siCon) and Rad18-depleted (siRad18) cells were infected with adenovirus encoding FLAG-Rad18 or HA-PCNA-Ub* fusion protein for 48 h. Soluble and chromatin extracts from the resulting cells were analyzed by SDS-PAGE and immunoblotting with antibodies against FANCA, Rad18 and PCNA. C, HCT116 cells were transfected with siPolη oligonucleotides or control nontargeting siRNA (siCon). The resulting cells were treated with UVC (10 J/m2). Cell extracts were prepared 2 h after UV treatment and analyzed by SDS-PAGE and immunoblotting using antibodies against FANCD2, FANCA, pol η, and phospho-Chk1 (Ser-317). D, GM6914 FANCA (−) fibroblasts or an isogenic FANCA (+) cell line stably reconstituted with wild type FANCA was infected with adenoviruses encoding Rad18 or PCNA-Ub* fusion protein or treated with 300 nm MMC. Soluble and chromatin extracts from the resulting cells were analyzed by SDS-PAGE and immunoblotting with antibodies against the indicated proteins. E, control and UBE2T-depleted H1299 cells were treated with 300 nm BPDE or 300 nm MMC for 6 h, or infected with AdRad18 for 48 h. Soluble and chromatin extracts from the resulting cells were analyzed by SDS-PAGE and immunoblotting with antibodies against FANCD2, UBE2T, phospho-Chk1, cyclin A, and PCNA.
Because PCNA mono-ubiquitination and TLS polymerases influenced FANCD2 mono-ubiquitination, we also determined the effect of PCNA-Ub* expression on the levels of chromatin-bound and soluble FANCA. As shown in Fig. 5B, expression of PCNA-Ub* stimulated accumulation of FANCA on chromatin but did not modify levels of soluble FANCA. Conversely, in pol η-depleted cells, genotoxin-induced association of FANCA with chromatin was attenuated (Fig. 5C). Taken together, these results suggest that efficient FANCD2 mono-ubiquitination is conferred by Rad18-mediated PCNA mono-ubiquitination and subsequent recruitment of TLS polymerases, leading to association of the FA core complex with chromatin.
FA Core Complex Is Necessary for the Rad18-induced Component of FANCD2 Ubiquitination
To test whether the FA core complex is necessary for Rad18-mediated FANCD2 mono-ubiquitination, FANCA (−) GM6914 cells and an isogenic FANCA (+) cell line complemented with wild type FANCA cDNA were infected with Ad-FLAG-RAD18, infected with AdHA-PCNA-Ub*, or treated with MMC as a positive control for FA pathway activation. As expected, MMC treatment induced mono-ubiquitinated FANCD2 on chromatin in FANCA (+) cells but not in FANCA (−) cells (Fig. 5D, 3rd and 4th lanes). Overexpression of Rad18 or of PCNA-Ub* increased accumulation of FANCD2 on chromatin in the FANCA (+) cells by ∼3-fold (Fig. 5D, 5th and 7th lanes), but these conditions did not affect chromatin binding of FANCD2 in FANCA-deficient cells (Fig. 5D, 6th and 8th lanes). Therefore, Rad18 expression or mono-ubiquitination of PCNA alone were insufficient for FANCD2 mono-ubiquitination in FANCA (−) cells, further suggesting that Rad18-mediated FANCD2 mono-ubiquitination is indirect and requires the FA-core complex.
UBE2T is the ubiquitin E2 ligase thought to mediate FANCD2 ubiquitylation. Therefore, we additionally tested whether UBE2T is necessary for Rad18-mediated mono-ubiquitination of FANCD2. We performed siRNA to deplete UBE2T expression. In those experiments, ablation of UBE2T was very toxic to H1299 cells (data not shown) and led to reduced expression levels of cyclin A and PCNA and reduced numbers of cells in S-phase (Fig. 5E). However, as shown in Fig. 5E, 7th and 8th lanes, RAD18-mediated FANCD2 mono-ubiquitination was compromised in UBE2T-depleted cells. Therefore, consistent with the results of Fig. 5D, RAD18-dependent mono-ubiquitination of FANCD2 likely occurs indirectly via activation of the FA core complex.
Genotoxin-induced PCNA Mono-ubiquitylation in FA Cells
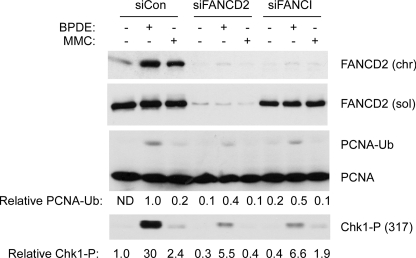
The results of Figs. 1–5 indicate that FA pathway activation occurs distal to Rad18-induced PCNA ubiquitylation, at least in the context of human cells that acquire bulky DNA lesions. In contrast, recent studies using replication-competent Xenopus cell-free extracts suggest that when replication forks converge on ICL, TLS events occur distal to the FA core complex and FANCD2-FANCI activation (17, 37). Therefore, we tested whether an intact FA pathway is required for PCNA ubiquitylation. We tested the effect of FANCD2 or FANCI depletion on the integrity of genotoxin-induced PCNA ubiquitylation. As shown in Fig. 6, we observed slightly reduced PCNA ubiquitylation in FANCD2- and FANCI-depleted H1299 cells relative to controls. However, we have found that FANCD2 deficiency results in reduced origin firing in many cultured cell lines, including H1299 (38). Therefore, the attenuated PCNA ubiquitylation observed following FANCD2/FANCI depletion likely results from reduced numbers of replicons in these cells. Indeed, DNA damage-inducible phosphorylation of Chk1 (which, similar to PCNA ubiquitylation, requires replication fork-dependent generation of replication protein A-coated ssDNA) was also attenuated in FANCD2- and FANCI-depleted cells (Fig. 6). We conclude that FANCD2 deficiency does not directly compromise Rad18-mediated PCNA mono-ubiquitination.
FIGURE 6.
Effect of FANCD deficiency on PCNA mono-ubiquitylation. H1299 cells were transfected with siRNA against FANCD2, FANCI, or nontargeting control siRNA. The resulting cells were treated with 300 nm BPDE, 300 nm MMC, or were left untreated for controls. Soluble and chromatin extracts from the resulting cells were analyzed by SDS-PAGE and immunoblotting with antibodies against FANCD2, phospho-Chk1 (Ser-317), and PCNA as indicated.
DISCUSSION
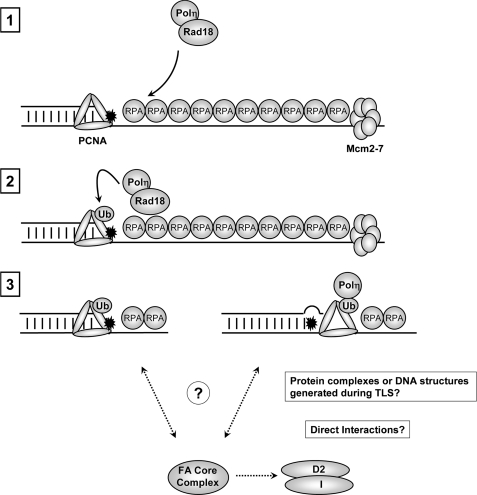
Here we have demonstrated that FANCD2 mono-ubiquitination is regulated by Rad18 in a manner that correlates closely with PCNA mono-ubiquitination. We have been unable to demonstrate that FANCD2 is a direct substrate for ubiquitylation by Rad18. However, we note that in vitro mono-ubiquitination of FANCD2 has been demonstrated successfully by only two groups, and it had been a very challenging task to reproduce complete mono-ubiquitination of FANCD2 in vitro (39, 40). Therefore, our inability to detect Rad18-dependent FANCD2 ubiquitylation in vitro does not necessarily exclude the possibility of a direct E3 ligase-substrate relationship between Rad18 and FANCD2. However, our results demonstrating a dependence of FANCD2 ubiquitylation on PCNA mono-ubiquitination and pol η strongly suggest that the mechanism by which Rad18 activates the FA pathway is indirect. Our results indicate that the Rad18-dependent mechanism of FA pathway activation requires the FA core complex. Moreover, the FA core complex component FANCA associates with chromatin in a manner that depends on Rad18 and TLS polymerase recruitment (Fig. 7).
FIGURE 7.
Hypothetical model for Rad18-dependent activation of the FA pathway in response to bulky DNA adducts. Bulky DNA adducts cause uncoupling of replicative helicase (Mcm2–7) and DNA polymerase activities (step 1). The ensuing accumulation of replication protein A-coated ssDNA recruits the Rad18-pol η complex leading to PCNA mono-ubiquitylation (step 2). Potentially, mono-ubiquitinated PCNA might promote FA pathway activation via pol η-independent mechanisms (step 3, pathway depicted on left-hand side). Alternatively, engagement of pol η or other DNA TLS polymerases with mono-ubiquitinated PCNA might generate protein complexes and/or DNA structures that facilitate the recruitment of FA core complex members to sites of replication stalling, promoting FANCD2-FANCI ubiquitination (step 3, pathway depicted on right-hand side).
In RAD18-deficient cells, activation of the FA pathway is attenuated but not entirely ablated, indicating that compensatory or partially redundant mechanisms exist for FA pathway activation in the absence of Rad18. Dutta and co-workers (41) recently showed that the CRL4Cdt2 complex supports a basal level of PCNA ubiquitylation and TLS, even in Rad18-deficient cells. Therefore, CRL4Cdt1-dependent TLS likely provides an additional mechanism for Rad18-independent activation of the FA pathway.
The FA pathway is generally activated during S-phase and in response to many replication fork-stalling lesions, yet FANC deficiency confers relatively specific ICL hypersensitivity, an observation attributed to redundant roles for FA pathway in repair of other lesions. Nevertheless, mild UV sensitivity of FA cells has been reported (30). Our results showing BPDE sensitivity of FANCD2(−) cells are also consistent with a role for the FA pathway in processing and repair of bulky adducts at stalled replication forks. Relatively little is known regarding the mechanisms dictating the modes of lesion bypass for replication forks that encounter damaged DNA. Nevertheless, it appears that both TLS- and homologous recombination-based mechanisms are available to cells bypassing bulky adducts, including BPDE (42). Therefore, coordinate activation of both TLS and FA pathways may be advantageous for tolerance of replication forks stalled by bulky DNA lesions.
Precisely how Rad18-dependent FA pathway activation is coupled with TLS may depend on the nature of the DNA lesion encountered by replication forks. In the case of bulky DNA adducts such as BPDE and cyclobutane pyrimidine dimers, recent models suggest that uncoupling of helicase and polymerase activities generates replication protein A-coated ssDNA that recruits Rad18 to sites of replication stalling (26, 43). Soluble Rad18 exists in a complex with pol η and is thought to chaperone pol η (and perhaps other TLS polymerases) to sites of replication fork stalling (8). Once recruited to stalled replication forks, Rad18 performs PCNA mono-ubiquitination (4). Depending on the identity of the DNA adduct, one or more TLS polymerases are thought to perform the insertion and extension steps of TLS.
Our results indicate that PCNA ubiquitination facilitates efficient activation of the FA pathway. The stimulatory effect of mono-ubiquitinated PCNA on the FA pathway activation may be mediated via pol η or other TLS DNA polymerases (Fig. 7, pathway depicted on right-hand side of step 3), a possibility that is supported by our pol η depletion experiments. According to this model, the TLS intermediate of mono-ubiquitinated PCNA in complex with pol η and possibly ssDNA or other DNA structures might be coupled to the activation of the FA core complex, leading to mono-ubiquitination of FANCD2 and FANCI. It is possible that the complex of mono-ubiquitylated PCNA with pol η (or other TLS polymerases) or replication structures generated during TLS provide a platform for FA proteins. Consistent with this possibility, Glover and co-workers (44) have identified a functional PCNA-interacting peptide-box in FANCD2.
Alternatively, putative effectors of PCNA-Ub other than pol η might contribute to efficient FA pathway activation (Fig. 7, pathway depicted on left-hand side of step 3). According to this scheme, the role of pol η is solely to promote efficient PCNA ubiquitination via mutually dependent formation of active Rad18-pol η complexes.
These results invite further questions regarding the role(s) of FA proteins in the tolerance and processing of bulky DNA adducts. Studies using yeast and DT40 cells indicate that PCNA ubiquitination and pol η are not required for ongoing replicon movement in UV-irradiated cells but instead play an important role in preventing accumulation of ssDNA gaps behind the replication fork (45, 46). Details of the putative re-priming mechanism that maintains leading strand synthesis on damaged templates are not well understood (47). Nevertheless, if the basic mechanisms of TLS are conserved in yeast, chicken (DT40), and mammalian cells and if TLS does indeed occur post-replicatively, it appears likely that Rad18-dependent activation of FA pathway is initiated behind re-primed replication forks.
It is possible that the FA pathway plays a role in the putative re-priming mechanism. Reportedly, the FA core complex is necessary for Rev1 recruitment (21, 27), and in DT40 cells, Rev1 is required for efficient replicon elongation in the presence of DNA damage (46). Therefore, because of its role in Rev1 recruitment, the FA core complex might be necessary for maintaining replicon elongation independently of FANCD2 ubiquitination.
Activation mechanisms of the TLS and FA pathways might be very different when replication forks encounter ICL and might differ between species and cell type. Elegant studies using replication-competent Xenopus cell-free extracts have demonstrated a requirement of the FANCD2-FANCI complex for lesion bypass by replication forks that converge on an ICL. Those studies demonstrated that essential factors for lesion bypass (including the TLS DNA polymerase pol ζ) are recruited in a manner that requires mono-ubiquitinated FANCD2-FANCI during replication-coupled ICL repair (17, 37). The roles of Rad18 and mono-ubiquitylated PCNA during replication-coupled ICL repair have not yet been reported in that experimental system. However, if Rad18 and PCNA ubiquitylations are required for FANCD2-mediated bypass when replication forks converge on ICL, it appears most likely that Rad18 functions downstream of the FA core complex.
Although replication forks converging simultaneously on cross-links in the context of a small plasmid template (17) require FANCD2-FANCI for replication-coupled bypass, it is possible that not all ICLs are processed via this mechanism in mammalian cells harboring genome-wide DNA damage. For instance, it is not clear whether the processing of ICL encountered by a replication fork in the context of a larger template (i.e. the mammalian genome) is delayed until arrival of a second converging fork. Thus, the elegant experimental system employed by Walter and co-workers (37) (which involves a single defined cross-link engineered into a small plasmid) is very different from cultured mammalian cells containing bulky adducts and direct comparison with results of our experiments may be inappropriate.
The study by Walter and co-workers is arguably the best defined example of cooperative actions of TLS and FA proteins, and it is clear from those studies that FANCD2 functions upstream of pol ζ. Our results indicate that there is not invariably a simple linear relationship between the TLS and FA pathways. However, this does not imply any disagreement between the two studies. We note also that different DNA repair proteins frequently have complex interactions in which clear proximal/distal relationships do not exist. For example, the p95/NBS1 protein is both an upstream component of the ataxia telangiectasia mutated signaling pathway and a target of ataxia telangiectasia mutated signaling.
In summary, our data provide a novel mechanism for cross-talk between the TLS and FA pathways. Taken together with previous results, it appears that signaling between FA and TLS mechanisms can be bi-directional and that multiple components of the FA and TLS pathways may communicate. Moreover, the nature of the DNA damage (bulky adducts, ICL) and perhaps other factors influence the mechanism and directionality of the cross-talk.
Acknowledgments
We thank Dr. Alan D'Andrea and Dr. Jungmin Kim for help and advice with these studies.
This work was supported, in whole or in part, by National Institutes of Health Grants ES09558 and ES12917 (to C. V.) and Grant R01 HL063776 (to G. M. K.).
- TLS
- translesion synthesis
- FA
- Fanconi anemia
- PCNA
- proliferating cell nuclear antigen
- pol
- polymerase
- MMC
- mitomycin C
- TEV
- tobacco etch virus
- ssDNA
- single strand DNA
- ICL
- interstrand cross-link
- BPDE
- benzo[a]pyrene dihydrodiol epoxide.
REFERENCES
- 1.Kastan M. B., Bartek J. (2004) Nature 432, 316–323 [DOI] [PubMed] [Google Scholar]
- 2.Zhou B. B., Elledge S. J. (2000) Nature 408, 433–439 [DOI] [PubMed] [Google Scholar]
- 3.Prakash S., Johnson R. E., Prakash L. (2005) Annu. Rev. Biochem. 74, 317–353 [DOI] [PubMed] [Google Scholar]
- 4.Kannouche P. L., Lehmann A. R. (2004) Cell Cycle 3, 1011–1013 [PubMed] [Google Scholar]
- 5.Kannouche P. L., Wing J., Lehmann A. R. (2004) Mol. Cell 14, 491–500 [DOI] [PubMed] [Google Scholar]
- 6.Ohmori H., Friedberg E. C., Fuchs R. P., Goodman M. F., Hanaoka F., Hinkle D., Kunkel T. A., Lawrence C. W., Livneh Z., Nohmi T., Prakash L., Prakash S., Todo T., Walker G. C., Wang Z., Woodgate R. (2001) Mol. Cell 8, 7–8 [DOI] [PubMed] [Google Scholar]
- 7.Bienko M., Green C. M., Crosetto N., Rudolf F., Zapart G., Coull B., Kannouche P., Wider G., Peter M., Lehmann A. R., Hofmann K., Dikic I. (2005) Science 310, 1821–1824 [DOI] [PubMed] [Google Scholar]
- 8.Watanabe K., Tateishi S., Kawasuji M., Tsurimoto T., Inoue H., Yamaizumi M. (2004) EMBO J. 23, 3886–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bi X., Slater D. M., Ohmori H., Vaziri C. (2005) J. Biol. Chem. 280, 22343–22355 [DOI] [PubMed] [Google Scholar]
- 10.Bi X., Barkley L. R., Slater D. M., Tateishi S., Yamaizumi M., Ohmori H., Vaziri C. (2006) Mol. Cell. Biol. 26, 3527–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim S. H., Michael W. M. (2008) Mol. Cell 32, 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang T. T., D'Andrea A. D. (2006) Nat. Rev. Mol. Cell Biol. 7, 323–334 [DOI] [PubMed] [Google Scholar]
- 13.Moldovan G. L., D'Andrea A. D. (2009) Annu. Rev. Genet. 43, 223–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papadopoulo D., Guillouf C., Mohrenweiser H., Moustacchi E. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 8383–8387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papadopoulo D., Guillouf C., Porfirio B., Moustacchi E. (1990) Prog. Clin. Biol. Res. 340A, 241–248 [PubMed] [Google Scholar]
- 16.Niedzwiedz W., Mosedale G., Johnson M., Ong C. Y., Pace P., Patel K. J. (2004) Mol. Cell 15, 607–620 [DOI] [PubMed] [Google Scholar]
- 17.Räschle M., Knipscheer P., Knipsheer P., Enoiu M., Angelov T., Sun J., Griffith J. D., Ellenberger T. E., Schärer O. D., Walter J. C. (2008) Cell 134, 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nijman S. M., Huang T. T., Dirac A. M., Brummelkamp T. R., Kerkhoven R. M., D'Andrea A. D., Bernards R. (2005) Mol. Cell 17, 331–339 [DOI] [PubMed] [Google Scholar]
- 19.Huang T. T., Nijman S. M., Mirchandani K. D., Galardy P. J., Cohn M. A., Haas W., Gygi S. P., Ploegh H. L., Bernards R., D'Andrea A. D. (2006) Nat. Cell Biol. 8, 339–347 [DOI] [PubMed] [Google Scholar]
- 20.Zhang J., Zhao D., Wang H., Lin C. J., Fei P. (2008) Cell Cycle 7, 407–413 [DOI] [PubMed] [Google Scholar]
- 21.Mirchandani K. D., McCaffrey R. M., D'Andrea A. D. (2008) DNA Repair 7, 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moldovan G. L., D'Andrea A. D. (2009) Annu. Rev. Genet. 43, 223–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiomi N., Mori M., Tsuji H., Imai T., Inoue H., Tateishi S., Yamaizumi M., Shiomi T. (2007) Nucleic Acids Res. 35, e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Izumi M., Yanagi K., Mizuno T., Yokoi M., Kawasaki Y., Moon K. Y., Hurwitz J., Yatagai F., Hanaoka F. (2000) Nucleic Acids Res. 28, 4769–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park W. H., Margossian S., Horwitz A. A., Simons A. M., D'Andrea A. D., Parvin J. D. (2005) J. Biol. Chem. 280, 23593–23598 [DOI] [PubMed] [Google Scholar]
- 26.Tsuji Y., Watanabe K., Araki K., Shinohara M., Yamagata Y., Tsurimoto T., Hanaoka F., Yamamura K., Yamaizumi M., Tateishi S. (2008) Genes Cells 13, 343–354 [DOI] [PubMed] [Google Scholar]
- 27.Hicks J. K., Chute C. L., Paulsen M. T., Ragland R. L., Howlett N. G., Guéranger Q., Glover T. W., Canman C. E. (2010) Mol. Cell. Biol. 30, 1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taniguchi T., Garcia-Higuera I., Andreassen P. R., Gregory R. C., Grompe M., D'Andrea A. D. (2002) Blood 100, 2414–2420 [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Higuera I., Taniguchi T., Ganesan S., Meyn M. S., Timmers C., Hejna J., Grompe M., D'Andrea A. D. (2001) Mol. Cell 7, 249–262 [DOI] [PubMed] [Google Scholar]
- 30.Tebbs R. S., Hinz J. M., Yamada N. A., Wilson J. B., Salazar E. P., Thomas C. B., Jones I. M., Jones N. J., Thompson L. H. (2005) DNA Repair 4, 11–22 [DOI] [PubMed] [Google Scholar]
- 31.Huang J., Huen M. S., Kim H., Leung C. C., Glover J. N., Yu X., Chen J. (2009) Nat. Cell Biol. 11, 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barkley L. R., Ohmori H., Vaziri C. (2007) Cell Biochem. Biophys. 47, 392–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gurtan A. M., D'Andrea A. D. (2006) DNA Repair 5, 1119–1125 [DOI] [PubMed] [Google Scholar]
- 34.Kennedy R. D., D'Andrea A. D. (2005) Genes Dev. 19, 2925–2940 [DOI] [PubMed] [Google Scholar]
- 35.D'Andrea A. D., Grompe M. (2003) Nat. Rev. Cancer 3, 23–34 [DOI] [PubMed] [Google Scholar]
- 36.Friedberg E. C. (2006) Mol. Cell 22, 150–152 [DOI] [PubMed] [Google Scholar]
- 37.Knipscheer P., Räschle M., Smogorzewska A., Enoiu M., Ho T. V., Schärer O. D., Elledge S. J., Walter J. C. (2009) Science 326, 1698–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song I. Y., Barkley L. R., Day T. A., Weiss R. S., Vaziri C. (2010) Cell Cycle 9, 2375–2388 [DOI] [PubMed] [Google Scholar]
- 39.Alpi A. F., Pace P. E., Babu M. M., Patel K. J. (2008) Mol. Cell 32, 767–777 [DOI] [PubMed] [Google Scholar]
- 40.Yuan F., El Hokayem J., Zhou W., Zhang Y. (2009) J. Biol. Chem. 284, 24443–24452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terai K., Abbas T., Jazaeri A. A., Dutta A. (2010) Mol. Cell 37, 143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adar S., Izhar L., Hendel A., Geacintov N., Livneh Z. (2009) Nucleic Acids Res. 37, 5737–5748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huttner D., Ulrich H. D. (2008) Cell Cycle 7, 3629–3633 [DOI] [PubMed] [Google Scholar]
- 44.Howlett N. G., Harney J. A., Rego M. A., Kolling F. W., 4th., Glover T. W. (2009) J. Biol. Chem. 284, 28935–28942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopes M., Foiani M., Sogo J. M. (2006) Mol. Cell 21, 15–27 [DOI] [PubMed] [Google Scholar]
- 46.Edmunds C. E., Simpson L. J., Sale J. E. (2008) Mol. Cell 30, 519–529 [DOI] [PubMed] [Google Scholar]
- 47.Yan S., Michael W. M. (2009) J. Cell Biol. 184, 793–804 [DOI] [PMC free article] [PubMed] [Google Scholar]